Abstract
We report the quantitative, label-free analysis of protein-protein interactions in free solution within picoliter volumes using backscatter interferometry (BSI). Changes in the refractive index are measured for solutions introduced on a PDMS microchip allowing determination of forward and reverse rate constants for two-mode binding. Time-dependent BSI traces are directly fit using a global analysis approach to characterize the interaction of the small heat-shock protein α-crystallin with two substrates: destabilized mutants of T4 lysozyme and the in vivo target βB1-crystallin. The results recapitulate the selectivity of αB-crystallin differentially binding T4L mutants according to their free energies of unfolding. Furthermore, we demonstrate that an αA-crystallin mutant linked to hereditary cataract has activated binding to βB1-crystallin. Binding isotherms obtained from steady-state values of the BSI signal yielded meaningful dissociation constants and establishes BSI as a novel tool for the rapid identification of molecular partners using exceedingly small sample quantities under physiological conditions. This work demonstrates that BSI can be extended to screen libraries of disease-related mutants to quantify changes in affinity and/or kinetics of binding.
INTRODUCTION
Protein-protein interactions play a critical role in the biochemical circuitry of the cell. Physiologic signals and responses are transmitted by protein complexes and controlled by the kinetics of their assembly. Of the thousands of proteins expressed, it has been estimated that 80% function in complexes.1 Often the affinity, cooperativity, and time-scales of protein-protein interactions are altered in a programmed regulatory manner to trigger the initiation of signaling pathways in response to cellular or environmental signals. Thus, quantitative characterization of the thermodynamics and kinetics of protein-protein interactions is critical to understand cellular activities such as the transfer of genetic information, protein folding, regulation of molecular machineries, and signal transduction.
Traditional methods for analysis of label-free in vitro molecular interactions such as isothermal titration calorimetry (ITC)2 and surface plasmon resonance (SPR)3 do not have the sensitivity and volume requirements for nanoscale analysis which is necessary in situations where the binding partners are rare or difficult to express. Furthermore, SPR requires immobilization of one of the binding partners, necessitating a priori knowledge of the system and possibly perturbing interactions due to the surface chemistry required for immobilization. Coupling microfluidic networks with label-free detection modalities overcome the shortcomings of sample requirement by SPR and ITC. Recently, small-angle X-ray scattering (SAXS) was combined with a microfluidic front-end to facilitate the structural analysis of proteins in a relatively high-throughput manner.4 Another emergent technology, back-scattering interferometry (BSI), was used for nanoscale, label-free, free-solution molecular interaction studies.5 BSI bridges the gap between the large samples required by ITC and the scale afforded by novel microfluidics 6, 7, while offering the sensitivity needed for extensive use in studying diverse biological systems. Employing a simple light propagation technique, Bornhop, et al.5 performed nanoliter-volume, ultra-high sensitivity interferometry to study analytes in free solution contained within a microfluidic device. The use of beam interference to measure changes in the local refractive index (RI) makes BSI a universal technique for analyte detection. The combination of BSI with microfluidic devices leads to faster analysis times, reduced sample size, increased throughput, and the added potential for multiplexing and mass production. Further, the multi-pass microfluidic–channel/laser beam interaction yields high finesse fringes facilitating high sensitivity within extremely small detection volumes (i.e. 24 zeptomoles in roughly 500 picoliters).5 Finally, the incorporation of an integrated mixer allows the simultaneous determination of reaction kinetics and binding affinity for interactions spanning six decades (µM to pM KD values) without the need for immobilization or labeling chemistries.
In this paper, we demonstrate that BSI can be used for quantitative analysis of the kinetics and thermodynamics of biologically important molecular interactions providing insights difficult to obtain from other techniques. The binding of various protein targets by the small heat-shock protein (sHSP) α-crystallin is monitored in real time using nanoliter volumes. Small heat shock proteins are a superfamily of molecular chaperones which play a central role in thermotolerance8, resistance to various forms of stress8, and are required for efficient protein folding9. Although the spontaneous self assembly of proteins into their native conformations occurs on a sub-second timescale10, aggregation-prone non native sates are populated along the folding pathway. Chaperones monitor these non-native states and guide aberrant proteins through refolding or degradation pathways. Protein misfolding or unfolding leads to aggregation which can be associated with cytotoxicity.11 Insoluble aggregates of misfolded proteins accumulate to form amyloid deposits in prevalent neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington’s diseases.12
Molecular chaperones are critical in maintaining the life-long transparency of the ocular lens. Proteins in the lens undergo extensive age-related post-translational modifications.13 Because the lens lacks the cellular machinery to degrade unstable proteins and synthesize new ones, modified proteins with reduced stability and/or solubility can nucleate the formation of aggregates that can scatter light.14 Age-related cataracts, the most common form of cataract, are characterized by the presence of cross-linked protein aggregates. The sHSP, α-crystallin, constitutes ~ 40% of the protein mass in the mammalian lens contributing to its refractive index as well as serving as a molecular chaperone.15 In vivo, α-crystallin is a protein-stability sensor that binds aggregation-prone proteins helping to maintain lens clarity and transparency.16 Knock-out of αA-crystallin leads to accelerated cataracts suggesting a direct relation between chaperone activity and transparency. Point mutants of αA-crystallin and αB-crystallin are associated with hereditary cataracts17 and desmin-related myopathy18 respectively.
BSI is used here to monitor the kinetics and analyze the thermodynamics of interactions between α-crystallin and a model substrate, T4 lysozyme (T4L), in a label-free, tether-free format. Traditional detection methodologies including fluorescence and calorimetry were also used in order to cross-validate BSI. The specificity of the detection methodology is highlighted by comparing the interaction of αB-crystallin with two mutants of T4L having similar structures but different free energies of unfolding. To demonstrate the potential use of BSI in screening libraries of structural analogs and mutant constructs, the interaction of a mutant α-crystallin genetically linked to autosomal dominant cataract17 with another lens protein, β-crystallin, was analyzed by BSI. Quantitative kinetic and thermodynamic results from BSI is compared to previous studies carried out using SPR.19
EXPERIMENTAL SECTION
Detailed descriptions of chip fabrication, BSI optical setup, thermodynamic and kinetic analyses, as well as statistical methods are provided in the Supporting Information (SI).
Protein Expression, Purification, and Labeling
Site-directed mutagenesis of T4L20, αA-crystallin 21, αB-crystallin22, and β-crystallin23 has been described previously. Briefly, protein expression was carried out in competent BL21 (sHSP) or K38 (T4L) cell lines. Cells were transformed with mutant plasmids and cultured in Luria-Burtani (LB) broth containing small concentrations of ampicillin overnight at 32°C. This seed culture was then increased in volume and incubated for 2 – 3 hours at 37ºC until mid-log phase was reached. After cooling to room temperature, protein expression was induced by the addition of 0.4 mM isopropyl β-D-thioglactopyranoside (IPTG). Protein expression was carried out for 3 hours at 32ºC and 2 hours at ~ 30ºC post-induction for sHSP and T4L respectively.
All T4L mutants were purified by a two step separation process. Cation exchange using a Resource S column was used as an initial clean-up step. Immediately following elution, T4L mutants were labeled with monobromobimane in a 10 fold stoichiometric ratio. Reaction of the fluorophore with cysteine at position 151 was allowed to proceed overnight to ensure complete derivatization. Although neither BSI nor calorimetry require a labeled analyte for detection, all T4L mutants were fluorescently derivatized and used throughout the experiments to ensure no variability existed between the ligands used in each detection method. Labeled solutions were further purified by size exclusion chromatography using a Superdex 75 column. The eluted analyte was concentrated using Amicon® centrifugal concentrators and characterized by UV-Vis spectroscopy. Labeling efficiency was determined by monitoring absorbance peaks at 280 and 380 nm. Mutant T4L concentrations were determined by absorbance at 280 nm using an extinction coefficient of 1.231 cm2/mg.
αB-crystallin was purified in a three step separation process. αB-crystallin was loaded onto a Source Q column for anion exchange and eluted with a sodium chloride gradient. After the eluant had been adjusted to a final concentration of 0.5M ammonium sulfate, the solution was loaded onto a phenyl-Sepharose column and eluted with a gradient transitioning from 1M to 0M ammonium sulfate. A final purification step was performed by size-exclusion chromatography using a Superose 6 column. No phenyl-Sepharose column was used in the purification of β-crystallin or αA-crystallin. Following anion exchange, purified β-crystallin was reacted with a 10-fold molar excess of bimane label and incubated for 2 hours at room temperature. Excess bimane label was removed and the fluorescently labeled β-crystallin was purified by size-exclusion chromatography on a Superdex 75 column. Solutions of sHSP were concentrated using centrifugal filters and then characterized by UV-Vis spectroscopy. The concentration of each sHSP construct was determined at 280 nm using the appropriate extinction coefficient.
BSI Preparation and Setup
Solutions of T4L and sHSP variants used in BSI experiments were buffered with 9mM Tris, 6mM MES, 50mM NaCl, and 0.02% sodium azide. The pH of each solution was adjusted by the addition of small amounts of 5N NaOH or 5N HCl and monitored by a standard pH electrode. All solutions were filtered and degassed prior to binding experiments. Solutions were kept on ice during the experiment and briefly allowed to warm to room temperature prior to their introduction into the microchannel. The mixing chip design used was a hybrid serpentine-hydrodynamic focusing mixer as described by Bornhop, et al.5 with microchip fabrication details provided in SI Section 1 and presented in SI Figure S1. Experiments were maintained at the desired temperature by a MELCOR temperature controller coupled to a Peltier device. Approximately 4 µL of each sHSP and T4L construct was used in obtaining a single association curve. Binding experiments were monitored in real-time at frequencies ca. 50–100 Hz and in detection volumes on the order of picoliters. A detailed description of BSI components, optical train, and working conditions are given in SI Section 4 and displayed in SI Figure S3.
ITC Preparation and Setup
A MicroCal VP-ITC was employed to cross validate results obtained by BSI. Solutions of αB-crystallin and T4L-L99A were buffered with 0.15M Na2H2PO4, 0.1M KCl, 0.1mM EGTA, and 0.1% sodium azide. αB-crystallin (~ 1.4 mL) was housed in the sample cell for ITC experiments and had an initial concentration of 12 µM. The buffer solution was kept in the reference cell. Approximately 260 µL of T4L mutant at a concentration equal to 120 µM was drawn into a syringe housed within an automated pipette system. 10 µL of the T4L mutant was injected into the sample cell containing αB-crystallin twenty-five times with ~ 7 minutes allowed between injections to bring the signal back to baseline. The heat evolved after each injection was recorded and experimental data was analyzed by Origin® software to calculate thermodynamic parameters for comparison to BSI and fluorescence experiments. Further ITC experimental parameters are provided in SI Section 3.
RESULTS AND DISCUSSION
Method Comparison and Benchmarking of BSI
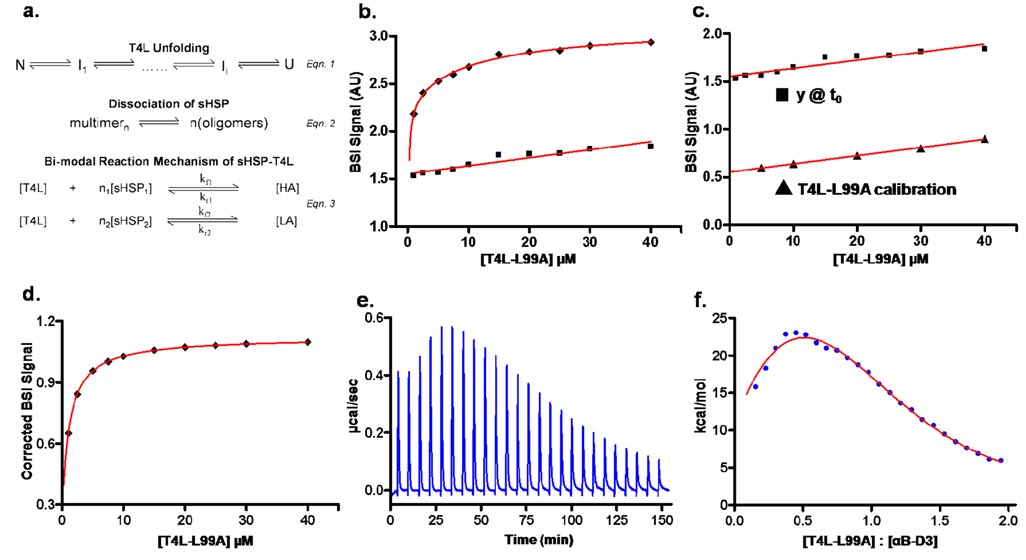
The interaction of sHSP with T4L can be represented by the coupled system shown in Figure 1a.24 T4L can partially or globally unfold (Figure 1a, eqn. 1), occupying different energy states thus altering the affinity for sHSP binding. sHSP form oligomers that rapidly exchange subunits and dissociate into different sized multimers. Previous studies of Hsp27/T4L interactions revealed that the Hsp27 dissociated species have higher affinities for substrate proteins (Figure 1a, eqn. 2).25 The binding of sHSP to T4L (Figure 1a, eqn. 3) has been shown to be bi-modal with sHSP having low and high affinity sites.24, 26 Here, we compare the apparent dissociation constants of T4L binding to αB-crystallin determined from steady-state ITC and fluorescence analyses to that determined by BSI.
Figure 1.
a) Binding of sHSP to their substrate is represented by three coupled equilibria: (1) T4L transition from native (N) to unfolded (U) states, (2) dissociation of the sHSP large oligomer into dimers or tetramers, and (3) formation of the sHSP•T4L complex. HA and LA refer to high affinity and low affinity complexes respectively. Steady-state BSI data (◆) shows that the magnitude of the binding signal increases upon the addition of increasing concentrations of T4L-L99A to a fixed concentration of αB-D3 (7.5 µM). [T4L-L99A] = 1.0, 2.5, 5.0, 7.5, 10, 15, 20, 25, 30, and 40 µM. The linear rise in starting values (■) reflects the response of BSI to increased concentrations of free L99A. c) The slope of the starting values of the traces in c) is identical to that obtained from direct injection of T4L without αB-D3. (d) The calibration curve was used as a baseline subtraction to obtain a corrected steady-state binding trace. Isothermal titration calorimetry (ITC) analysis of αB-D3•T4L-L99A. (e) Heat evolved after each 10 µL injection of T4L-L99A (120 µM) into a reservoir containing 1.4 mL of αB-D3 (12 µM) was detected for 25 injections. f) The area under the curve was extracted and plotted against the molar ratio to obtain a binding isotherm. Non-linear least-squares analysis (red curve) was used to determine thermodynamic parameters.
The αB-crystallin variant used in this study is a triply substituted analog of the native protein in which serine residues 19, 45, and 59 are mutated to aspartic acids. At physiological pH, the acidic residues create a triply charged αB-crystallin species that mimics phosphorylation. Referred to as αB-D3, this αB-crystallin mutant has increased affinity for non-native proteins presumably due to a shift in the equilibrium of oligomer dissociation (Figure 1a).24 The substrate, bimane-labeled T4L-L99A, is a mutant which has been exhaustively characterized against multiple sHSP.22, 24–28 With an alanine substituted for leucine at the hydrophobic core position 99, the buried surface area of T4L-L99A is reduced causing enlargement of preexisting cavities.29 The structure of T4L-L99A is nearly identical to that of the wild-type (WT) T4L with only minor rearrangement in the core. The mutation lowers the Gibb’s free energy of unfolding (ΔGunf) by 5.1 kcal/mol relative to WT.22, 28
Titration of a constant amount of αB-D3 with increasing T4L-L99A concentrations (Figure 1b) increases the absolute BSI signal. A binding curve is obtained by plotting the steady-state amplitude of the BSI signal versus the concentration of T4L-L99A. This is equivalent to the fluorescence-based binding isotherms24 except that the concentration of T4L is being varied rather than sHSP. There are two distinguishing characteristics of the BSI isotherm. First, the end point values do not appear to reach saturation, rather they increase linearly at high concentrations of ligand (◆ – Figure 1b). Second, a similar pattern was also observed for the starting values (■ – Figure 1b) for each αB-D3•T4L-L99A binding run. The linear increase in signal at t = 0 was found to reflect increases in the free T4L concentration. This trend was not observed in previous BSI-monitored binding assays since the reactant concentrations used were small. Therefore changes in the amount of free ligand led to signal contributions that were near or below the sensitivity of the instrument.5 However, the span of high concentrations for the relatively large ligand, T4L (16.7 kDa), produces a linear response with increased amounts of free T4L-L99A. To confirm this interpretation, a calibration curve of T4L-L99A was constructed to detect the response of BSI to free concentrations of ligand (Figure 1c). The slope of the calibration curve matches (Δm = 2.3%) the slope of the ascending starting values from αB-D3•T4L-L99A binding traces (y @ t0 – Figure 1c). This indicates the linear rise of end point values at high concentrations is due to an increase in free T4L-L99A. Accounting for this slope in the steady-state data amounts to a baseline subtraction and was performed prior to further data analysis (SI Section 2).
Thermodynamic analysis of binding was carried out on the baseline-corrected data using a two-mode binding model as described by Sathish, et al.24 (SI Section 2). The detected signal was modeled as arising from two components: free T4L and the sHSP•T4L complex. Binding depletes the free ligand pool and increases the contribution of the complex to the signal. Using the two-mode binding formalism, steady-state data can be fit to obtain the dissociation constants, the number of binding sites, and the BSI signal arising from each mode. A single-site binding analysis was also performed to serve as a null hypothesis. F ratio comparison of the single and two-site binding formalisms led to a rejection of the null hypothesis at α = 0.05 signifying that the BSI signal arises from a bi-modal molecular interaction. Further confirmation of two-mode binding was obtained from a phenomenological analysis of the real-time binding data. All kinetic traces were fit to both a single and double exponential by nonlinear least squares regression. A comparison of the square of residuals from each fitting routine confirmed the double exponential nature of the data (SI Figure S2). The observed rates, as determined from the iterative fitting process, are linear functions of L99A concentration. This dependence is predicted by the law of mass action and additionally suggests at least two separate kinetic events contribute to the BSI signal change.
Thermodynamic parameters determined by BSI were compared to results obtained from ITC experiments (Figure 1e and 1f) and fluorescent binding measurements in Table 1. The experimental low affinity dissociation constant (KD2) is very similar across platforms. However, the KD for high affinity binding obtained from ITC and fluorescence studies are close to 20-fold larger when compared to BSI analysis. It is likely that the overestimation of the affinity by BSI reflects the limited data set in the range of large molar ratios between αB-D3 and T4L in Figure 1c. A thorough analysis using determination of forward and reverse kinetic rates yield more consistent parameters as shown below. For the purpose of this comparison, however, steady state data was exclusively used because kinetic data cannot be obtained from traditional ITC.
Table 1.
Comparison of binding parameters obtained by BSI, ITC and fluorescence. Also highlighted is the substantial reduction in material needed for BSI experiments.
| T = 37°C and pH = 7.2 | |||||
|---|---|---|---|---|---|
| Data | |||||
| ITC | error bounds |
Fluorescence24 | BSI | error bounds |
|
| moles of αB-D3 used | 24 × 10−9 | ~ 50 × 10−9 | 1.5 × 10−9 | ||
| moles of L99A used | 54 × 10−9 | ~ 50 × 10−9 | 2.9 × 10−9 | ||
| n1 | 0.25 | 0.24 | 0.24 | ||
| KD1 (µM) | 0.015 | + 0.04 | 0.020 | 0.001 | + 0.04 |
| −0.04 | −NWD | ||||
| n2 | 1.1 | 1.1 | 1.1 | ||
| KD2 (µM) | 2.44 | + 31.3 | 1.00 | 3.11 | + 2.59 |
| −31.3 | −2.31 | ||||
NWD ≡ not well defined
Overall the three methods report consistent levels of binding since the high affinity binding sites are saturated and low affinity binding is comparable. However, BSI analysis was carried out with considerably lower amounts of reagent. ITC and fluorescence required roughly 100 nanomoles each of sHSP and T4L while BSI experiments consumed nearly 20 times less. The reduced consumption of analytes is an intrinsic property of BSI and is valuable in molecular interaction studies. Different constructs or mutated sequences often have lower expression yields, limiting the number of analyses that can be performed. In fact, the level of T4L expression for double mutants is enough to run only two experiments via ITC. The same amount of protein is sufficient for approximately forty experiments with BSI.
Specificity of BSI-detected Binding: Increased α-crystallin Affinity for Destabilized Proteins
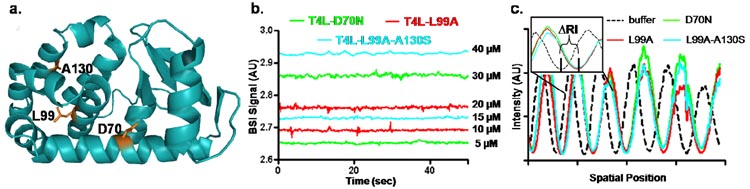
The coupled equations in Figure 1a predict that the affinity of sHSP to their substrates reflect the propensity of the latter to occupy non-native states. This prediction was verified for a number of sHSP using a set of T4L mutants differing in their ΔGunf but having similar overall structure to the native state.30 In order to establish the specificity of detection by BSI and highlight the additional kinetic dimension provided by this technique, we investigated the interaction of αB-D3 to two T4L mutants (Figure 2a) having different ΔGunf. The double mutant, T4L-L99A-A130S, has a ΔGunf equal to 3.5 kcal/mole. T4L-D70N is a more stable mutant with a ΔGunf equal to 6.8 kcal/mol.28
Figure 2.
a) Structure of T4L highlighting the mutation sites. b) Time-dependence of BSI signals following the injection of T4L mutants without sHSP. Increasing T4L concentration shifts the baseline but does not lead to a time-dependent change, ruling out mixing artifacts. c) BSI fringe patterns of the T4L mutants demonstrate BSI is insensitive to differences in their stabilities. A zoomed in region of the interference patterns is shown and compared to a fringe pattern from a buffer solution demonstrating the sensitivity of the instrument to changes in refractive index.
To monitor possible mixing artifacts, mutants of T4L were mixed in channels with buffer solution and the kinetic traces recorded by BSI (Figure 2b). Increasing T4L concentration results in an offset but the traces are not time-dependent. Thus, time-dependent BSI signals observed in the presence of αB-D3 and T4L are not an artifact of mixing. Figure 2c shows the interference pattern observed for each T4L mutant and the buffer solution while contained within the microfluidic channel. The zoomed-in region of the fringe pattern demonstrates the sensitivity of BSI to refractive index, with a substantial shift seen between the interference patterns of the T4L mutants and buffer. The fringe patterns for all T4L mutants studied precisely overlap with point-to-point minima matching. Thus, differences in BSI signals for different αB-crystallin/T4L-mutants must reflect changes in their interactions and not merely different BSI signals for different mutants.
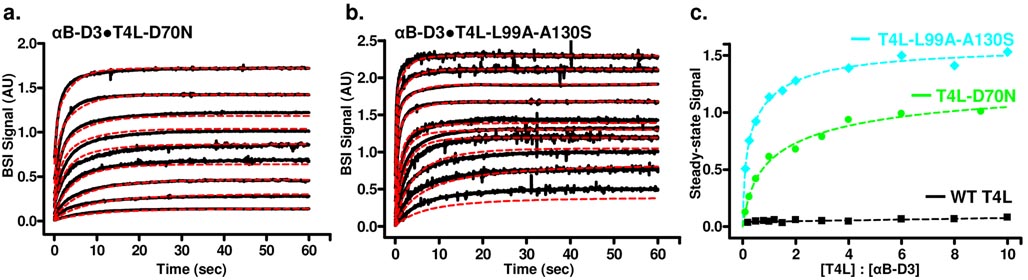
The association of αB-D3 with multiple concentrations of T4L was monitored in real-time by BSI analysis (Figure 3a and 3b). As described above, T4L calibration curves were calculated for each mutant and used for baseline subtraction of the initial starting values. Comparison of corrected steady-state data for αB-D3•T4L interactions reveals a qualitative difference between the BSI signals of each mutant (Figure 3c). An ~ 40% reduction in magnitude is observed for T4L-D70N relative to T4L-L99A-A130S at equivalent αB-D3 to T4L molar ratios suggesting lower levels of binding. Furthermore, saturation is observed at lower molar ratios for the T4L-L99A-A130S mutant. The lack of interaction between αB-D3 and WT-T4L serve as a control. Given the larger ΔGunf of the WT, the equilibrium fraction of unfolded or non-native conformations is negligible (Figure 1a, eqn. 1). Therefore marginal binding is predicted across the entire concentration range. Experimentally, BSI signals for αB-D3•T4L-WT were within the experimental noise level.
Figure 3.
Kinetics of αB-D3 (10 µM) binding to multiple concentrations of T4L-D70N (a) and T4L-L99A-A130S (b) were monitored by BSI (black) with kinetic traces fit via global analysis (red). Analysis of the steady-state data (c) shows that the magnitude of binding as detected by BSI for αB-D3•T4L-L99A-A130S is significantly greater than seen with αB-D3•T4L-D70N. As a control, αB-D3 was assayed against multiple concentrations of WT-T4L, exhibiting no binding across the concentration range. [T4L-D70N] = 1, 2.5, 5, 10, 20, 30, 40, 60, and 90 µM. [T4L-L99A-A130S] = 1, 2.5, 5, 10, 15, 20, 40, 60, 80, and 100 µM.
In order to quantitatively describe the kinetics of the αB-D3•T4L system observed by BSI, a more general approach was developed which overcome the limitations of the kinetic analysis performed in earlier BSI studies. Bornhop, et al.5 fit real-time traces to a single exponential based on pseudo-first order kinetics under conditions of excess ligand concentration. Given the two-mode binding of T4L by α-crystallin, such a restricted parametric approach is not adequate for quantitative data analysis. Complex systems are more accurately described by higher ordered functions, generally a double exponential for interaction studies. Therefore, the BSI traces were fit simultaneously to one set of parameters that minimizes a global error function, χ2. Analysis of the kinetic data is based on simple rate equations derived from the binding equilibria (SI Section 2). The time dependent BSI signal was fit to obtain forward (kf) and reverse (kr) rate constants. The global analysis31 strategy employed reduces the effects of instrumental artifacts in a particular kinetic trace on the final parameters and overcomes the uneven sensitivity to the parameters at either extreme of molar ratios.
Results from the global analysis of BSI experiments (Table 2) indicate that the affinity of αB-D3 binding in the high affinity mode increases by a factor of 4 for the more destabilized mutant, T4L-L99A-A130S. The interaction of αB-D3 with both T4L mutants appears similar and weak in the low affinity mode. These results clearly indicate that BSI is capable of measuring molecular interactions with high specificity, resolving binding of a sHSP to structurally similar T4L mutants differing only in their ΔGunf. Furthermore, comparison of the data obtained from the global analysis of BSI kinetic data is consistent with previously published fluorescent analyses of the same sHSP•T4L binding systems.22, 24
Table 2.
Kinetic parameters acquired from the global analysis of BSI data for the interaction of αB-D3 with destabilized mutants of T4L.
| T4 Lysozyme | ||||||
|---|---|---|---|---|---|---|
| Data | ||||||
| WT | D70N | error bounds |
L99A-A130S | error bounds |
||
| ΔGunf (kcal/mol) | 14 | 9.2 | 7.9 | |||
| kf1 (M−1s−1) | UND | 25545 | + 4455 | 58515 | + 11485 | |
| − 3545 | + 11515 | |||||
| kr1 (s−1) | UND | 1.52 × 10−2 | + 0.008 | 5.03 × 10−6 | NWD | |
| − 0.007 | NWD | |||||
| KD1 | UND | 0.595 µM | + 0.450 | 0.132 µM | NWD | |
| − 0.328 | NWD | |||||
| kf2 (M−1s−1) | UND | 2342 | + 458 | 2970 | + 1030 | |
| − 442 | − 770 | |||||
| kr1 (s−1) | UND | 0.068 | + 0.017 | 0.115 | NWD | |
| − 0.013 | − 0.25 | |||||
| KD2 | UND | 29.1 µM | + 15.6 | 33.0 µM | NWD | |
| + 9.5 | − 2.70 | |||||
UND ≡ undetectable ; NWD ≡ not well defined
Interaction of an αA-crystallin Mutant with the Lens Protein βB1-crystallin
The molecular targets of α-crystallin are lens proteins such as β and γ-crystallins.14 These proteins are long lived and undergo extensive post-translational modifications; some of which reduce the free energy of unfolding and hence are expected to trigger binding to α-crystallin. A detailed study of the energetics of α-crystallin interaction with bimane-labeled β-crystallin has been reported32 demonstrating that α-crystallins have relatively low affinity even for highly destabilized β-crystallins.
Here we use BSI to evaluate binding of a αA-crystallin mutant genetically-linked to hereditary cataract to β-crystallin. Mackay, et al.17 identified a missense mutation in the HspB4 gene on chromosome 21q causing the formation of nuclear cataract. The point mutation underlying cataractogenesis was determined to be a replacement of arginine for cysteine at residue 49 (αA-R49C). Evidence from cellular studies17 suggest the deleterious effect of the mutated protein is caused by a toxic gain of function. αA-R49C was reported to have substantially higher affinity to destabilized mutants of T4L suggesting that this mutant acts to unfold cellular proteins forming insoluble substrate-saturated complexes.27 Furthermore, interaction of αA-R49C with thiolated substrates leads to formation of disulfide-linked α-crystallin dimers.33
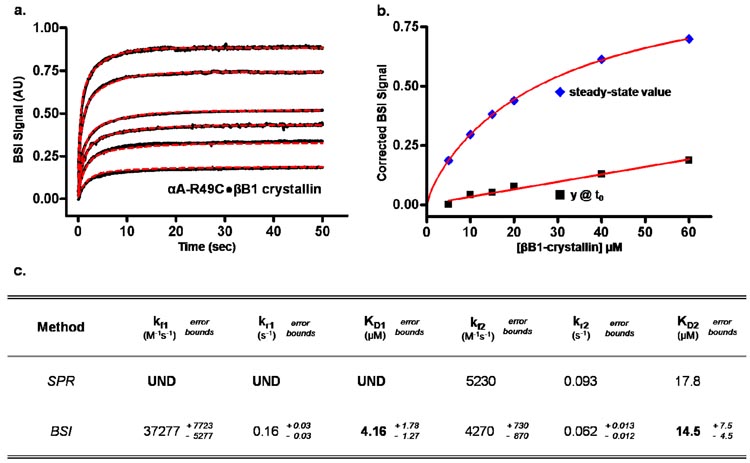
To highlight the utility of BSI in monitoring native protein-protein interactions, binding studies of the cataract-linked mutant, αA-R49C, were performed with the lens protein βB1-crystallin at physiological pH and at 37 °C. To mimic cysteine thiolation in the lens, βB1-crystallin was labeled with a bimane probe via a disulfide linkage. Other experimental conditions and the optical setup were kept consistent with the sHSP studies described above. Previously, Kamei, et al.19 used SPR to characterize the interaction of αA-crystallin with β-crystallin from human lens. αA-crystallin was immobilized onto a gold substrate using a carboxylated dextran matrix and activated by the 7 minute injection of an imide solution. β-crystallin was passed over the substrate bound αA-crystallin for 25 minutes at a flow rate of 2 µL/min. Binding was detected from changes in the local refractive index near the gold surface. Regeneration of the SPR surface was accomplished by introduction of a 15 µL solution of 0.1M sodium acetate and 0.15M sodium chloride at an extreme pH of 2.3. This procedure was repeated using various concentrations of β-crystallin.
BSI signals were collected for multiple concentrations of βB1-crystallin while keeping the concentration of αA-R49C constant (Figure 4a). The resulting time-dependent traces were simultaneously fit to yield the rate constants. BSI analysis reveals two-mode binding in contrast to the single mode reported by SPR. As seen in Figure 4c, values calculated for the second equilibrium dissociation constant (KD2) via SPR and BSI are within experimental variance. The second binding mode not detected by SPR presumably reflects the activation of αA-R49C previously reported using T4L as a substrate.27 In addition, the time and amount of protein needed for the SPR experiment far exceeds that used with BSI. In fact, a single molecular interaction run would take ~25 minutes longer using SPR. Assuming six runs were needed to complete an interaction assay, a SPR experiment would take almost 3 hours while the same BSI experiment would be completed in less than 20 minutes. The potential automation of sample introduction, on-chip dilution, and multiple detection channels made available by microfluidics can further reduce the amount of time needed to perform molecular interaction assays with BSI, enhancing the high-throughput nature of the technique.
Figure 4.
(a) Interaction of a constant concentration of αA-R49C-crystallin (15 uM) with multiple concentrations of βB1-crystallin at physiologically relevant conditions was detected by BSI. [βB1-crystallin] = 5, 10, 15, 20, 40, 60. Corrected steady-state values ( - b) were obtained by background subtraction of a βB1 calibration curve (■ - b). Global analysis of BSI kinetic data was compared to published kinetics from SPR experiments (c).21
- b) were obtained by background subtraction of a βB1 calibration curve (■ - b). Global analysis of BSI kinetic data was compared to published kinetics from SPR experiments (c).21
Conclusions
Until recently, requirements for large sample quantities and/or poor sensitivity have limited label-free molecular interaction studies. BSI’s sensitivity, ultra-small volumes, label-free and free-solution format has the potential to eliminate many of the problems associated with molecular immobilization and modification through labeling. Observations of proteins in their native state can facilitate a more accurate assessment of molecular interactions. Here we further demonstrate the utility of BSI, showing it is a practical, robust and quantitative method for characterizing protein-protein interactions. By implementing a global analysis of time-resolved data the simultaneous determination of kinetic and thermodynamic parameters using just a few nanomoles of proteins was possible. Cross validation of BSI was performed with established methods based on fluorescence and calorimetry. ITC and fluorescent experiments provided comparable outcomes to those obtained by BSI for the interaction of a sHSP construct with a mutant T4 lysozyme, yet required either labeling with a fluorescent probe and/or large sample quantities to be consumed. The specificity of BSI was highlighted by the differential binding of the chaperone to T4L mutants of varying stability. Lastly, physiologically relevant investigations were characterized in picoliter volumes facilitating the thermodynamic and kinetic evaluation of a mutated sHSP genetically linked to autosomal dominant cataract.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Vanderbilt Institute for Integrative Biosystems and Education (VIIBRE) for use of their cleanroom. Financial support was provided by the National Eye Institute grants T32-EY07135 to Joey C. Latham, R01-12018 to Hassane S. Mchaourab and R01-EB353701 to Darryl J. Bornhop.
REFERENCES
- 1.Berggard T, Linse S, James P. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 2.Velazquez Campoy A, Freire E. Biophys Chem. 2005;115:115–124. doi: 10.1016/j.bpc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Boozer C, Kim G, Cong SX, Guan HW, Londergan T. Current Opinion in Biotechnology. 2006;17:400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Toft KN, Vestergaard B, Nielsen SS, Snakenborg D, Jeppesen MG, Jacobsen JK, Arleth L, Kutter JP. Analytical Chemistry. 2008;80:3648–3654. doi: 10.1021/ac800011y. [DOI] [PubMed] [Google Scholar]
- 5.Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sorensen HS. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 6.Shekhawat G, Tark SH, Dravid VP. Science. 2006;311:1592–1595. doi: 10.1126/science.1122588. [DOI] [PubMed] [Google Scholar]
- 7.Torres FE, Kuhnt P, De Bruyker D, Bell AG, Wolkin MV, Peeters E, Williamson JR, Anderson GB, Schmitz GP, Rechtx MI, Schweizer S, Scott LG, Ho JH, Elrod SA, Schultz PG, Lerner RA, Bruce RH. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9517–9522. doi: 10.1073/pnas.0403573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 9.Jakob U, Gaestel M, Engel K, Buchner J. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 10.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 11.Bucciantini M, Giannoni1 E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 12.Forman MS, Lee VM, Trojanowski JQ. Trends Neurosci. 2003;26:407–410. doi: 10.1016/S0166-2236(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 13.Derham BK, Harding JJ. Prog Retin Eye Res. 1999;18:463–509. doi: 10.1016/s1350-9462(98)00030-5. [DOI] [PubMed] [Google Scholar]
- 14.Tardieu A, Delaye M. Annual Review of Biophysics and Biophysical Chemistry. 1988;17:47–70. doi: 10.1146/annurev.bb.17.060188.000403. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz J. Semin Cell Dev Biol. 2000;11:53–60. doi: 10.1006/scdb.1999.0351. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz J. Proc Natl Acad Sci U S A. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay DS, Andley UP, Shiels A. Eur J Hum Genet. 2003;11:784–793. doi: 10.1038/sj.ejhg.5201046. [DOI] [PubMed] [Google Scholar]
- 18.Chavez Zobel AT, Loranger A, Marceau N, Theriault JR, Lambert H, Landry J. Hum Mol Genet. 2003;12:1609–1620. doi: 10.1093/hmg/ddg173. [DOI] [PubMed] [Google Scholar]
- 19.Kamei A, Matsuura N. Biol Pharm Bull. 2002;25:611–615. doi: 10.1248/bpb.25.611. [DOI] [PubMed] [Google Scholar]
- 20.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 21.Berengian AR, Parfenova M, McHaourab HS. J Biol Chem. 1999;274:6305–6314. doi: 10.1074/jbc.274.10.6305. [DOI] [PubMed] [Google Scholar]
- 22.Koteiche HA, McHaourab HS. J Biol Chem. 2003;278:10361–10367. doi: 10.1074/jbc.M211851200. [DOI] [PubMed] [Google Scholar]
- 23.Sathish HA, Koteiche HA, McHaourab HS. J Biol Chem. 2004;279:16425–16432. doi: 10.1074/jbc.M313402200. [DOI] [PubMed] [Google Scholar]
- 24.Sathish HA, Stein RA, Yang G, McHaourab HS. J Biol Chem. 2003;278:44214–44221. doi: 10.1074/jbc.M307578200. [DOI] [PubMed] [Google Scholar]
- 25.Shashidharamurthy R, Koteiche HA, Dong J, McHaourab HS. J Biol Chem. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Koteiche HA, McHaourab HS, Stewart PL. J Biol Chem. 2006;281:40420–40428. doi: 10.1074/jbc.M608322200. [DOI] [PubMed] [Google Scholar]
- 27.Koteiche HA, McHaourab HS. J Biol Chem. 2006;281:14273–14279. doi: 10.1074/jbc.M512938200. [DOI] [PubMed] [Google Scholar]
- 28.McHaourab HS, Dodson EK, Koteiche HA. J Biol Chem. 2002;277:40557–40566. doi: 10.1074/jbc.M206250200. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson AE, Baase WA, Matthews BW. J Mol Biol. 1993;229:747–769. doi: 10.1006/jmbi.1993.1077. [DOI] [PubMed] [Google Scholar]
- 30.Matthews BW. Faseb J. 1996;10:35–41. doi: 10.1096/fasebj.10.1.8566545. [DOI] [PubMed] [Google Scholar]
- 31.Beechem JM. Biophys J. 1998;74:2141. doi: 10.1016/S0006-3495(98)77922-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McHaourab HS, Kumar MS, Koteiche HA. FEBS Lett. 2007;581:1939–1943. doi: 10.1016/j.febslet.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar MS, Koteiche HA, Claxton DP, McHaourab HS. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.11.047. In Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.