Summary
High-content screening (HCS), historically limited to drug-development companies, is now a powerful and affordable technology for academic researchers. Through automated routines, this technology acquires large datasets of fluorescence images depicting functional states of thousands to millions of cells. Information on shapes, textures, intensities, and localizations is then used to create unique representations, or “phenotypic signatures,” of each cell. These signatures quantify physiologic or diseased states, for example, dendritic arborization, drug response, or cell coping strategies. Live-cell imaging in HCS adds the ability to correlate cellular events at different points in times, thereby allowing sensitivities and observations not possible with fixed endpoint analysis. HCS with live-cell imaging therefore provides unprecedented capability to detect spatiotemporal changes in cells and is particularly suited for time-dependent, stochastic processes such as neurodegenerative disorders.
Introduction
Biological research is entering a new era. Molecular biology will be combined with novel engineering technologies and increased computational power to examine living systems in exciting new ways. We are only beginning to understand the benefits—in fact, the necessity—of studying biological systems with large-scale unbiased screens[1]. Here we focus on high-content screening (HCS) and considerations needed to use this method effectively to study normal and disease physiology in primary cells, currently the most biologically relevant models.
Why high-content screening?
HCS is a multiplexed, functional screening method based on extracting multiparametric fluorescence data from multiple targets in intact cells [2,3]. By temporally and spatially resolving fluorescent readouts within individual cells, HCS yields an almost unlimited number of kinetic and morphometric outputs. HCS was developed to facilitate drug-target validation and lead optimization before costly animal testing [4]. Today it is broadly used to catalog cellular, subcellular, and intercellular responses to multiple systematic perturbations and is applicable to basic science, translational research, and drug development. We distinguish HCS from high-content analysis (HCA). HCA refers to extracting information from image data. HCS is the automated, high-throughput application of HCA.
HCS can fill a gap in academic research. Our growing awareness of biological complexity underscores the need to examine more than one variable at a fixed point in time. Traditional low-throughput methods have severe limitations. For complex systems with many interacting genes, measuring any single perturbation is not very informative. For gain-of-function diseases, especially those with late onset, a toxic gain-of-function may not be related to a protein’s normal function. Unbiased screens therefore identify potential pathogenic mechanisms faster and more comprehensively, and the large datasets are less prone to sampling error when analyzing stochastic events.
HCS assays capture cell-system dynamics and exploit typically confounding cell-to-cell variability. For example, a recent study used simultaneous tracking of ~1000 proteins in lung carcinoma cells after drug treatment to detect time-dependent proteomic changes that predicted individual cell fate [5]. Hypotheses in HCS are used to design tracked variables and outputs that maximize the likelihood of meaningful results. We labeled mutant huntingtin and measured cell survival to determine the role of inclusion bodies in Huntington’s disease (HD)[6], a question unanswered by 10 years of time-invariant, low-throughput approaches. HCS provides large datasets that unveil multiple, often nonintuitive, correlations that seed subsequent lines of thought. Thus, HCS accelerates the iterative process of classical hypothesis-driven research [7].
Primary cells or cell lines?
Choosing the best cell type for a particular HCS assay is challenging. Each option comes with inherent benefits and drawbacks (Table 1). Primary cells provide high-quality models for several reasons. They are more physiologically relevant than immortalized cell lines [8]. They form synapses, thus incorporating significant neuromodulatory and trophic inputs. Neuronal physiology and disease are also notoriously cell-type specific, and neurons differentiated in vivo best recapitulate actual neuronal subpopulations. One study found hepatoma cell lines differ profoundly from primary hepatocytes, consistent with a shift from oxidative to anaerobic metabolism, upregulation of mitotic proteins, and downregulation of typical hepatocyte functions [9]. High attrition rates for candidate neuropharmacologics (Fig. 1) suggest even more striking differences in neurons.
Table 1. Neuronal cell models for HCS.
The advantages and disadvantages of different cell types are summarized for their use in HCS. Adapted from Eglen et al [10].
| Property | Immortalized cells | Primary Neurons | Embryonic stem cells | Induced pluripotent stem cells |
|---|---|---|---|---|
| Current use in HCS | Ubiquitous | Limited | Differentiation screens | Differentiation screens |
| Ready for HCS | Yes | Yes | No | No |
| Source | Specific to cell line | Animal tissue
|
Established or new cell line
|
Established or new cell line
|
| Freeze/Thaw | Yes | Once | Yes | Yes |
| Proliferative capacity | Very High | Post-mitotic | High
|
High
|
| Differentiation required | In some cases | No | Yes | Yes |
| Population type | Clonal or Heterogeneous |
Heterogeneous | Clonal → Heterogeneous | Clonal → Heterogeneous |
| Handling | Durable | Sensitive | Sensitive | Sensitive |
| Ability to be engineered | High | Limited | Medium to high | Medium to high |
| Cost | Low | High | Medium | Medium |
| Physiologic relevance | Low | High | Medium to high | Medium to high |
| Major challenge for HCS | • Physiologic relevance |
• Limited human source • Labor intensive |
• Limited human source • Differentiation • Quality control |
• Dedifferentiation • Differentiation • Quality control |
| Major benefits for HCS | • Quantity • Engineering |
• Physiologic relevance |
• Quantity • Diversity of cell types |
• Quantity • Diversity of cell types • Patient-specific screening |
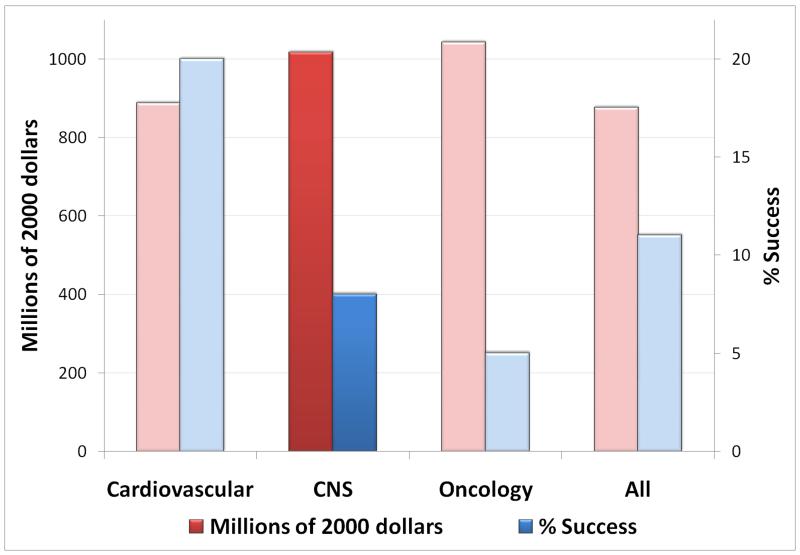
Figure 1. Success rates and millions of dollars spent from first-in-man to registration by therapeutic area.
The overall clinical success rate is 11% with ~900 million dollars spent. However when the analysis is carried out by therapeutic area, big differences emerge with central nervous system (CNS) and oncology trailing far behind cardiovascular diseases in the % success rate compared to the dollars spent [48,49].
Most screenings have involved cell lines, but future screenings will use primary and stem cells [10,11]. Embryonic stem (ES) cells can be differentiated into motor neurons in large numbers [12]. Mouse and human induced pluripotent stem (iPS) cells [13,14] may better predict in vivo drug side effects and are particularly attractive for disease-focused HCS [15-17]. For example, iPS cells from patients with spinal muscular atrophy differentiated into motor neurons retained pathological deficits and drug responses consistent with the disease. More work is needed to characterize iPS cell lines, and better dedifferentiation protocols will avoid viral vectors and oncogenes [17-20]. Ultimately, HCS will place additional demands on dedifferentiation and redifferentiation, including high efficiency and reproducibility. High throughput screens are already helping to address these needs [21,22].
Despite technical challenges in isolating, culturing, and transfecting primary neurons, their use decreases false negatives and saves time and money wasted on pursuing false positives. Until protocols are improved for differentiating ES and iPS cells into many neuronal cell types, primary cells will remain the most physiologically relevant model for large-scale screens.
HCS planning for live-cell imaging
Assay development encompasses selecting fluorophores and proteins to label, choosing a transfection method, migrating to 96- or 384-well formats, upgrading automation, and completing preliminary experiments to determine robustness of readouts. None of these steps are trivial. Migrating to a new format alone requires re-optimizing labware, intra- and inter-well cell distributions, and transfection and image-acquisition protocols. During this time, a lab data management system must also be integrated.
Fluorophores
Excellent reviews describe fluorophores for HCA [23,24]. Notably, mKate [25] (now mKate2), mOrange2 and TagRFP-T [26], and EBFP2 [27] provide improved brightness and photostability. After balancing these features, the best options for live-cell imaging are listed in Table 2. HCS allows up to four fluorophores with sufficient spectral separation to avoid crosstalk. In the future, more channels will be simultaneously acquired with spectral imaging [28].
Table 2. Recommended fluorescent proteins.
Physical properties for fluorescent proteins (FPs) in each spectral class. Adapted from Shaner et. al. [23,26].
| Fluorescent Protein* |
Spectral Class |
Excitation peak (nm) |
Emission peak (nm) |
Brightness† | Photostability‡ | pKa‡ | Association state‡ |
Filter Set§ |
|---|---|---|---|---|---|---|---|---|
| EBFP2 | Blue | 383 | 448 | 18 | 55 | 5.3 | Weak dimer | DAPI/BFP |
| mCerulean | Cyan | 433/445 | 475/503 | 27/24 | 36 | 4.7 | Monomer | CFP |
| mEGFP | Green | 488 | 507 | 34 | 174 | 6.0 | Monomer | FITC/GFP |
| mEmerald | Green | 487 | 509 | 39 | 101¶ | 6.0 | Monomer | FITC/GFP |
| EYFP | Yellow | 514 | 527 | 51 | 60 | 6.9 | Weak dimer | FITC/YFP |
| mCitrine | Yellow | 516 | 529 | 59 | 49 | 5.7 | Monomer | FITC/YFP |
| mOrange2 | Orange | 549 | 565 | 35 | 228 | 6.5 | Monomer | TRITC/DsRed |
| TagRFP-T | Orange | 555 | 584 | 33 | 337 | 4.6 | Monomer | TRITC/DsRed |
| mCherry | Red | 587 | 610 | 17†† | 96 | <4.5 | Monomer | TxRed |
| mKate2 | Far-red | 588 | 633 | 25 | 118 | 5.4 | Monomer | TxRed |
Common literature FP abbreviation.
Product of the molar extinction coefficient and the quantum yield (mM × cm)-1.
Literature values except as noted.
Specialized applications may require choosing filter combinations that closely match the spectral profiles [50].
Measured in live cells with mEGFP (t1/2=150 seconds) as a control.
Averages of literature values.
Transfection
Lipid-based methods, Ca2+-phosphate co-precipitatation, viral infection, electroporation, and nucleofection all have benefits and drawbacks [29]. Primary neurons pose additional challenges: they are susceptible to transfection toxicities and plagued by low transfection efficiency [30]. We found Lipofectamine 2000 (Invitrogen) best for efficiency, cell viability, and automation in assays that require transfection after cell plating. With this reagent, most transfection variability results from cell-plating density, total mass of DNA, and ratio of transfection reagent to DNA. These factors must be optimized for specific cells and DNAs. Reverse transfection with this reagent now makes arrayed libraries of transfection-ready DNA and siRNA a reality for HCS [31,32]. Although biochemical assays of large numbers of pooled cells rely on high transfection efficiencies, this actually complicates microscopy-based screening of individual cells. Identifying the same cell over time can be confounded by cell movement. The researcher must strike a balance between maximizing transfected cell number per field and verifying the ability of image-analysis algorithms to accurately track the cells.
Automation
Automation can be applied to each step of HCS, including sample preparation, image acquisition and analysis, quality-control measures, and data reporting. Highly capable liquid-handling robots are increasingly affordable for individual labs. They represent scalable options for liquid aspiration and dispensing of large and small volumes. Multiple high-content microscopy systems are now available [33]. The most popular use confocal or wide-field microscopes, and all offer hardware autofocus, options for environmental control, and data management and image-analysis software. They provide out-of-the-box access to HCS for many scientific applications. Downsides to these solutions include expense, proprietary image formats and algorithms, and the inability to write ground-level scripts for true user customization. Lab automation upgrades should be integrated early into low-throughput assay development so quality measures are determined from datasets reflecting the automation.
Robustness
Minimizing assay variability is essential for HCS. The Z’-factor is a useful way to estimate assay quality and is calculated as a signal detection window between positive and negative controls scaled by the dynamic range [34]. It is an excellent measure of single-output assays. Since HCS allows powerful multiparametric analyses with potentially hundreds of quantified parameters, a Z’-factor can be calculated individually for each parameter [35]. Alternatively, multivariate criteria without informational losses due to averaging can be instituted from the beginning [36]. In either case, large data sets from positive and negative controls should be used to determine assay quality before initiating screening.
Data Management
HCS datasets are large. Live-cell imaging of a single 96-well plate with three channels and nine images per well yields ~30 GB of raw image data. A reliable informatics infrastructure is needed. Data should flow seamlessly from acquisition to storage on a server where it can be accessed for offline image analysis. Initially, hierarchal file structures can be used, but optimal management should include a central database for storing images and metadata that can be accessed by both acquisition and image-analysis software.
Image analysis, the new bottleneck
Automation advancements have been valuable for HCS, but extracting meaningful data from complex image sets poses major challenges. These challenges arise from a combination of microscopy and image-processing limitations and the need for new statistical tools. Neuroscience poses particular difficulties due to complexities in neuronal morphology and subcellular trafficking. Most laboratories use image-analysis algorithms and manual labor to analyze images, but the throughput is too low for HCS. More robust and accurate image-analysis algorithms that can be applied to entire data sets with minimal user intervention are necessary[37]. Zhang et al. published a neurite extraction algorithm [38] for HCS, and multiple commercial packages quantify neuronal bodies and neurites. To understand HCS informatics problems more fully, we refer you to excellent reviews [39-41].
HCA uniquely provides multiplexed quantification of individual cell features with temporal and spatial resolution. Image analysis comprises image segmentation and cell tracking, extraction of individual cell features, and data modeling and classification [41]. Image-analysis programs routinely measure size, shape, intensity, texture, moments, and subcellular localization that, when combined, yield hundreds of parameters that characterize a specific cell phenotype [42]. For example, Loo et al. used ~300 unbiased parameters and a multivariate clustering algorithm to determine separation between drug-treated HeLa cells and controls [36]. The redundancy of this parameter set was reduced, resulting in a minimal phenotypic signature of the treated cells at various drug dosages. With the signatures, a drug class could be predicted, and therapeutic windows could also be deduced. The close relationship of neuronal morphology and functional state [43] holds promise for similar phenotypic signatures to emerge from HCS focused on neuronal development, physiology, and disease. For instance, an HCS study of cultured rat primary cortical neurons identified Aβ1-42 induced reduction in neurite outgrowth with no apparent effect on neuron number, pointing to more subtle morphological changes that can precede cell death. These studies used fixed-cell imaging, however, the full potential of HCS will be realized with imaging live cells over time[44].
HCS and live-cell imaging of primary neurons: putting it all together
HCS with live-cell imaging in relevant neuronal models promises to elucidate physiologic and pathophysiologic processes with unprecedented sensitivity and correlative power. Live-cell imaging captures cell phenotype changes. Thus, previously static features are transformed into dynamic features where timed occurrences and rates of change generate more informative phenotypic signatures. Imaging in live cells also permits cause-and-effect relationships to be determined. We use this novel approach to investigate pathogenic mechanisms of neurodegenerative disorders, including HD, Parkinson’s disease, and frontotemporal dementia. Our system (Fig. 2) allows us to correlate events in thousands of neurons to individual cell fates—enabling us to determine if the events are adaptive, maladaptive, or incidental to disease progression [45]. For instance, we used live-cell imaging in a primary neuron model of HD to establish a mitigating role for inclusion bodies [6] and reveal the interplay between ubiquitin-proteasome system function and inclusion body formation [46]. Such studies necessitate large sample sizes and the ability to follow individual neurons over time. They highlight the power of HCS, when coupled with live-cell imaging, to reveal causal relationships in biological processes.
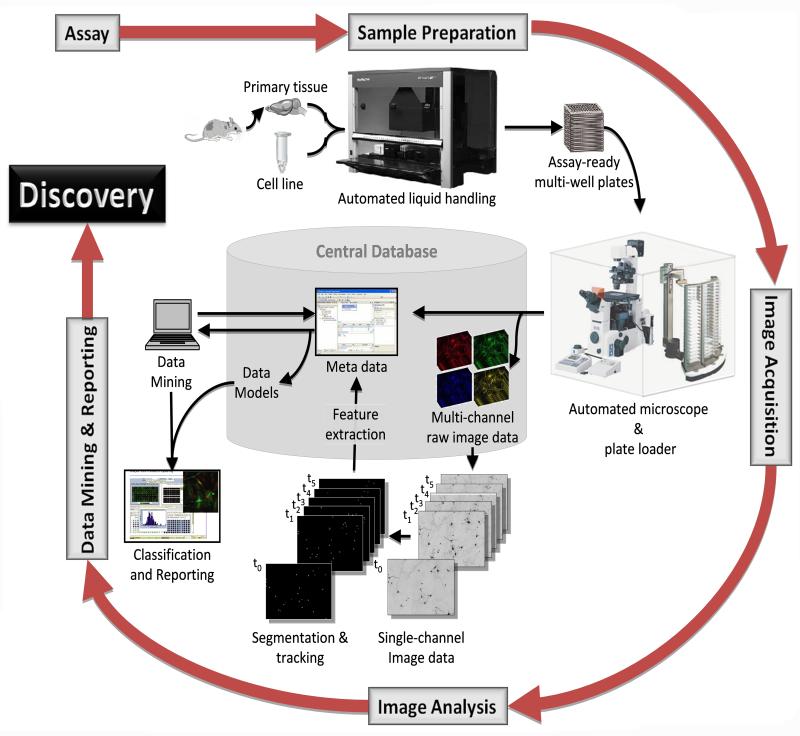
Figure 2. Workflow of our second generation high content screening system for live-cell imaging.
Our system uses primary neurons from embryonic mice. A Microlab STARlet (Hamilton, Reno, CA) automated pipetting workstation prepares and transfects cells in 96-well plates, which are then transferred to the plate stacker of a KiNEDx 4-axis robot (Peak Robotics, Colorado Springs, CO). The plates are loaded onto an MS-2000 stage (Applied Scientific Instruments, Eugene, OR) fixed to a Nikon TE-2000 (Nikon, Melville, NY) microscope. The robot and microscope are enclosed in an environmental chamber (InVivo Scientific, St Louis, MO) to enable around-the-clock imaging for 6-7 days. Widefield images are acquired according to in-house scripts written in Image-Pro Plus (MediaCybernetics, Silver Spring, MD). At each time point, montage images are generated for each well and fluorophore channel. Image analysis algorithms then extract cell-based information. Metadata generated from image acquisition and analysis flows into a central database for data modeling, mining and classification.
Repeated measures of individual cells by automated microscopy facilitates use of powerful statistical techniques, such as Cox proportional hazards (CPH) analysis[47]. CPH integrates a user-defined number of parameters to determine whether they explain time-to-event outcomes, for instance cell survival. Much as in a prospective cohort study, we allow cells, through stochastic diversification, to “take on” certain traits and then retrospectively determine how significant these traits are in predicting outcomes. Our goal is to find robust, disease-specific phenotypic signatures for screening small-molecule pharmacological agents and genome-wide siRNA libraries. CPH takes advantage of inherent cell-to-cell heterogeneity, and the increased sensitivity resulting from temporal analysis permits fewer cells to be analyzed. We therefore avoid two main drawbacks of screening in primary cells—decreased transfection efficiency and lack of cell homogeneity.
Conclusion
HCS is a technology with vast potential for academic researchers and particularly neuroscience. Large-scale screens are strategically essential in understanding complex biological systems and gain of function diseases. HCS can be applied to an incredible diversity of assay types depending on the experimental conditions and labeled proteins. Challenges still remain in image analysis and data interpretation, and new statistical tools will be necessary to analyze time-dependent processes of millions of cells across thousands of conditions. Advances in HCS will result from new microscopy techniques, such as spectral imaging, better fluorescence proteins, and the maturation of stem cell technology. Greater knowledge of what proteins to probe for particular physiologic and disease processes will increase HCS sensitivity. HCS with live-cell imaging in primary neurons is practical and will likely contribute to some of the most elusive questions in neurobiology and related disease.
Acknowledgements
We thank members of the Finkbeiner lab for their generous support and advice. We thank G. Howard and S. Ordway for editorial assistance and K. Nelson for administrative assistance. This work was supported by the Consortium for Frontotemporal Dementia Research, the Taube-Koret Center for Huntington’s Disease Research, National Institutes of Health (NIH) grants 2R01 NS039074 and 2R01045491 from the National Institutes of Neurological Disorders and Stroke and 2P01 AG022074 from the National Institutes of Aging and by the J. David Gladstone Institutes (to S.F.). Support was also provided by the NIH-NIGMS UCSF Medical Scientist Training Program (to A.C.D.) and the California Institute of Regenerative Medicine (P.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Giuliano KA, DeBiasio RL, Dunlay RT, Gough A, Volosky JM, Zock J, Pavlakis GN, Taylor DL. High-Content Screening: A New Approach to Easing Key Bottlenecks in the Drug Discovery Process. J Biomol Screen. 1997;2:249–259. [Google Scholar]
- 3.Krausz E. High-content siRNA screening. Mol Biosyst. 2007;3:232–240. doi: 10.1039/b616187c. [DOI] [PubMed] [Google Scholar]
- 4.Giuliano KA, Taylor DL. Fluorescent-protein biosensors: new tools for drug discovery. Trends Biotechnol. 1998;16:135–140. doi: 10.1016/S0167-7799(97)01166-9. [DOI] [PubMed] [Google Scholar]
- 5 ••.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. Dynamic Proteomics of Individual Cancer Cells in Response to a Drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165.Live-cell imaging of individual cancer cells was used to measure the dynamics of ~1000 proteins after drug treatment. Cell-cell variation in the expression of DDX5, an RNA helicase, and DNA replication factor RFC1 correlated with the emergence of drug resistant subpopulations. The study demonstrates a novel HCS method to observe real-time proteomics.
- 6.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 7.Smalheiser NR. Informatics and hypothesis-driven research. EMBO Rep. 2002;3:702. doi: 10.1093/embo-reports/kvf164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan GP. What’s wrong with drug screening today. Nat Chem Biol. 2007;3:187–191. doi: 10.1038/nchembio0407-187. [DOI] [PubMed] [Google Scholar]
- 9 •.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8:443–450. doi: 10.1074/mcp.M800258-MCP200.The authors used stable isotope labeling and mass spectrometry to compare the proteomes of cell lines to primary cells. The Hep1-6 liver cell line showed downregulation of proteins involved in complement and coagulation factor production along with the important P450 family of enzymes. There was also a drastic shift from oxidative to anaerobic metabolism.
- 10.Eglen RM, Gilchrist A, Reisine T. An overview of drug screening using primary and embryonic stem cells. Comb Chem High Throughput Screen. 2008;11:566–572. doi: 10.2174/138620708785204108. [DOI] [PubMed] [Google Scholar]
- 11.Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132:549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 12 •.Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885.An in vitro model based on ES cells is presented for studying amyotrophic lateral sclerosis (ALS). ES cells cultured from SOD1G23A transgenic mice were efficiently differentiated into motor neurons and exhibited decreased survival when compared to differentiated motor neurons without the transgene. Co-culture with SOD1G23A glial cells exacerbated death for both motor neuron types. The study exemplifies the increasing role ES cells will play in disease-focused HCS.
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 16.Ebert AD, Yu J, Rose FF, Jr., Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of Mouse Induced Pluripotent Stem Cells Without Viral Vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 20.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 22.Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 24.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The Fluorescent Toolbox for Assessing Protein Location and Function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 25.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Meth. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 26 •.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Meth. 2008;5:545–551. doi: 10.1038/nmeth.1209.A novel screening method is presented that increased the photostability of bright red and orange fluorescent proteins TagRFP and mOrange to create TagRFP-T and mOrange2. Through a combination of random and site directed mutagenesis, the new proteins became 9 and 25 times more photostable, respectively. More photostable proteins are necessary to increase sampling rate in live-cell imaging.
- 27.Ai H-w, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of New Chromophore Structures Leads to the Identification of Improved Blue Fluorescent Proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann T. Spectral imaging and linear unmixing in light microscopy. Adv Biochem Eng Biotechnol. 2005;95:245–265. doi: 10.1007/b102216. [DOI] [PubMed] [Google Scholar]
- 29.Zeitelhofer M, Vessey JP, Xie Y, Tubing F, Thomas S, Kiebler M, Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat Protoc. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- 30.Halterman MW, Giuliano R, DeJesus C, Schor NF. In-tube transfection improves the efficiency of gene transfer in primary neuronal cultures. Journal of Neuroscience Methods. 2009;177:348–354. doi: 10.1016/j.jneumeth.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R. Reverse transfection on cell arrays for high content screening microscopy. Nat Protoc. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- 32.Erfle H, Neumann B, Rogers P, Bulkescher J, Ellenberg J, Pepperkok R. Work Flow for Multiplexing siRNA Assays by Solid-Phase Reverse Transfection in Multiwell Plates. J Biomol Screen. 2008;13:575–580. doi: 10.1177/1087057108320133. [DOI] [PubMed] [Google Scholar]
- 33.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 35.Abraham VC, Towne DL, Waring JF, Warrior U, Burns DJ. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. J Biomol Screen. 2008;13:527–537. doi: 10.1177/1087057108318428. [DOI] [PubMed] [Google Scholar]
- 36 ••.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4:445–453. doi: 10.1038/nmeth1032.The authors present an HCS and informatics approach to better detect drug class and toxicity. Unbiased feature sets were extracted from individual cells treated with titrated drug dosages to form multivariate descriptions of cell phenotypes. A unique, quantitative descriptor for each drug dosage was generated that could predict drug class and toxicity and give insight into mechanism of action.
- 37.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhou X, Degterev A, Lipinski M, Adjeroh D, Yuan J, Wong ST. Automated neurite extraction using dynamic programming for high-throughput screening of neuron-based assays. Neuroimage. 2007;35:1502–1515. doi: 10.1016/j.neuroimage.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmer C, Bo Z, Dufour A, Thebaud A, Berlemont S, Meas-Yedid V, Marin JCO. On the digital trail of mobile cells. Signal Processing Magazine, IEEE. 2006;23:54–62. [Google Scholar]
- 40.Meijering E, Smal I, Danuser G. Tracking in molecular bioimaging. Signal Processing Magazine, IEEE. 2006;23:46–53. [Google Scholar]
- 41.Xiaobo Z, Wong STC. Informatics challenges of high-throughput microscopy. Signal Processing Magazine, IEEE. 2006;23:63–72. [Google Scholar]
- 42.Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. doi: 10.1016/j.devcel.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Rocchi MB, Sisti D, Albertini MC, Teodori L. Current trends in shape and texture analysis in neurology: aspects of the morphological substrate of volume and wiring transmission. Brain Res Rev. 2007;55:97–107. doi: 10.1016/j.brainresrev.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Meth. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 45.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci U S A. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitra S, Tsvetkov AS, Finkbeiner S. Single Neuron Ubiquitin-Proteasome Dynamics Accompanying Inclusion Body Formation in Huntington Disease. J. Biol. Chem. 2009;284:4398–4403. doi: 10.1074/jbc.M806269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein JP, Moeschberger, Melvin L. Survival Analysis. edn 2 Springer; 2005. [Google Scholar]
- 48.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 49.Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health Aff (Millwood) 2006;25:420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 50.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]