Abstract
The Culex pipiens complex in Asia includes a temperate subspecies, Culex pipiens pallens, of uncertain taxonomic status. The shape of the male genitalia suggests it is a hybrid between Cx. pipiens and Cx. quinquefasciatus. We studied populations of Cx. p. pallens in Japan, Korea, and China and compared them to local populations of Cx. quinquefasciatus and Cx. p. pipiens. We examined variation in a nuclear intron in the acetylcholinesterase-2 gene [ACE] and eight microsatellite loci. We found a distinct microsatellite signature for Cx. p. pallens indicating restricted gene flow between Eastern and Western populations of Cx. pipiens, supporting the existence of two subspecies. Furthermore, a multilocus genotype analysis revealed current hybridization between Cx. p. pallens and Cx. quinquefasciatus in southern Japan, Republic of Korea, and China but not in Hokkaido, in northern Japan. Surprisingly, however, we found that the sex-linked ACE locus in chromosome I has introgressed asymmetrically through the males such that all male Cx. p. pallens have a copy of the Cx. quinquefasciatus ACE locus. This result highlights some of the potential consequences of hybridization between local and introduced species to disease transmission worldwide.
Keywords: HYBRIDIZATION, GENETIC INTROGRESSION, SPECIATION, INVASIVE SPECIES, DISEASE VECTORS, ASIA, SEX-LINKED
INTRODUCTION
Natural hybridization, defined as “Successful matings in nature between individuals from two populations, or groups of populations, that are distinguishable on the basis of one or more heritable characters”(Harrison 1990, Arnold 2004b), can provide favorable conditions for rapid evolution and even lead to speciation (Arnold 1997, 2004a, Jiggins et al. 2008). Although hybridization processes are harder to observe when they involve populations of the same species or cryptic taxa, the advent of molecular techniques has allowed their detection (Ellstrand and Schierenbeck 2000, Meixner et al. 2002, Kovarik et al. 2005, Fonseca et al. 2006). We can now further our studies of hybridization as an evolutionary force and, in particular, examine the contribution of the horizontal transfer of advantageous genes (Arnold 2004b, Currat et al. 2008). Nonetheless, the consequences of natural hybridization can be difficult to study since each is a “natural experiment” often unique or with multiple events locally correlated. Exceptionally in the Culex pipiens complex a group of important disease vectors, hybrid zones appear to have been created repeatedly and independently across the World, as the two main species in the complex, Cx. pipiens and Cx. quinquefasciatus were introduced into new areas with migrating people and commercial traffic (Mattingly et al. 1951).
Recurring hybridization occurs in the Culex pipiens complex mostly between the two widespread species in the complex, Culex (Culex) pipiens L. 1758 and Cx. (Cx.) quinquefasciatus Say 1823. Hybrids have been documented in North America, Argentina, as well as in Madagascar (Barr 1957, Urbanelli et al. 1995, Urbanelli et al. 1997, Humeres et al. 1998), and have been hypothesized in East Asia (Smith and Fonseca 2004, Cui et al. 2007). This is therefore a system where comparative analyses are potentially feasible. With that aim, we have been examining the Cx. pipiens complex in East Asia. A temperate sub-species of Cx. pipiens, Cx. pipiens pallens Couquillett 1898 is restricted to Eastern Asia where it vectors lymphatic filariasis and canine dogworm and has been well studied especially under the threat of introduction of West Nile virus (Choi et al. 2002, Jang et al. 2002, Oda et al. 2002, Yang et al. 2003). Its relationship to the remaining members of the complex has, however, remained a mystery. Because the male genitalia (phallosome) of Cx. p. pallens has a shape very similar to that of hybrids of Cx. pipiens and Cx. quinquefasciatus, which is intermediate between the two parent species (Barr 1957, Tanaka et al. 1979), Cx. p. pallens has often been described as a hybrid (Bekku 1956, Laven 1967, Cornel et al. 2003). Bekku (1956) found a north-south gradient in the morphology of the phallosome of Cx. p. pallens in which specimens from northern Japan were more like the temperate species, Cx. pipiens, and specimens from southern Japan were more like Cx. quinquefasciatus, the tropical species. In Japan, Cx. quinquefasciatus are restricted to the Ryukyu Islands (Kasai et al. 2008), and while they have not been reported in the Republic of Korea (commonly referred to as “South Korea” and from now on abbreviated ROK), this species occurs in southern China (Cui et al. 2007). Furthermore, Culex p. pipiens, has two epidemiologically distinct forms: form “pipiens” and form “molestus” (Harbach et al. 1984). The urban form of Cx. p. pipiens (form molestus) was introduced into northern East Asia likely during WWII (Mattingly et al. 1951), but the feral form (Cx. p. pipiens form pipiens) has not been found in Asia (Cui et al. 2007, Kasai et al. 2008).
While mitochondrial DNA has failed to show informative variation, possibly as a result of selective sweeps driven by Wolbachia pipientis (Guillemaud et al. 1997), members of the Cx. pipiens complex can be identified by rapid assays using nuclear loci (Smith and Fonseca 2004, Bahnck and Fonseca 2006, Kasai et al. 2008). The objective of this study was to test the hypothesis that populations of Cx. p. pallens are the result of hybridization between the western Cx. pipiens (Cx. p. pipiens) and Cx. quinquefasciatus as well as examine the overall population structure of this subspecies in eastern Asia (China, ROK, and Japan). To do so we used variation in the acetylcholinesterase-2 locus [ACE] previously found to be useful in examining hybridization in the Cx. pipiens complex (Smith and Fonseca 2004, Kothera et al. 2009), as well as a panel of microsatellite loci developed for the Cx. pipiens complex.
MATERIAL AND METHODS
We examined variation at a nuclear intron (the ACE locus) and at eight microsatellite loci. The microsatellite loci were developed from other members of the Cx. pipiens complex but were optimized in Culex pipiens pallens (Smith et al. 2005). Figure 1 depicts a geographic map of the relative positions of the collection sites. The specimens used are listed in Table 1 and were obtained by us and from local entomologists. DNA was extracted using a standard phenol/chloroform method (Fonseca et al. 2000), and the presence of diagnostic ACE bands for members of the Cx. pipiens complex was scored (Smith and Fonseca 2004). Specimens identified as Cx. p. pipiens were further examined with a second rapid assay (Bahnck and Fonseca 2006) to identify the form (“pipiens” vs. “molestus”). We also compared the microsatellite signature of Asian Cx. p. pallens populations to those of a sample of Cx. p. pipiens form pipiens from Northern Europe previously published (Fonseca et al. 2004).
Figure 1.
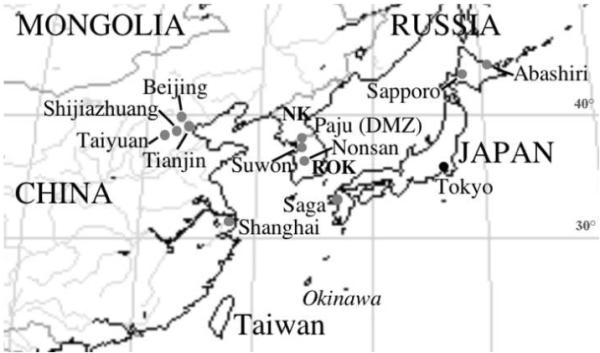
Schematic geographic map of parts of northeastern Asia depicting the relative positions of the sources of specimens of Culex pipiens pallens examined in this study. NK= Democratic People’s Republic of Korea; ROK= Republic of Korea. Please refer to Smith and Fonseca (2004) for a world map depicting the approximate distributions of the various members of the Culex pipiens complex.
Table 1.
Populations of Culex pipiens complex mosquitoes analyzed in this study. Nm and NACE are the numbers of specimens from each population examined with microsatellite markers and the ACE locus, respectively
| Species | Location | Latitude (deg min) | Nm | NAce | Collection date | Source* |
|---|---|---|---|---|---|---|
| Culex pipiens pallens and hybrids | Abashiri, Japan | 44 01 N | 14 | 3 | 07/2004 | M. Mogi |
| Sapporo, Japan | 43 05 N | 22 | 3 | 06/2001 | M. Mogi | |
| Saga, Japan | 43 05 N | 16 | 4 | 06/1999 | M. Mogi | |
| Paju, ROK | 37 80 N | 8 | 2 | 08/2003 | M. Turrell | |
| Suwon, ROK | 37 17 N | 7 | 2 | 11/2003 | H-C. Kim | |
| Nonsan, ROK | 36 12 N | 16 | 3 | 06/2004 | H-C. Kim | |
| Beijing, China | 39 55 N | 8 | 1 | 10/2002 | T. Zhao/C. Curtis | |
| Tianjin, China | 39 08 N | 7 | 1 | 10/2002 | T. Zhao/C. Curtis | |
| Shijiazhuang, China | 38 03 N | 5 | 2 | 10/2002 | T. Zhao/C. Curtis | |
| Taiuman, China | 37 55 N | 5 | 1 | 10/2002 | T. Zhao/C. Curtis | |
| Shanghai, China | 31 14 N | 7 | 0 | 04/2004 | P. L. M. Rueda | |
| Cx. quinquefasciatus | Okinawa, Japan | 26 20 N | 22 | 06/1999 | I. Miyagi | |
| Shanghai, China | 31 14 N | 2 | 04/2004 | P. L. M. Rueda | ||
| Cx. pipiens pipiens | Seoul, ROK | 37 33 N | 8 | 11/2003 | H-C. Kim | |
| Busan, ROK | 32 06 N | 15 | 10/2003 | H-C. Kim | ||
Dr. Mike Turrell, U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Maryland 21702; Dr. Tongyan Zhao, Beijing Institute of Microbiology & Epidemiology, Beijing, China; Dr. Chris Curtis, London School of Hygiene & Tropical Medicine, London, UK; Dr. Pollie L. M. Rueda, WRBU, WRAIR/Smithsonian Institution, Washington, DC, USA; Dr. Ichiro Miyagi, University of the Ryukyus, Okinawa, Japan.
The ACE locus
A small part of exons 2 and 3 and the entire intron II of the ace-2 gene (the ACE locus,(Bourguet et al. 1998) were amplified using a PCR+1 protocol to prevent polymerase errors and heteroduplex formation (Borriello and Krauter 1990). First, an asymmetric amplification was performed in 50-μL reactions containing 0.25 μM of the primer F1457 (5′-GAGGAGATGTGGAATCCCAA-3′), 2.5 μM of B1246 (5′-TGGAGCCTCCTCTTCACGGC-3′ ), 1X Easy-A reaction buffer, 500 μM of each dNTP, 2.5 units Easy-A high-fidelity PCR cloning enzyme (Stratagene, La Jolla, CA), and approximately 6 ng of DNA template. The amplification program consisted of 95°C for 2 min, 30 cycles of 95°C for 40 sec, 54°C for 30 sec, 72°C for 1 min, followed by 72°C for 7 min. Twenty microliters of the PCR product were used in a new 50-μl reaction for one additional cycle (the PCR+1 step, times and temperatures identical to those above) with 2.5 μM of a third primer F1457MluI (5′-ACGCGTGAGGAGATGTGGAATCCCAA-3′) with an extra 5μl of 1 X Easy-A buffer, additional dNTPs (to 500μM), and 2.5 units of Easy-A. The primer F1457MluI contains a cut-site for the restriction enzyme Mlu I.
The ACE locus of Cx. p. pallens was amplified using the PCR+1 protocol and cloned with a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Clones were amplified with the sequencing primers M13F(-20) and M13R. Reactions were carried out in 50-μL volumes containing 0.75 μM of each primer, 1X PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl), 500 μM of each dNTP, and 1.5 units Taq polymerase (Applied Biosystems, Foster City, CA). The M13 product was cleaned using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and digested with the restriction enzyme Mlu I (New England Biolabs, Beverly, MA) according to manufacturer’s instructions. The digestion product was electrophoresed on 1% agarose gel to detect clones that contained the cut site from the PCR+1 amplification step. Only clones with the cut site (less than 30% of the digested clones) were sequenced since it ensured they were produced during the last PCR step using the third primer and therefore did not result from priming by partially extended DNA fragments, which can lead to PCR cloning artifacts. Two clones per specimen were sequenced using standard cycle sequencing conditions (Big Dye, ABI, Foster City, CA). The resulting fragments were analyzed by electrophoresis in a slab gel (ABI 377) automated sequencer (Applied Biosystems, Foster City, CA). Sequences were aligned in Sequencher version 4.1 (GeneCodes, Ann Arbor, MI).
Microsatellite analysis
We used eight of the twenty microsatellite loci currently optimized for the Cx. pipiens complex (CQ11F2R2, CQ26FR, CxqGT4F3R, CxqGT6bFR, EmmaFR, CxpGT12F2R2, CxpGT46FR, and GT9FR) since they appeared to amplify consistently in Cx. p. pallens (Smith et al. 2005). The loci were multiplexed and amplified in 20-μL reactions containing 0.2 μM of each primer, 1X PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl), 200 μM of each dNTP, 250 μM MgCl2, 150 μg/mL of bovine serum albumin, 0.5 units of Taq polymerase (Applied Biosystems, Foster City, CA), and approximately 6 ng of the DNA template. The amplification program consisted of one cycle of 94°C for 5 min, 30 cycles of 94°C for 30 sec, 52°C or 54°C (depending on primer, (Smith et al. 2005) for 30 sec, 72°C for 30 sec, and one cycle of 72°C for 5 min. A positive control with the clone used to design the microsatellite primers and a water negative control were included in each batch of samples. Microsatellite regions were sized in an ABI3100 automatic sequencer (Applied Biosystems, Foster City, CA) and analyzed with GeneMapper 3.7 (Applied Biosystems, Foster City, CA).
Statistical analyses
The ACE locus sequence data was analyzed in Arlequin (Schneider et al. 2000) to obtain measures of molecular diversity, calculate FSTvalues, and their significance, as well as obtain exact tests of population differentiation. The microsatellite data was first examined for compliance with Hardy-Weinberg equilibrium GENEPOP 1.2. (Raymond and Rousset 1995), and then pair-wise FSTvalues and their significance, were obtained using Arlequin (Schneider et al. 2000). Further, we assigned individuals to clusters based on their microsatellite multilocus genotypes with a maximum likelihood algorithm implemented in the program Structure 2.0 (Pritchard et al. 2000b). This method combines all the individual multilocus genotypes and separates them into distinct clusters analogous to the hierarchical branching of tree diagrams (Rosenberg et al. 2001). We used 20,000 burn-in steps and 1,000,000 runs with a model of uncorrelated allele frequencies allowing admixture (lambda = 0.64, calculated at K = 1 (Pritchard et al. 2000a). In this analysis, the origin of each specimen is not disclosed but the number of clusters (K) is decided a priori for each run. To assess the consistency of the analysis we performed an exhaustive comparison of 10 runs at each K scoring the similarity coefficient described in Rosenberg and others (2002).
RESULTS
We examined 115 specimens of Culex pipiens pallens, 24 of Cx. quinquefasciatus, and 23 of Cx. pipiens pipiens form molestus (Table 2). All specimens of Cx. p. pipiens from Japan and ROK examined had the genetic signature of the “molestus” form.
Table 2.
Polymorphisms in the second intron of the ace-2 gene. Of the 612 bp sequenced only the variable positions are shown. Position 1 corresponds to position 1 in GenBank accession #AY497524.(Smith and Fonseca 2004) Deletions were not included unless the position was also polymorphic, and are indicated by a dash (-). JPN, SKO, and CHN, refer to the frequency of each allele in Japan, ROK, and China, respectively
| 0 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 3 | 3 | 3 | 4 | 4 | J | S | C | |
| 6 | 1 | 5 | 6 | 7 | 2 | 3 | 7 | 1 | 8 | 8 | 5 | 7 | P | K | H | |
| 6 | 5 | 1 | 4 | 5 | 1 | 7 | 5 | 2 | 4 | 7 | 1 | 2 | N | O | N | |
| A1 | C | C | C | G | - | C | T | C | G | T | T | A | C | 2 | 0 | 0 |
| A2 | . | . | . | . | . | . | . | . | . | . | . | G | . | 1 | 0 | 0 |
| A3 | . | . | . | A | T | . | . | T | . | . | . | G | . | 1 | 1 | 0 |
| A4 | T | A | . | . | A | . | . | . | . | . | . | . | . | 6 | 1 | 0 |
| A5 | . | . | . | . | T | . | . | . | T | . | C | G | . | 1 | 0 | 0 |
| A6 | . | . | . | . | T | . | . | . | T | . | . | G | . | 3 | 3 | 3 |
| A7 | T | A | . | . | . | . | . | . | . | . | . | . | T | 1 | 5 | 0 |
| A8 | . | . | . | . | . | A | A | . | . | C | . | G | . | 0 | 0 | 1 |
| A9 | . | . | T | . | . | . | . | . | . | . | . | . | . | 0 | 0 | 1 |
| A10 | . | . | . | . | A | . | . | . | . | . | . | . | . | 0 | 2 | 0 |
| A11 | . | . | T | . | A | . | . | . | . | . | . | . | . | 0 | 1 | 0 |
| A12 | . | . | . | . | . | . | . | . | . | C | . | G | . | 1 | 0 | 0 |
The ACE locus was polymorphic across and within the populations of Cx. p. pallens examined (gene diversity = 0.87±0.03 with 13 polymorphic sites/612 total basepairs examined, Table 2). We excluded from this analysis sequences that matched those obtained from Cx. quinquefasciatus (Smith and Fonseca 2004) since we had independent evidence of hybridization (see below) and wished to examine just Cx. p. pallens sequences for phylogeographic purposes. To increase sample sizes, we grouped sequences by country (Japan, ROK, and China) and found that on average one quarter of the ACE sequences were unique to each country (Table 2), although we did not find evidence of population differentiation based on this locus (all pair-wise FST values failed to be statistically significant, data not shown).
The rapid assay based on diagnostic bands in the ACE locus revealed complete hybridization between Cx. p. pallens and Cx. quinquefasciatus as all male specimens had the “quinques” diagnostic band as well the “pallens” band (Table 3, Figure 3). However, this assay did not reveal extensive hybridization in females as they have overwhelmingly only the “pallens” ACE diagnostic band (Table 3). A few males and females in very southern populations had only the “quinques” diagnostic band (Table 3).
Table 3.
Comparison of rapid assay (ACE) results and gender in Culex pipiens pallens. Names between quotes refer to the presence of a single ACE diagnostic band
| “pallens” | “quinquefasciatus” | both bands | Total | |
|---|---|---|---|---|
| male | 0 | 8 | 38 | 46 |
| female | 58 | 5 | 7 | 70 |
χ2 probability of gender parity <0.001
Figure 3.
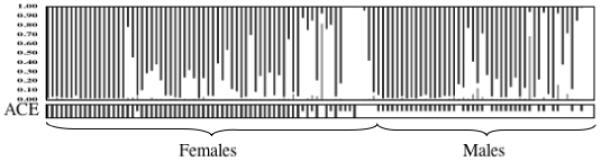
Comparison of the multilocus genotype signatures of male and female Culex pipiens pallens. The specimens are in the same latitudinal order as in Figure 2 but sorted by sex. Only the multilocus genotype analysis using five microsatellite loci is depicted.
One of the eight-microsatellite loci used in the analyses (CxpGT9) amplified very poorly in all populations of Cx. pipiens pallens and was excluded from further analyses although it is in Hardy-Weinberg (H-W) equilibrium in all populations of Cx. p. pipiens examined to date (Fonseca et al. 2004). There were also departures from H-W equilibrium in CQ11 and CxqGT6b in some populations (Supplementary Information). Since a multilocus genotype analysis assumes all loci are in H-W (Pritchard et al. 2000a), we ran analyses with all seven loci, as well as excluding CQ11 (the locus with the most significant departures from H-W), and both CQ11 and CxqGT6b (Figure 2). Irrespective of the number of loci used, the results of the multilocus genetic structure analysis, which combines all the individual multilocus genotypes and separates them into most likely clusters, show hybridization between Cx. p. pallens and Cx. quinquefasciatus (Figure 2). Both males and females have a similar hybrid microsatellite signature (Figure 3). The uniformity of results across 10 replicates was high (0.9-0.99). Microsatellite alleles unique to Cx. quinquefasciatus (see GT4 in Supplementary Materials) are not found in populations from the northernmost island in Japan (Hokkaido) while they are particularly common in Chinese populations and even dominant in Shanghai specimens resulting in specimens indistinguishable from “pure” Cx. quinquefasciatus (Figure 2). Furthermore, two specimens, one in Suwon, ROK, and another in Tianjin, near Beijing, China, show strong ancestries from Cx. pipiens pipiens form molestus (Figure 2).
Figure 2.
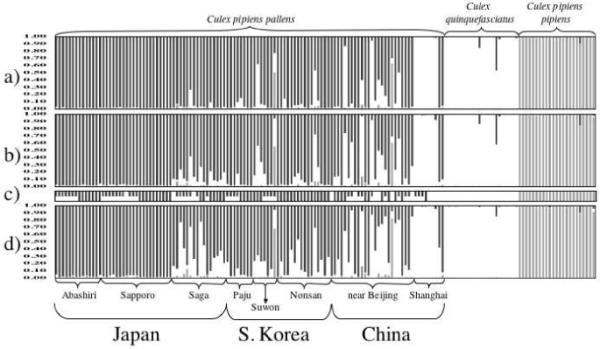
Comparison of the results of multilocus genotype analyses of Japanese, ROKn, and Chinese populations of mosquitoes in the Culex pipiens complex using (a) 7, (b) 6, and (d) 5 microsatellite loci. Each of the individuals is represented by a column partitioned into three shaded segments that represent the individual’s probability of ancestry from one of three genetic clusters (dark grey, Culex pipiens pallens, white, Cx. quinquefasciatus, light grey, Cx. p. pipiens form molestus). For this representation individuals were grouped by location (bracketed). Populations of Cx. p. pallens are ordered from North to South within each country. (c) Summary of the individual ACE rapid assays. The diagnostic bands are represented by a bar of a single shade or, in the case of hybrids, by a bar divided into two shades (the shades are the same as in the multilocus genotype analyses).
Analysis of population differentiation using the five loci that fit H-W expectations in all populations revealed significant differentiation mostly between hybrid and non-hybrid populations irrespective of geographic distance (Table 4) although the hybrid zone has a geographic component with more hybrids in the southernmost locations (Figure 2). The small sample sizes of the populations from Paju and Suwon likely decreased the power of the comparisons to other populations. Since Shijiazhuang, Beijing, Tianjin, and Taiuman are all locations on or very near Beijing, we grouped them to increase the sample size. A pair-wise comparison using the 5 microsatellite loci revealed significant differentiation between the western and eastern subspecies of Cx. pipiens (FST values ranged from 0.148 - 0.253, Table 5), while among northern European Cx. p. pipiens populations the FST values ranged from only 0.001 to 0.138 and were not different from zero, indicating considerable gene flow.
Table 4.
Pairwise FST values (upper diagonal) among populations of Culex pipiens pallens and respective p-values (lower diagonal) based on the allelic frequency distributions at 5 microsatellite loci
| Abashiri | Sapporo | Saga | Paju | Suwon | Nonsan | Beijing | Shanghai | |
|---|---|---|---|---|---|---|---|---|
| Abashiri | 0.038 | 0.115 | 0.060 | 0.062 | 0.052 | 0.062 | 0.270 | |
| Sapporo | 0.008 | 0.195 | 0.136 | 0.144 | 0.082 | 0.122 | 0.328 | |
| Saga | 0.001* | 0.001* | 0.091 | 0.019 | 0.041 | 0.026 | 0.189 | |
| Paju | 0.003 | 0.001* | 0.001* | 0.043 | 0.031 | 0.065 | 0.295 | |
| Suwon | 0.012 | 0.001* | 0.135 | 0.057 | 0.001 | 0.019 | 0.158 | |
| Nonsan | 0.001* | 0.001* | 0.004 | 0.058 | 0.420 | 0.020 | 0.169 | |
| Beijing | 0.001* | 0.001* | 0.026 | 0.003 | 0.169 | 0.048 | 0.137 | |
| Shanghai | 0.001* | 0.001* | 0.001* | 0.001* | 0.002 | 0.001* | 0.001* |
significant p-values after Bonferroni correction
Beijing=specimens from Beijing, Tianjin, Shijiazhuang, and Taiuman, China; Shan= Shanghai, China.
Table 5.
Pairwise FST values (upper diagonal) among populations of Culex pipiens pallens from Abashiri and Sapporo in Hokkaido, the northernmost island where hybridization based on microsatellite analysis is not evident, and Cx. p. pipiens (form pipiens) from several Northern European populations. In the lower diagonal are the p-values based on the allelic frequency distributions at 5 microsatellite loci
| Abashiri | Sapporo | Mens | Camb | Wed | Lough | Alsace | Germ | |
|---|---|---|---|---|---|---|---|---|
| Abashiri | 0.038 | 0.164 | 0.193 | 0.167 | 0.148 | 0.163 | 0.180 | |
| Sapporo | 0.008 | 0.220 | 0.253 | 0.227 | 0.202 | 0.225 | 0.246 | |
| Mens | 0.001* | 0.001* | 0.138 | 0.008 | 0.009 | 0.026 | 0.024 | |
| Camb | 0.001* | 0.001* | 0.135 | 0.008 | 0.001 | 0.012 | 0.017 | |
| Wed | 0.001* | 0.001* | 0.189 | 0.180 | 0.001 | 0.027 | 0.017 | |
| Lough | 0.001* | 0.001* | 0.315 | 0.730 | 0.694 | 0.019 | 0.012 | |
| Alsace | 0.001* | 0.001* | 0.036 | 0.225 | 0.009 | 0.108 | 0.010 | |
| Germ | 0.001* | 0.001* | 0.054 | 0.072 | 0.036 | 0.234 | 0.405 |
significant p-values after Bonferroni correction
Please refer to Fonseca and colleagues (2004) for more detailed location, sample, and source information. Mens=Menstrie, Scotland; Camb=Cambridge, England; Wed=Wedmore, England;
Lough=Loughborough, England; Alsace= Staffelfelden and Cernay, France; Germ= Nonnenweier and Altrip, Germany.
DISCUSSION
Although our analyses have uncovered extensive hybridization between Culex pipiens pallens and Cx. quinquefasciatus in Eastern Asia, they also provide strong evidence that Cx. p. pallens is not simply a hybrid of the European Cx. p. pipiens and Cx. quinquefasciatus. The multilocus genotype analysis based on the microsatellite loci identified two specimens with a mixed signature involving Cx. p. pipiens but all of Cx. p. pipiens tested revealed the diagnostic Cx. p. pipiens form molestus CQ11-band (Bahnck and Fonseca 2006), corroborating the hypothesis that only the molestus form has been introduced to Japan. Further our extensive testing and sequencing of the ACE locus across the Cx. p. pallens range did not recover the diagnostic pipiens-ACE sequence, only the “pallens” as well as “quinquefasciatus” bands were present (Smith and Fonseca 2004). The ACE sequences we obtained from Cx. p. pallens specimens are diagnostic and Cx. p. pallens populations have a unique microsatellite genotype signature when compared to populations from Northern Europe, both indicating lack of gene flow. We therefore conclude that Cx. p. pallens differs from hybrids of Cx. p. pipiens and Cx. quinquefasciatus.
However, we also found many hybrids of Cx. p. pallens and Cx. quinquefasciatus, an observation that explains morphological clines (Bekku 1956). As expected, a detailed analysis found evidence of a north-south population differentiation most likely because of differences in the extent of hybridization with Cx. quinquefasciatus. Interestingly, although the ACE locus was considerably polymorphic, we did not find a significant geographic assortment of alleles. These results, as well as the low FST values among northern European populations (Table 5), agree with expectations of extensive movement in mosquitoes belonging to the Culex pipiens complex possibly as a result of their close association with humans (Vinagradova 2000).
The north-south microsatellite signature of Cx. p. pallens in Japan (Figure 2) resembles the situation in North America where Cx. p. pipiens and Cx. quinquefasciatus hybridize extensively (Kothera et al. 2009, Fonseca and others unpublished data). However, while the ACE rapid assay in North America is a good measure of the extent of the hybridization between the two species (Smith and Fonseca 2004, Kothera et al. 2009), such is not the situation in Japan (Figure 2). In Japan, all males identified as Cx. p. pallens have two DNA bands, one diagnostic of “pallens” the other of “quinquefasciatus” (Figure 3). A few exceptions occur in the southernmost populations where some males have just a “quinquefasciatus” band and are likely either males of Cx. quinquefasciatus or the result of extensive hybridization with Cx. quinquefasciatus. Our survey failed to detect a male with just the “pallens” band (Table 3). Females, in contrast, have overwhelmingly only the “pallens” band, although specimens with both bands and just the “quinquefasciatus” band also occur (Table 3), again mostly in the southernmost populations. Indeed, such ACE-quinques males and ACE-hybrid females are found exclusively in areas of high hybridization detected using microsatellites e.g. Saga (88% hybrids based on multilocus genotype, where a hybrid is being defined as a specimen with more than 5% probability of ancestry from two or more taxa) and China (76% hybrids). We therefore conclude there has been asymmetric introgression of Cx. quinquefasciatus genetic material across chromosomal regions into Cx. p. pallens, similar to that found in birds (Parsons et al. 1993), where traits under selection (male sexual plumage) introgressed further than neutral markers like microsatellite loci. These results expand the findings of Smith and Fonseca (2004) and Kasai and colleagues (2008).
Gender in the Cx. pipiens complex is thought to be determined by a male determining locus (MDL), where males are heterozygous, Mm, and females are homozygous, mm (Gilchrist and Haldane 1947). The ACE locus is within the ace-2 gene, which is physically linked (distance calculated at ≤ 0.8 cMorgans) to the MDL (Malcolm et al. 1998). Our results suggest that the Cx. quinquefasciatus ACE locus and possibly its associated M allele have introgressed to fixation in the Cx. p. pallens populations examined. If so, all male Cx. p. pallens have a “pallens”, ACEP, and a “quinquefasciatus”, ACEQ, allele at the MDL, while females are homozygous for ACEP. If ACE and MDL are linked, then females are homozygous for (m/ACE)P, while male Cx. p. pallens must be (m/ACE)P + (M/ACE)Q. This suggests that the male determining locus of Cx. quinquefasciatus may have replaced that of Cx. p. pallens in all the populations we examined. The altered phallosome morphology that distinguishes Cx. p. pallens from Cx. p. pipiens may be the result.
An alternative to this scenario is that because of the way male Cx. p. pallens are identified (by the shape of their phallosome), we have missed the “true” pallens males in our analyses (i.e. our analysis includes only female Cx. p. pallens and hybrid males). However, the unique “pallens” microsatellite signature is shared between males and females in the two populations from the northern island of Hokkaido and those specimens (males and females) do not have a hybrid microsatellite signature. The specimens with the Cx. p. pipiens phallosome that we examined in Asia (Japan and Korea) all had a Cx. p. pipiens f. molestus microsatellite signature (Fonseca et al. 2004). It is apparent therefore that we are examining a representative sample of the taxon. There is the additional possibility that the current gender difference in introgression of the ACE locus may involve negative selection on the heterozygous females. The discrepancy between microsatellite and ACE loci, and the similarity in microsatellite signature between males and females, however, do not support that hypothesis.
The function of ace-2 is still unknown (Malcolm et al. 1998) since in Cx. pipiens it has been shown that AchE insecticide insensitivity is linked to ace-1, a paralogous gene (Weill et al. 2002). It is therefore still unclear if the asymmetric introgression we uncovered is related to the MDL, the ace-2 locus, or some other gene in this region. We do not yet know what are the selective forces driving or maintaining this apparent introgression of genetic material but are currently examining Single Nucleotide Polymorphisms (SNPs) in the chromosome 1 to understand this phenomenon. Although the transfer of advantageous traits across hybrid zones has been proposed as an important consequence of hybridization (for reviews see Arnold 2004b, Currat et al. 2008), the proposed complete swap of a Cx. p. pallens nuclear region by that of Cx. quinquefasciatus may be an extreme example of the power of hybridization to affect the evolution of an organism. The need to examine this possibility is made more pressing by the medical importance of these vectors of human diseases.
Supplementary Material
Acknowledgments
we are indebted to the many collaborators that provided us with invaluable mosquito samples; Carolyn Bahnck, Kenli Okada, and Andrea Widdel at the time in the Fonseca lab at the Academy of Natural Sciences in Philadelphia, for comments and suggestions on an earlier version of this manuscript; Tovi Lehmann, Robert Fleischer, and an anonymous reviewer for enlightened comments and suggestions; as well as Tapan Ganguly, and the DNA Sequencing Facility, University of Pennsylvania, for technical assistance.
Financial support: NIH R01GMO63258
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dina M. Fonseca, Center for Vector Biology, Rutgers University, 180 Jones Av. New Brunswick, NJ 08901
Julie L. Smith, Genetics Program, Smithsonian Institution, 3001 Connecticut Av. NW, Washington, DC, current address: University of Delaware Graduate College of Marine Studies, Lewes, DE, 19958
Heung-Chol Kim, 5th Medical Detachment, 18th Medical Command, U.S. Army, APO AP 96205-5247, Republic of Korea.
Motoyoshi Mogi, Department of Microbiology, Saga Medical School, Nabeshima 5-1-1, Saga 849-8501, Japan.
REFERENCES
- Arnold ML. Natural Hybridization and Evolution. Oxford University Press; Oxford: 1997. [Google Scholar]
- Arnold ML. Natural hybridization and the evolution of domesticated, pest and disease organisms. Mol Ecol. 2004a;13:997–1007. doi: 10.1111/j.1365-294X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- Arnold ML. Transfer and origin of adaptations through natural hybridization: were Anderson and Stebbins right? Plant Cell. 2004b;16:562–570. doi: 10.1105/tpc.HistPersp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahnck CM, Fonseca DM. Rapid assay to identify the two genetic forms of Culex (Culex) pipiens L. (Diptera: Culicidae) and hybrid populations. American Journal of Tropical Medicine and Hygiene. 2006;75:251–255. [PubMed] [Google Scholar]
- Barr AR. The distribution of Culex p. pipiens and Culex p. quinquefasciatus in North America. Am J Trop Med Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- Bekku H. Studies on the Culex pipiens group of Japan. Nagasaki Medical Journal. 1956;31:956–966. [Google Scholar]
- Borriello F, Krauter K. Reactive site polymorphism in the murine protease inhibitor gene family is delineated using a modification of the PCR reaction (PCR+1) Nucleic Acids Research. 1990;18:5481–5487. doi: 10.1093/nar/18.18.5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet D, Fonseca D, Vourch G, Dubois MP, Chandre F, Severini C, Raymond M. The acetylcholinesterase gene Ace: a diagnostic marker for the Pipiens and Quinquefasciatus forms of the Culex pipiens complex. J Am Mosq Control Assoc. 1998;14:390–396. [PubMed] [Google Scholar]
- Choi WS, Park BS, Ku SK, Lee SE. Repellent activities of essential oils and monoterpenes against Culex pipiens pallens. J Am Mosq Control Assoc. 2002;18:348–351. [PubMed] [Google Scholar]
- Cornel AJ, Mcabee RD, Rasgon J, Stanich MA, Scott TW, Coetzee M. Differences in extent of genetic introgression between sympatric Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in California and South Africa. J. Med. Entomol. 2003;40:36–51. doi: 10.1603/0022-2585-40.1.36. [DOI] [PubMed] [Google Scholar]
- Cui F, Qiao CL, Shen BC, Marquine M, Weill M, Raymond M. Genetic differentiation of Culex pipiens (Diptera: Culicidae) in China. Bull Entomol Res. 2007;97:291–297. doi: 10.1017/S0007485307004968. [DOI] [PubMed] [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00413.x. in press. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci U S A. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Fleischer RC, Mehmet C, Schaffner F, Mogi M, Wilkerson RC. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, LaPointe DA, Fleischer RC. Bottlenecks and multiple introductions: population genetics of the vector of avian malaria in Hawaii. Molecular Ecology. 2000;9:1803–1814. doi: 10.1046/j.1365-294x.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Smith JL, Wilkerson RC, Fleischer RC. Pathways of expansion and multiple introductions illustrated by large genetic differentiation among worldwide populations of the southern house mosquito. Am J Trop Med Hyg. 2006;74:284–289. [PubMed] [Google Scholar]
- Gilchrist BM, Haldane JBS. Sex linkage and sex determination in a mosquito, Culex molestus. Hereditas. 1947;33:175–190. [Google Scholar]
- Guillemaud T, Pasteur N, Rousset F. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc Biol Sci. 1997;264:245–251. doi: 10.1098/rspb.1997.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach RE, Harrison BA, Gad AM. Culex (Culex) molestus Forskal (Diptera: Culicidae): neotype designation, description, variation, and taxonomic status. Proc. Entomol. Soc. Wash. 1984;86:521–542. [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. Oxford Surveys in Evolutionary Biology. 1990;7:69–128. [Google Scholar]
- Humeres SG, Almiron WR, Sabattini MS, Gardenal CN. Estimation of genetic divergence and gene flow between Culex pipiens and Culex quinquefasciatus (Diptera: Culicidae) in Argentina. Mem Inst Oswaldo Cruz. 1998;93:57–62. doi: 10.1590/s0074-02761998000100011. [DOI] [PubMed] [Google Scholar]
- Jang YS, Baek BR, Yang YC, Kim MK, Lee HS. Larvicidal activity of leguminous seeds and grains against Aedes aegypti and Culex pipiens pallens. J Am Mosq Control Assoc. 2002;18:210–213. [PubMed] [Google Scholar]
- Jiggins CD, Salazar C, Linares M, Mavarez J. Review. Hybrid trait speciation and Heliconius butterflies. Philos Trans R Soc Lond B Biol Sci. 2008 doi: 10.1098/rstb.2008.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai S, Komagata O, Tomita T, Sawabe K, Tsuda Y, Kurahashi H, Ishikawa T, Motoki M, Takahashi T, Tanikawa T, Yoshida M, Shinjo G, Hashimoto T, Higa Y, Kobayashi M. PCR-Based Identification of Culex pipiens Complex Collected in Japan. Jpn J Infect Dis. 2008;61:184–191. [PubMed] [Google Scholar]
- Kothera L, Zimmerman EM, Richards CM, Savage HM. Microsatellite characterization of subspecies and their hybrids in Culex pipiens complex (Diptera: Culicidae) mosquitoes along a north-south transect in the central United States. J Med Entomol. 2009;46:236–248. doi: 10.1603/033.046.0208. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, Lim KY, Sherwood AM, Matyasek R, Rocca J, Soltis DE, Soltis PS. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven H. Speciation and evolution in Culex pipiens. In: Wright JW, Pal R, editors. Genetics of insect vectors of disease. Elsevier; Amsterdam: 1967. pp. 251–275. [Google Scholar]
- Malcolm CA, Bourguet D, Ascolillo A, Rooker SJ, Garvey CF, Hall LMC, Pasteur N, Raymond M. A sex-linked Ace gene, not linked to insensitive acetylcholinesterase-mediated insecticide resistance in Culex pipiens. Insect Mol Biol. 1998;7:107–120. doi: 10.1046/j.1365-2583.1998.72055.x. [DOI] [PubMed] [Google Scholar]
- Mattingly PF, Rozeboom LE, Knight KL, Laven H, Drummond FH, Christophers SR, Shute PG. The Culex pipiens Complex. Transactions of the Royal Entomological Society of London. 1951;102:331–382. [Google Scholar]
- Meixner MD, McPheron BA, Silva JG, Gasparich GE, Sheppard WS. The Mediterranean fruit fly in California: evidence for multiple introductions and persistent populations based on microsatellite and mitochondrial DNA variability. Mol Ecol. 2002;11:891–899. doi: 10.1046/j.1365-294x.2002.01488.x. [DOI] [PubMed] [Google Scholar]
- Oda T, Eshita Y, Uchida K, Mine M, Kurokawa K, Ogawa Y, Kato K, Tahara H. Reproductive activity and survival of Culex pipiens palllens and Culex quinquefasciatus (Diptera: Culicidae) in Japan at high temperature. J Med Entomol. 2002;39:185–190. doi: 10.1603/0022-2585-39.1.185. [DOI] [PubMed] [Google Scholar]
- Parsons TJ, Olson SL, Braun MJ. Unidirectional spread of a secondary sexual plumage traits across an avian hybrid zone. Science. 1993;260:1643–1646. doi: 10.1126/science.260.5114.1643. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000a;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000b;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop ver. 1.2: population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MA, Hillel J, Maki-Tanila A, Tixier-Boichard M, Vignal A, Wimmers K, Weigend S. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics. 2001;159:699–713. doi: 10.1093/genetics/159.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin ver. 2000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva; Geneva, Switzerland: 2000. [Google Scholar]
- Smith JL, Fonseca DM. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae) American Journal of Tropical Medicine and Hygiene. 2004;70:339–345. [PubMed] [Google Scholar]
- Smith JL, Keyghobadi N, Matrone MA, Escher R, Fonseca DM. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Molecular Ecology Notes. 2005;5:697–700. [Google Scholar]
- Tanaka K, Mizusawa K, Saugstad ES. A revision of the adult and larval mosquitoes of Japan (including the Ryukyu archipelago and the Ogasawara islands) and Korea (Diptera: Culicidae) Contrib. Amer. Ent. Inst. 1979;16:1–987. [Google Scholar]
- Urbanelli S, Silvestrini F, Reisen WK, De Vito E, Bullini L. Californian hybrid zone between Culex pipiens pipiens and Cx. p. quinquefasciatus revisited (Diptera:Culicidae) J Med Entomol. 1997;34:116–127. doi: 10.1093/jmedent/34.2.116. [DOI] [PubMed] [Google Scholar]
- Urbanelli S, Silvestrini F, Sabatinelli G, Raveloarifera F, Petrarca V, Bullini L. Characterization of the Culex pipiens complex (Diptera: Culicidae) in Madagascar. J. Med. Entomol. 1995;32:778–786. doi: 10.1093/jmedent/32.6.778. [DOI] [PubMed] [Google Scholar]
- Vinagradova EB. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. Pensoft; Moscow: 2000. [Google Scholar]
- Weill M, Fort P, Berthomieu A, Dubois MP, Pasteur N, Raymond M. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proc. R. Soc. Lond. 2002;269:2007–2016. doi: 10.1098/rspb.2002.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YC, Lim MY, Lee HS. Emodin isolated from Cassia obtusifolia (Leguminosae) seed shows larvicidal activity against three mosquito species. J Agric Food Chem. 2003;51:7629–7631. doi: 10.1021/jf034727t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


