Abstract
Altering the subcellular localization of signal transducing proteins is a novel approach for therapeutic intervention. Mislocalization of tumor suppressors, oncogenes, or factors involved in apoptosis result in aberrant functioning of these proteins, leading to disease. In the case of chronic myelogenous leukemia (CML), cytoplasmic Bcr-Abl causes oncogenesis/proliferation. On the other hand, nuclear entrapment of endogenous Bcr-Abl (in K562 human leukemia cells) causes apoptosis. The goal of this study was to determine whether plasmid expressed Bcr-Abl could cause apoptosis of K562 cells when specifically directed to the nucleus via strong nuclear localization signals (NLSs). A single NLS from SV40 large T-antigen or four NLSs were subcloned to Bcr-Abl (1NLS-Bcr-Abl or 4NLS-Bcr-Abl). When transfected into K562 cells, only 4NLS-Bcr-Abl translocated to the nucleus. Bcr-Abl alone was found to localize in the cell cytoplasm, colocalizing with actin due to its actin binding domain. 1NLS-Bcr-Abl also localized with actin. Apoptosis induced by 4NLS-Bcr-Abl was evaluated 24 hours post-transfection by morphologic determination, DNA staining, and caspase-3 assay. This is the first demonstration that altering the location of plasmid expressed Bcr-Abl can kill leukemia cells. Multiple NLSs are required to overcome Bcr-Abl binding to actin, thus driving it into the nucleus and causing apoptosis.
Keywords: Bcr-Abl, apoptosis, nuclear localization, protein switch, CML
INTRODUCTION
The causative agent for 95% of all CML cases, Bcr-Abl, is derived from the fusion of the breakpoint cluster region (Bcr) gene on chromosome 22 and the Abelson leukemia oncogene (Abl) on chromosome 9. This reciprocal translocation results in an abnormal, shortened chromosome - “Philadelphia chromosome” or Ph (+) phenotype[1, 2]. The resulting Bcr-Abl fusion protein acts as an oncoprotein, and the constitutive activation of tyrosine kinase activity of Abl leads to cell proliferation. Although Gleevec is currently the “gold standard” drug of choice for Bcr-Abl-positive CML[3–5], resistance to treatment with Gleevec occurs. This is mostly due to mutations in the Bcr-Abl kinase domain that render it unable to bind to Gleevec[6–8]. Some mutations create a more potent Bcr-Abl oncogene and accelerate disease progression[9]. Other mechanisms for resistance include Bcr-Abl amplification or overexpression, clonal evolution, a decrease in Gleevec bioavailability or cell exposure, and upregulation of drug efflux pumps[6, 10]. Many other tyrosine kinase inhibitors (TKIs) are being studied and developed, with a few already approved nonetheless, Bcr-Abl also has the potential to develop resistance to these molecules. In addition, since FDA approval, potentially fatal side effects of Gleevec have been uncovered. These include cardiotoxicity[11], the possibility of developing other cancers (due to blockade of tumor suppressor p63)[12], and acute renal failure[13]. Therefore, finding alternative strategies to Gleevec therapy are necessary.
In the cytoplasm Bcr-Abl acts as an oncogene by interacting with multiple signal transduction pathways that transmit anti-apoptotic and mitogenic signals[14]. The key pathways involve ras, MAP kinases, the STAT family, PI3 kinase, and myc, among others[15]. In the nucleus, Bcr-Abl may cause apoptosis due to nuclear Abl’s ability to stabilize p73 and activate its pro-apoptotic functions. Vigneri and Wang have previously shown that nuclear entrapment of Bcr-Abl in K562 cells results in apoptosis, and requires an active tyrosine kinase domain to do so[2]. They used Gleevec to stimulate Bcr-Abl to go to the nucleus (by unknown mechanism), followed by nuclear entrapment by leptomycin B (LMB), a general inhibitor of nuclear export. After washout of Gleevec, Bcr-Abl’s tyrosine kinase activity is re-activated, and the cells undergo spontaneous apoptosis. Unfortunately LMB causes neuronal toxicity and cannot be used therapeutically. This study attempts to address a fundamental question that remains: Does the apoptosis caused by nuclear entrapment of Bcr-Abl require depletion of Bcr-Abl from the cytoplasm, or is it sufficient to send plasmid expressed Bcr-Abl to the nucleus to cause apoptosis?
MATERIALS AND METHODS
Subcloning and Construction of Plasmids
Full length Bcr-Abl was removed from pEYK3.1 retroviral vector (a kind gift from Dr. George Daley, Harvard Medical School, Boston) using EcoRI and cloned into pEGFP-C1 (Clontech, Mountainview, CA) at the EcoRI site to make EGFP-Bcr-Abl.
The oligonucleotides 5’- CCGGAAGCCCAAAGAAGAAGAGAAAAGTAGAAT-3’ and 5’- CCGGATTCTACTTTTCTCTTCTTCTTTGGGCTT-3’ were ligated to pEGFP-Bcr-Abl at the BspEI site. The oligonucleotide insert encodes for the nuclear localization signal (NLS) from SV40 large T-antigen (amino acids PKKKRKV) and is flanked with the BspEI digested sequence. The ligation resulted in the formation of pEGFP-1NLS-Bcr-Abl and pEGFP-4NLS-Bcr-Abl (a concatemer consisting of four nuclear localization signals).
The bases encoding key residues (T65A, Y66A) in the EGFP chromophore [16, 17] of pEGFP-4NLS-Bcr-Abl were mutated using the QuikChange II Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA) to eliminate EGFP fluorescence (for use in some co-transfection experiments). The primers used for the mutagenesis were 5’- CTCGTGACCACCCTGGCCGCCGGCGTGCAGTGCTTC-3’ and its reverse compliment. This plasmid encodes 4NLS-Bcr-Abl and non-fluorescent EGFP.
Cell Line and Culture Conditions
Bcr-Abl positive K562 cells (human chronic myelogenous leukemia cell line), from our collaborator Dr. K. Elenitoba-Johnson (Univ. of Michigan), were cultured as suspension cells in RPMI 1640 supplemented with 10% FBS (Hyclone laboratories, Logan, UT), 1% penicillin-streptomycin (100U/ml, GIBCO BRL, Grand Island, NY), 0.1% gentamycin (Hyclone), and 1% L-glutamine (Hyclone). Cells were maintained in a 5% CO2 incubator at 37°C. Cells were split at a density of 0.5 × 105/ml two days before transfection.
Transfection
Transient transfections were performed using Amaxa Nucleofector II according to the Amaxa protocol for K562 cells. Briefly, 2 × 106 cells were pelleted from a cell density of 1–5 × 105 cells/mL, and then resuspended in 100 µL Amaxa Solution V. This solution was then added to 10 µg DNA and transfected in an Amaxa cuvette using program T-013. Following, 500 µL RPMI was added to the cuvette and cells were transferred to 15 mL RPMI and plated in a 75 cm2 flask for caspase-3 assays. Small aliquots (200 µL) of cells were plated into 4-well live-cell chambers for fluorescent microscopy (Lab-tek chamber slide system, 2 mL, Nalge NUNC International, Naperville, IL) for determination of transfection efficiency. For co-localization experiments where 2 plasmids were transfected simultaneously, 5µg of each plasmid was used; 5µg of a single plasmid was used for comparison studies. Cells were incubated for ~20–24 hours before any other assays were performed.
To calculate transfection efficiency, four or more fields of cells were counted under the 40x objective. The number of transfected cells (as indicated by EGFP expression; see methods below for microscope settings) was divided by the total number of cells to obtain transfection efficiency.
Caspase-3 Activity Assay
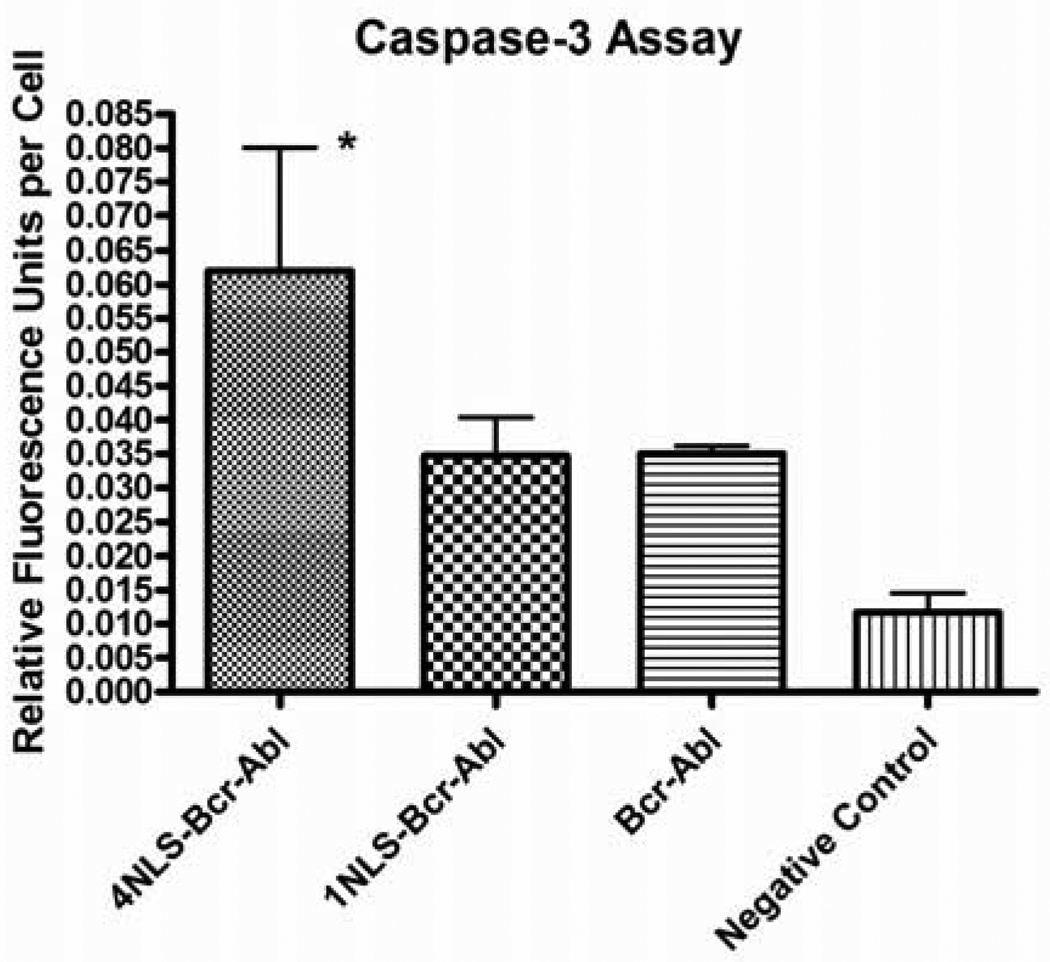
The induction of apoptosis was monitored through the enzymatic activity of caspase-3 using the EnzChek Caspase-3 Assay Kit #1 (Molecular Probes, Eugene, OR) following the manufacturer’s protocol. Briefly, 1.5 × 106 cells were pelleted and resuspended in 50 µL 1x cell lysis buffer followed by a freeze-thaw cycle. The lysed cells were centrifuged for five minutes at 2100 × g. As a control, 1 µL of 1 mM caspase-3 inhibitor was added to one of the lysates of untransfected K562 cells. 2x substrate (50 µL) solution was then added to 50 µL of lysate and incubated at room temperature for 30 minutes. A standard curve was made using known amounts of 7-amino-4-methylcoumarin (AMC). Fluorescence was then measured at an excitation wavelength of 355 nm and an emission wavelength of 460 nm. Caspase assays were performed three or more separate times (n = 3). Data was represented as relative fluorescence units per cell, taking transfection efficiencies into account (figure 5).
Figure 5.
Caspase-3 activity in K562 cells transfected with EGFP-4NLS-Bcr-Abl, EGFP-1NLS-Bcr-Abl, EGFP-Bcr-Abl, or pGL3basic plasmid (a negative control; plasmid is promoter- and enhancer-less and therefore nothing is expressed). *p<0.05; n = 3. Statistical significance determined using one-way Anova with Tukey’s correction.
Actin Staining and Microscopy
K562 cells transfected with EGFP-Bcr-Abl were fixed with formaldehyde and stained for actin using BODIPY 558/568 Phalloidin from Molecular Probes (Eugene, OR) according to manufacturer’s protocol. Briefly, cells were washed with PBS, pelleted, and resuspended in 4% formaldehyde (in PBS). Cells were incubated in formaldehyde solution for ten minutes at room temperature and subsequently washed in PBS. Cells were then placed in 1 mL of 0.1% Triton X-100 (in PBS) for five minutes followed again by washing with PBS. The cells were then incubated with staining solution for 20 minutes at room temperature and then washed with PBS. Following staining the cells were viewed at 40x with an Olympus IX701F inverted fluorescence microscope (Scientific Instrument Company, Aurora, CO) using the HQ:TRITC filter to detect actin, and a high-quantity narrow band GFP filter (excitation HQ480/20nm, emission HQ510/20nm, with beam splitter Q4951p) to detect EGFP. Cells were photographed using a F-View Monochrome CCD camera.
DNA staining and EGFP microscopy
K562 cell nuclei were stained by the addition of 0.8 µL Hoechst 33342 (10mg/ml) (Invitrogen, Carlsbad, CA) to 1 mL of cells in a Lab-Tek®II 4-well live-cell chamber (Nalge NUNC) and incubated for 30 min at 37°C. Pictures were taken approximately 24 hours after transfection using a fluorescence microscope with high-quantity narrow band GFP filter (to detect EGFP) and Cyan GFP v2 filter (excitation D436/20nm, emission D480/40nm, with beam splitter 455dclp) to detect H33342. To minimize photobleaching of EGFP chromophore, cells were imaged using neutral density filters in combination with short exposure times. An air stream incubator (Nevtek ASI 400, Burnsville, VA) with variable temperature control was used to maintain the microscope stage and the live-cell chambers at 37°C. All filters were purchased from Chroma Technology (Brattleboro, VT).
Cell Tracing
Quantitation of Bcr-Abl in the nucleus and cytoplasm was carried out by measuring the fluorescence intensity of EGFP, tagged to the Bcr-Abl, as previously described[18]. All the images were analyzed using analySIS® software (Soft Imaging System, Lakewood, CO).
Statistical Analysis
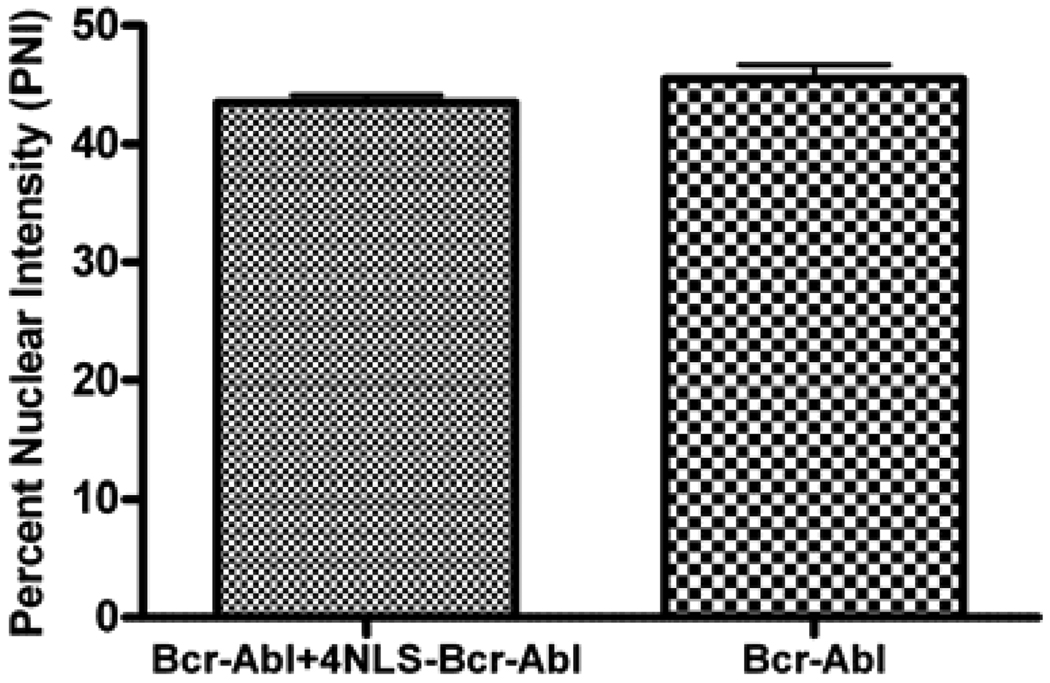
All experiments were done at least in triplicate (n≥3). The difference between the percent nuclear intensity values (for figure 4) was analyzed using an unpaired t-test with Welch’s correction. One-way ANOVA with Tukey’s multiple comparisons post-test was used to assess the differences between relative fluorescence intensity values from the caspase-3 assay (figure 5). All statistics were calculated using GraphPad Prism (San Diego, CA).
Figure 4.
Percent nuclear intensity for cells transfected with EGFP-Bcr-Abl alone (right column) or EGFP-Bcr-Abl and 4NLS-Bcr-Abl (left column). n = 5 cells for each group.
RESULTS
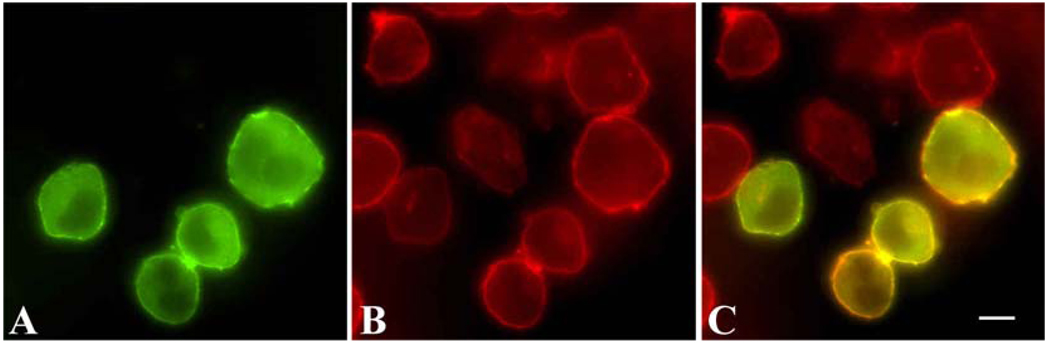
When transfected into K562 cells, Bcr-Abl (pEGFP-Bcr-Abl) localizes in the cytoplasm and forms a distinctive ring around the cell (figure 1). This localization is indicative of binding to actin, and is expected due to previous reports of Bcr-Abl localization with actin[19]. Colocalization of EGFP-Bcr-Abl and actin is shown in figure 1.
Figure 1.
EGFP-Bcr-Abl localization in K562 cells. A. Localization of Bcr-Abl via EGFP. B. Actin staining of cells. C. Overlay of EGFP-Bcr-Abl localization with actin. White scale bar, 5µm.
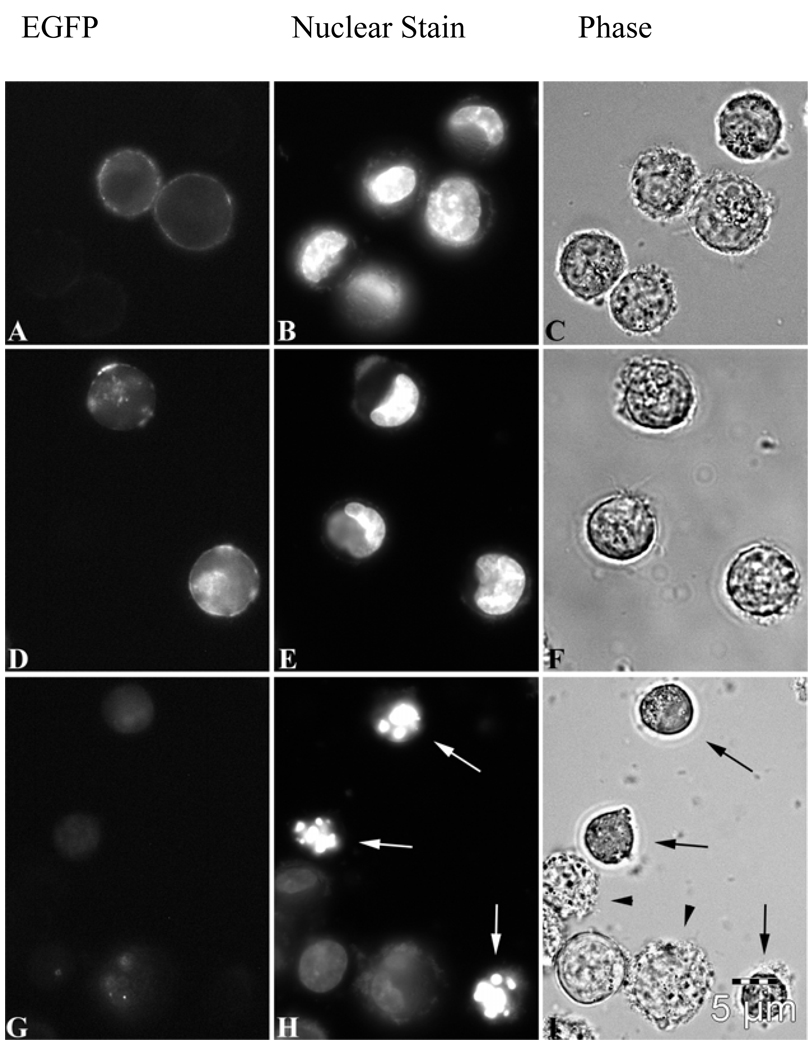
Additional nuclear localization signal(s) were added to EGFP-Bcr-Abl to attempt to overcome Bcr-Abl binding to actin. Two plasmids were created, one with 1 NLS subcloned to EGFP-Bcr-Abl, and one with 4 NLSs subcloned to EGFP-Bcr-Abl. When transfected into K562 cells, only the EGFP-4NLS-Bcr-Abl localized to the nucleus (figure 2G) with the distinct absence of the actin ring surrounding the cell (compare to 2A, wt-Bcr-Abl). The 1NLS construct, while somewhat dispersed throughout the cell, still formed a peripheral actin ring (figure 2D), just like wt-Bcr-Abl (figure 2A and figure 1).
Figure 2.
A–C. EGFP-Bcr-Abl transfected into K562 cells; fluorescent (EGFP), nuclear staining with H33342, and phase contrast images, respectively. D–F. EGFP-1NLS-Bcr-Abl transfected into K562 cells; fluorescent (EGFP), nuclear staining with H33342, and phase contrast images, respectively. G–H. EGFP-4NLS-Bcr-Abl transfected into K562 cells; fluorescent (EGFP), nuclear staining with H33342, and phase contrast images, respectively. White arrows (H) indicate apoptotic cells undergoing massive DNA segmentation. Black arrows (I) indicate that these same cells have undergone cell shrinkage, a morphological hallmark of apoptosis[20–24]. Black arrowheads (I) show cells undergoing zeotic/cytoplasmic blebbing. Scale bar (5µm) representative for all panels (A–I).
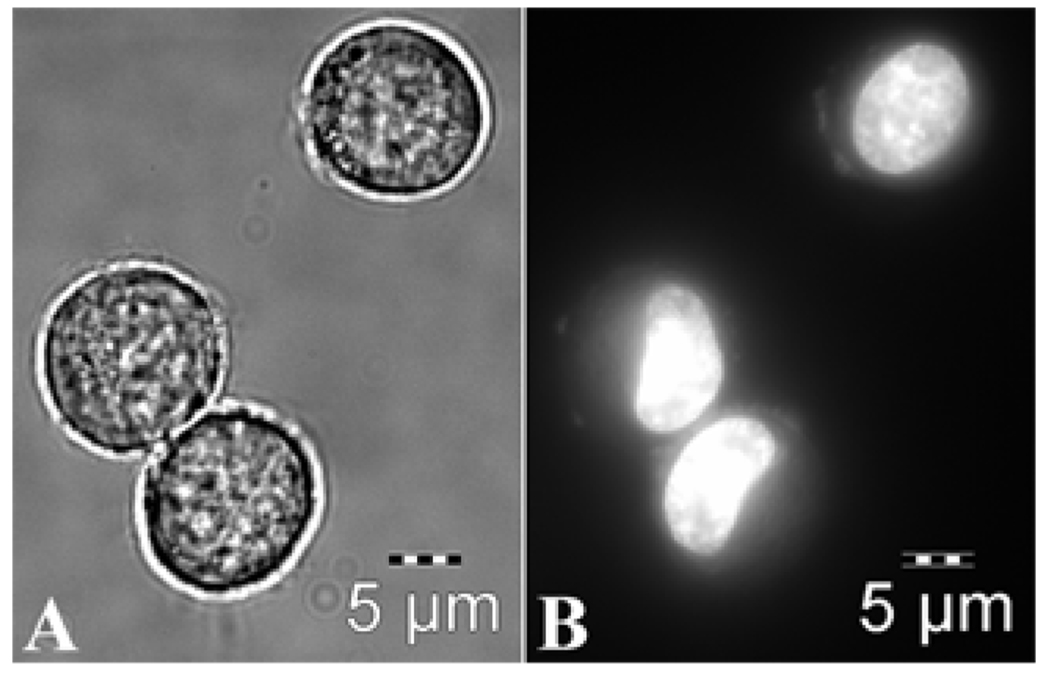
Next, cell studies demonstrating apoptosis were performed on Bcr-Abl, 4NLS-Bcr-Abl, and 1NLS-Bcr-Abl. For comparison, figure 3 shows healthy K562 cells. Phase contrast images show round, healthy cells (figure 3, left panel), while H33342 staining indicates normal (unsegmented) nuclei (figure 3, right panel). Nuclei in healthy K562 cells are round/oval or “kidney” shaped (figure 3, right panel).
Figure 3.
Healthy K562 cells. Left panel, phase contrast image. Right panel, nuclear staining with H33342. Nuclei of healthy cells are either round/oval or “kidney” shaped.
Twenty-four hours after transfection, cytochemical analyses of apoptosis, including overall cell morphology (figure 2; C, F, and I) and DNA morphology/segmentation (figure 2; B, E, and H), were examined.
Morphological changes indicating apoptosis include zeotic and cytoplasmic blebbing, cell shrinkage, and fragmentation of cells into smaller bodies[20–24]. These morphological changes are seen obviously in cells transfected with 4NLS-Bcr-Abl, comparing figure 2I (apoptosis) to figures 2C and 2F (no apoptosis). Black arrows indicate shrunken cells (with cell shrinkage indicating apoptosis). Black arrowheads (2I) show cells undergoing zeotic/cytoplasmic blebbing.
The characteristic change in nuclear morphology (DNA segmentation) is “the most accurate indicator of the involvement of apoptosis in the death of a cell”[22], where the nucleus changes shape from the normal round/oval shape into smaller, non-homogeneous segments. As shown in figure 2H, cells transfected with 4NLS-Bcr-Abl show this characteristic DNA segmentation into smaller pieces (white arrows), not seen in other cells (compare to figures 2B and 2E), including healthy cells (figure 3, right panel).
Transfected cells were also counted for DNA segmentation or nuclear morphology changes, an accurate indicator of apoptosis[22]. As shown in table 1, cells transfected with EGFP-4NLS-Bcr-Abl had a higher percentage of cells with apoptotic nuclei than EGFP-1NLS-Bcr-Abl and EGFP-Bcr-Abl. Only cells with obvious DNA segmentation (more than two nuclear fragments formed) were counted as segmented.
Table 1.
DNA segmentation in K562 cells transfected with EGFP-Bcr-Abl, EGFP-1NLS-Bcr-Abl, or EGFP-4NLS-Bcr-Abl.
| Construct transfected | No. of cells transfected |
No. transfected cells with segmented DNA |
Percentage of cells with segmented DNA |
|---|---|---|---|
| EGFP-Bcr-Abl | 178 | 7 | 3.9 |
| EGFP-1NLS-Bcr-Abl | 154 | 5 | 3.2 |
| EGFP-4NLS-Bcr-Abl | 375 | 46 | 12.3 |
Bcr-Abl is known to form homodimers (tetramers)[25, 26]. To determine if the 4NLS-Bcr-Abl was capable of multimerizing with Bcr-Abl, and altering its localization, a co-transfection (colocalization) experiment was performed. EGFP-4NLS-Bcr-Abl with mutated EGFP chromophore (not fluorescent) was transfected with fluorescent EGFP-Bcr-Abl. If the 4NLS-Bcr-Abl protein is able to drag Bcr-Abl into the nucleus, EGFP fluorescence will be detected in the nucleus. If 4NLS-Bcr-Abl does not drag Bcr-Abl into the nucleus, EGFP-Bcr-Abl will remain in the cytoplasm. For this analysis, a semi-quantitative assay was used that measured the fluorescence intensity in the nucleus in the co-transfected cells vs. the singly transfected (with EGFP-Bcr-Abl) cells, normalized to each cell’s cytoplasmic intensity. These nuclear to cytoplasmic intensities were termed “percent nuclear intensity.” 4NLS-Bcr-Abl did not “capture” wt-Bcr-Abl and drag it into the nucleus, since the nuclear intensity of wt-EGFP-Bcr-Abl did not change in the presence of the 4NLS-Bcr-Abl construct (figure 4). This result is not unexpected since the presence of strong NLSs would presumably drive any protein it was attached to immediately into the nucleus. From this result, it was determined that cytoplasmic depletion of Bcr-Abl by 4NLS-Bcr-Abl was not occurring.
Next, the caspase-3 assay was used to determine mid-stage apoptosis. Caspases, from cysteine-aspartate proteases, are a group of proteolytic enzymes that perpetuate the apoptotic signal by cleavage of proteins such as actin, lamins, fodrin, ICAD, and PARP. Prior to receiving the apoptotic signal, caspases are present in cells in an inactive pro-enzyme form. The caspase cascade starts with activation of initiator caspases (caspase-2, -8, - 9, and -10), which then activate effector caspases (caspase-3, -6, and -7). Both caspase-3 and caspase-7 cleave the peptide sequence DEVD, which makes it an ideal substrate for use in measuring apoptosis as a majority of apoptotic signaling pathways converge on these effector caspases. The caspase-3 assay used measures the cleavage of a substrate with the amino acid sequence DEVD. Caspase-3 activity (figure 5) was determined in several treatment groups, including EGFP-4NLS-Bcr-Abl, EGFP-1NLS-Bcr-Abl, EGFP-Bcr-Abl, or a negative control (tranfected with pGL3 basic plasmid which has no promoter or enhancer and hence is not expressed). As shown in figure 5, only cells transfected with EGFP-4NLS-Bcr-Abl showed a statistically significant increase in caspase activity compared to the negative control.
DISCUSSION/CONCLUSIONS
Nuclear localization of Bcr-Abl converts it from an oncogene to an apoptotic factor. Nuclear localization could not be achieved without adding multiple NLSs to Bcr-Abl. Once additional NLSs were added to Bcr-Abl, this caused apoptosis of CML cells (K562 cells) measured by morphologic changes (cytoplasmic blebbing, cell shrinkage), DNA segmentation (nuclear morphology changes), and the caspase-3 assay. This critical finding shows that the change in location of Bcr-Abl from cytoplasmic to nuclear dramatically alters its function.
Nuclear entrapment and cytoplasmic depletion of wt-Bcr-Abl may have a synergistic effect on cell death. Nuclear entrapment results in apoptosis via the Abl domain which can cause cell death by stabilizing p73 and activating its pro-apoptotic functions[2, 27, 28]. Cytoplasmic depletion of wt-Bcr-Abl will remove its ability to interact with signal transduction proteins involved in gene transcription, mitochondrial processing of apoptotic responses, cytoskeletal organization, and degradation of inhibitory proteins[29]. Here we report that plasmid expressed Bcr-Abl directed to the nucleus is sufficient to induce apoptosis with out cytoplasmic depletion. However, it may be speculated that coupling cytoplasmic depletion with nuclear entrapment will synergistically enhance the apoptotic signal. To this end, our ultimate goal is to deplete wt-Bcr-Abl from the cytoplasm, and direct it to the nucleus by our previously described “protein switch” technology[18, 30]. We exploit known dimerization domains (DD) of proteins to capture them and move them to a different subcellular compartment. An plasmid expressed “protein switch” can be designed to target any protein with a DD. The protein switch also contains inducible signal sequences to localize to a starting compartment in the cell (e.g., cytoplasm). Upon addition of ligand, the protein switch can move to a different subcellular compartment (e.g., nucleus). Our ultimate goal is to use the protein switch [18, 30] to control the subcellular location of Bcr-Abl, and convert Bcr-Abl from an oncogene to an apoptotic factor.
Alternative therapies for CML are needed, despite the “blockbuster” results with Gleevec. Gleevec is not a cure for CML, and patients need to take it for the rest of their lives. Gleevec is also known to have severe side effects including cardiotoxicity [11] and the possibility of developing other cancers (due to blockade of tumor suppressor p63)[12]. Importantly, Gleevec resistance mostly occurs due to different point mutations in Bcr-Abl that make traditional small molecule inhibitors (like Gleevec) unable to bind/inhibit Bcr-Abl. In some cases, a decrease in Gleevec bioavailability/cell exposure and upregulation of drug efflux pumps [6, 10] have led to resistance.
The protein switch concept is also being tested in our laboratory with the anti-apoptotic factor survivin, and the tumor suppressor p53 (mislocalized in cancers). Changing localization to alter function may prove to be a new therapeutic strategy for many types of cancer, including CML[31].
ACKNOWLEDGEMENTS
Funding provided by NIH R01 CA129528. We would like to thank David W. Woessner for technical assistance. We also thank S.W. Kim, K. Elenitoba-Johnson, J. Yockman, T. Cheatham, J.R. Davis, M. Mossalam, and other lab members.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Vigneri P, Wang JY. Induction of apoptosis in chronic myelogenous leukemia cells through nuclear entrapment of BCR-ABL tyrosine kinase. Nat Med. 2001;7(2):228–234. doi: 10.1038/84683. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 4.Oldham R, Dillman R. Gold standard or wrong standard? Cancer Biother Radiopharm. 2004;19(3):271–272. doi: 10.1089/1084978041425034. [DOI] [PubMed] [Google Scholar]
- 5.Peggs K. Imatinib mesylate--gold standards and silver linings. Clin Exp Med. 2004;4(1):1–9. doi: 10.1007/s10238-004-0032-5. [DOI] [PubMed] [Google Scholar]
- 6.Baccarani M, Saglio G, Goldman J, Hochhaus A, Simonsson B, Appelbaum F, Apperley J, Cervantes F, Cortes J, Deininger M, Gratwohl A, Guilhot F, Horowitz M, Hughes T, Kantarjian H, Larson R, Niederwieser D, Silver R, Hehlmann R. Evolving concepts in the management of chronic myeloid leukemia. Recommendations from an expert panel on behalf of the European Leukemianet. Blood. 2006 doi: 10.1182/blood-2006-02-005686. [DOI] [PubMed] [Google Scholar]
- 7.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 8.Shah NP, Sawyers CL. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22(47):7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- 9.Roumiantsev S, Shah NP, Gorre ME, Nicoll J, Brasher BB, Sawyers CL, Van Etten RA. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc Natl Acad Sci U S A. 2002;99(16):10700–10705. doi: 10.1073/pnas.162140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melo JV, Hughes TP, Apperley JF. Chronic myeloid leukemia. Hematology (Am Soc Hematol Educ Program) 2003:132–152. doi: 10.1182/asheducation-2003.1.132. [DOI] [PubMed] [Google Scholar]
- 11.Kerkela R, Grazette L, Yacobi R, Iliescu C, Patten R, Beahm C, Walters B, Shevtsov S, Pesant S, Clubb FJ, Rosenzweig A, Salomon RN, Van Etten RA, Alroy J, Durand JB, Force T. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12(8):908–916. doi: 10.1038/nm1446. [DOI] [PubMed] [Google Scholar]
- 12.Ongkeko WM, An Y, Chu TS, Aguilera J, Dang CL, Wang-Rodriguez J. Gleevec suppresses p63 expression in head and neck squamous cell carcinoma despite p63 activation by DNA-damaging agents. Laryngoscope. 2006;116(8):1390–1396. doi: 10.1097/01.mlg.0000225941.60901.0f. [DOI] [PubMed] [Google Scholar]
- 13.Francois H, Coppo P, Hayman JP, Fouqueray B, Mougenot B, Ronco P. Partial fanconi syndrome induced by imatinib therapy: a novel cause of urinary phosphate loss. Am J Kidney Dis. 2008;51(2):298–301. doi: 10.1053/j.ajkd.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Calabretta B, Perrotti D. The Biology of CML Blast Crisis. Blood. 2004 doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 15.Goldman JM, Melo JV. Chronic myeloid leukemia--advances in biology and new approaches to treatment. N Engl J Med. 2003;349(15):1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 16.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32(5):1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 17.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273(5280):1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 18.Kakar M, Davis JR, Kern SE, Lim CS. Optimizing the protein switch: altering nuclear import and export signals, and ligand binding domain. J Control Release. 2007;120(3):220–232. doi: 10.1016/j.jconrel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantschel O, Wiesner S, Guttler T, Mackereth CD, Rix LL, Mikes Z, Dehne J, Gorlich D, sattler M, Superti-Furga G. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol Cell. 2005;19(4):461–473. doi: 10.1016/j.molcel.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Schwartzman RA, Cidlowski JA. Apoptosis: the biochemistry and molecular biology of programmed ell death. Endocr Rev. 1993;14(2):133–151. doi: 10.1210/edrv-14-2-133. [DOI] [PubMed] [Google Scholar]
- 21.Vermes I, Haanen C. Apoptosis and programmed cell death in health and disease. Adv Clin Chem. 1994;31:177–246. doi: 10.1016/s0065-2423(08)60336-4. [DOI] [PubMed] [Google Scholar]
- 22.Willingham MC. Cytochemical methods for the detection of apoptosis. J Histochem Cytochem. 1999;47(9):1101–1110. doi: 10.1177/002215549904700901. [DOI] [PubMed] [Google Scholar]
- 23.Bortner CD, Cidlowski JA. Apoptotic volume decrease and the incredible shrinking cell. Cell Death Differ. 2002;9(12):1307–1310. doi: 10.1038/sj.cdd.4401126. [DOI] [PubMed] [Google Scholar]
- 24.Bortner CD, Cidlowski JA. Uncoupling cell shrinkage from apoptosis reveals that Na+ influx is equired for volume loss during programmed cell death. J Biol Chem. 2003;278(40):39176–39184. doi: 10.1074/jbc.M303516200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. Structure of the Bcr-Abl oncoprotein ligomerization domain. Nat Struct Biol. 2002;9(2):117–120. doi: 10.1038/nsb747. [DOI] [PubMed] [Google Scholar]
- 26.McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13(12):7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aloisi A, Di Gregorio S, Stagno F, Guglielmo P, Mannino F, Sormani MP, Bruzzi P, Gambacorti-Passerini C, Saglio G, Venuta S, Giustolisi R, Messina A, Vigneri P. BCR-ABL nuclear entrapment kills human CML cells: ex vivo study on 35 patients with the combination of imatinib mesylate and leptomycin B. Blood. 2006;107(4):1591–1598. doi: 10.1182/blood-2005-05-2123. [DOI] [PubMed] [Google Scholar]
- 28.Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000;19(49):5643–5650. doi: 10.1038/sj.onc.1203878. [DOI] [PubMed] [Google Scholar]
- 29.Dai Z, Kerzic P, Schroeder WG, McNiece IK. Deletion of the Src homology 3 domain and C-terminal proline-rich sequences in Bcr-Abl prevents Abl interactor 2 degradation and spontaneous cell migration and impairs leukemogenesis. J Biol Chem. 2001;276(31):28954–28960. doi: 10.1074/jbc.M101170200. [DOI] [PubMed] [Google Scholar]
- 30.Kakar M, Cadwallader AB, Davis JR, Lim CS. Signal sequences for targeting of gene therapy products to subcellular compartments: the role of CRM1 in nucleocytoplasmic shuttling of the protein switch. Pharm Res. 2007;24(11):2146–2155. doi: 10.1007/s11095-007-9333-1. [DOI] [PubMed] [Google Scholar]
- 31.Davis JR, Kakar M, Lim CS. Controlling protein compartmentalization to overcome disease. Pharm Res. 2007;24(1):17–27. doi: 10.1007/s11095-006-9133-z. [DOI] [PubMed] [Google Scholar]