Abstract
An important approach for developing a safer smallpox vaccine is to identify naturally processed immunogenic vaccinia-derived peptides rather than live whole vaccinia virus. We used two-dimensional liquid chromatography coupled to mass spectrometry to identify 116 vaccinia peptides, encoded by 61 open reading frames, from a B-cell line (homozygous for HLA class I A*0201, B*1501, and C*03) after infection with vaccinia virus (Dryvax). Importantly, 68 of these peptides are conserved in variola, providing insight into the peptides that induce protection against smallpox. Twenty-one of these 68 conserved peptides were 11 amino acids long or longer, outside of the range of most predictive algorithms. Thus, direct identification of naturally processed and presented HLA peptides gives important information not provided by current computational methods for identifying potential vaccinia epitopes.
Keywords: vaccinia virus, HLA class I, antigen presentation, smallpox vaccine
1. Introduction
Despite the success of the smallpox (vaccinia) vaccine in eradicating smallpox world-wide, the targets of cell-mediated immunity (CD8+ and CD4+ T cell epitopes) are mostly unknown. From an adaptive immunity standpoint, the most important requirement for an immunogenic smallpox vaccine is the ability of the immune system to process and present specific peptides to T cells in the context of specific human leukocyte antigen (HLA) molecules. Antigen specificity of both CD8+ and CD4+ T cells is strongly controlled by HLA and other gene polymorphisms, and the immunogenicity (and ultimately the efficacy) of vaccines are dependent on particular HLA allelic polymorphisms [1] that restrict which peptides can be bound and hence presented [2, 3]. Both class I and class II HLA polymorphisms control antigen and peptide epitope specificity of T cell responses and have been associated with differences in smallpox vaccine response outcome [4-6]. Therefore, the immune response to smallpox vaccine is determined in large part by multigenic influences controlling the presentation of specific sets of peptides [7]. The existence of HLA allelic “supertypes” (for example, the HLA-A2, A3, B7, and B62 supertypes), which are functionally defined by their ability to bind similar sets of, and in some cases, identical peptides [8-10], allow for the identification of “promiscuous” or shared presented peptides that possess enormous potential for future peptide-based smallpox vaccine development.
To date, the majority of known T-cell epitopes from vaccinia have been discovered using predictive computer algorithms, followed by assays that measure immunoproliferative response to synthesized peptides of the predicted sequences [11-13]. While algorithms have some success at predicting peptides that bind to HLA, there are still gaps in being able to predict which peptides will be produced by the proteasome and transported to HLA class I molecules in the endoplasmic reticulum via the transporter associated with antigen processing (TAP), and which exogenous peptides will be displayed on the HLA class II molecules [14].
Alternatively, mass spectrometry (MS) has been used for directly sequencing peptides presented by HLA molecules from infected cells, as pioneered by Hunt and co-workers [15], and has been used to identify naturally processed class I and class II epitopes from pathogens [16-20]. An imposing challenge of this approach has been the complexity of the peptide mixture isolated from HLA molecules and the often relatively low abundance of HLA ligands of pathogenic origin. Another targeted approach to identifying HLA ligands of pathogenic origin takes advantage of both predictive algorithms and direct identification by MS. The ‘predict-calibrate-detect’ strategy uses predictive algorithms to generate target sequences predicted to be epitopes. MS methods are then optimized for peptide sequences predicted by the HLA-binding algorithm [21-23].
Recently, proteomics-driven advances in MS sensitivity and speed of acquiring data-directed MS/MS data, along with automated data processing, database searching, and statistical evaluation of large sets of search results have provided an unprecedented ability to rapidly characterize the range of peptides presented by the immune system. This approach has the potential advantages of being able to detect post-translationally modified ligands, or ligands of atypical length or binding motif.
In previous research studying measles virus, we identified HLA class II (DRB1*0301) bound measles-derived peptides using orthogonal dimensions of liquid chromatography to separate the complex pool of peptides, followed by tandem mass spectrometry (MS/MS) to generate peptide sequence information [24, 25]. This “reverse immunogenetics” approach represents a promising solution to the identification of vaccine candidates for many diseases [20, 26, 27], including vaccines against agents of bioterrorism, such as smallpox.
The goal of the present study was to identify the range of vaccinia virus-derived peptides presented by HLA-A*0201 (A2 supertype), HLA-B*1501 (B62 supertype), and HLA-C*03 alleles using orthogonal chromatography separations and MS/MS, with high mass accuracy measurement of precursor ions, similar to methods applied in previous work that identified class II peptides from measles virus [25]. In addition, we compared a set of vaccinia-derived peptides, identified after natural infection with vaccinia, with those predicted by computer algorithms. We believe the identification of these naturally processed and presented peptides resulting from vaccinia virus infection complements predictive methods and will aid in understanding the immune process, perhaps leading to effective, integral components of a new generation of vaccines based upon cocktails of synthetic peptides.
2. Materials and Methods
2.1. Cell culturing and vaccinia virus infection
Epstein-Barr virus (EBV)-transformed B cells (European Collection of Cell Cultures – ECACC, number 86052111, Salisbury, Wiltshire, UK), homozygous for the HLA-A*0201, B*1501, and C*03 were of human origin and used as APC. The New York City Board of Health (NYCBOH, Dryvax®) vaccine-strain of vaccinia virus was cultured in HeLa cells in Dulbecco’s modified Eagle’s medium, supplemented with 5% fetal calf serum (FCS; Life Technologies, Gaithersburg, Maryland). Cells were infected with live vaccinia virus at a multiplicity of infection (moi) of 0.1 PFU/cell for 2 hours and further maintained for 24-30 hrs in RPMI-1640 supplemented with 8% FCS. These uninfected and vaccinia-infected cells (approximately 1 × 109 cells each) were used for obtaining cell lysates for the class I HLA molecules purification.
2.2. Isolation of HLA-associated peptides from uninfected and vaccinia-infected cells
Class I HLA molecules were purified from homozygous B cells using an immunoaffinity approach, and their associated peptides were extracted as previously described [28, 29]. In brief, cells were lysed with a buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, 1% CHAPS and protease inhibitors (1 mM Pefabloc SC, Roche Applied Science, Indianapolis, IN). The clarified supernatants were passed over a protein A-sepharose 4B (Sigma) column containing the monoclonal antibody (mAb) W6/32 specific for HLA-A, B and C [30, 31]. The HLA molecules (1.2 mg/mL) were dissociated from their bound class I peptides using 0.2 N acetic acid, pH 2.7 and peptides were separated from the HLA by filtration through a 10-kDA molecular mass cutoff filter (Millipore, Bedford, MA).
2.3. Strong cation exchange fractionation
Strong cation exchange (SCX) fractionation of the sample was performed after desalting the peptide pool with a reversed phase 1mm by 8mm peptide trap (Michrom BioResources, Auburn, CA). SCX chromatography used a gradient of 5 mM KH2PO4, pH 3.0 to 0.4 M KCl of 5 mM KH2PO4, pH 3.0. Acetonitrile was added to each mobile phase to 20% by volume. Desalted peptides were loaded onto a polysulfoethyl aspartamide column (Michrom BioResources) at 0.5% mobile phase A and a gradient was developed to 20% B over 20 minutes at a flow rate of 200 μL/min, then from 20% B to 80% B over the next 10 minutes. Two minute fractions were collected; each fraction was vacuum centrifuged to dryness before analyzing the fractions by nLC-MS/MS.
2.4. LTQ-Orbitrap nLC-MS/MS analyses
Automated nLC-MS/MS analyses were performed on a commercial linear ion trap-Fourier transform hybrid mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific, Waltham, MA r), interfaced to a nano-scale liquid chromatograph and autosampler (Eksigent NanoLC 1D, Dublin, CA), using a 15cm by 75 μm i.d. column packed with Magic C18AQ (5 μm particles, 200 Å pore size, Michrom BioResources). The autosampler loaded 5-20 μL onto a 0.25μL pre-column (Optimize Technologies, Oregon City, OR), custom-packed with Magic C8, 5μm, 300 Å (Michrom BioResources). Mobile phase A consisted of water/acetonitrile/formic acid (98/2/0.2 by volume) and mobile phase B was acetonitrile/water/formic acid (90/10/0.2 by volume). A 90 minute LC method employed a gradient from 2-40% B over 60 minutes, followed by a second segment to 90% B at 85 minutes, with a column flow of 0.4μL/min. A third pump was used to load peptides from the autosampler to the pre-column, with 0.05% TFA and 0.15% formic acid in water at 15μL/min.
SCX fractions were analyzed multiple times by nLC-MS/MS using data-dependant acquisition of tandem mass spectra. The first experiment targeted singly charged precursors between 750 and 1500 on the m/z (molecular mass m divided by charge z) scale. A second experiment targeted doubly and triply charged precursors between m/z 375 and 750, consistent with MHC Class I peptides which are predominantly 9-11 amino acids long.
The LTQ-Orbitrap was operated in a data-dependant mode, first acquiring an Orbitrap survey scan with 60000 resolving power (FWHM at m/z 400), a target cell population of 1×106 ions, and a maximum ion fill time of 300 ms. The preview Fourier transform was used to select the five most abundant ions for MS/MS experiments in the LTQ. LTQ MS/MS spectra were acquired with a 2.5 mass unit isolation width, target ion population of 1×104 ions, one microscan, maximum ionization fill time of 100 ms, normalized collision energy of 35%, activation Q of 0.25, and activation time of 30 ms. Once ions were selected for MS/MS, they were subsequently excluded for 45 seconds. The exclusion window was 1 m/z below, and 1.6 m/z above the exclusion mass.
2.5. Peptide identification from MS/MS data
Database searching was performed using the SWIFT workflow tool developed in-house. SWIFT coordinates the generation of database search files (using extract_msn software from ThermoFisher Scientific), initiates database searches using MASCOT, Sequest, and X!Tandem search engines, and integrates these search results using Scaffold (Ver. 1_07_00, Proteome Software, Portland, OR). Database searches were done against a subset of the SwissProt database (January, 2007) obtained using the Bioworks 3.2 (Thermo Fisher) database utility to select human, bovine, and vaccinia proteins. Bovine proteins were included in the database since cell cultures were supplemented with fetal calf serum. Database searches were performed with a precursor mass tolerance of 7 parts-per-million (ppm), fragment ion mass tolerance of 0.6 mass units, and without any protease specificity. Single oxidation on methionine residues was considered as a variable modification. The database was appended with decoy protein entries consisting of randomized proteins sequences (MASCOT utility) for estimating the false positive rate resulting in a database of 43,400 entries (including the randomized decoy entries). Results from all analyses of all SCX fractions were combined by Scaffold and exported to an Excel spreadsheet.
The Scaffold program (ProteomeSoftware) was used to combine search results from all of the LC-MS/MS analyses and to calculate peptide identification probabilities using Scaffold’s implementation of PeptideProphet [32, 33]. An export function within Scaffold (Spectrum Report) was used to create a text file of all search results passing a lenient filter threshold of 80% protein probability, with at least 1 peptide identified above an 80% peptide probability threshold; 23,800 search results met those criteria, 1,521 of these were from decoy peptides. Additional filtering was then applied to this dataset while estimating the false-positive rate (FPR) of identifications from the incidence of identifications from the decoy database using the formula: 2×# matches to decoy peptides / (number of true positives + number of false positives) as described by Elias and Gygi [34]. The FPR was calculated as a function of thresholds for the following scoring parameters: Sequest cross correlation score (XCorr), the difference between the top two normalized cross-correlation scores (ΔCn), Mascot Ion Score, and mass error of the precursor mass.
3. Results
3.1. HLA class I peptide identification by tandem mass spectrometry
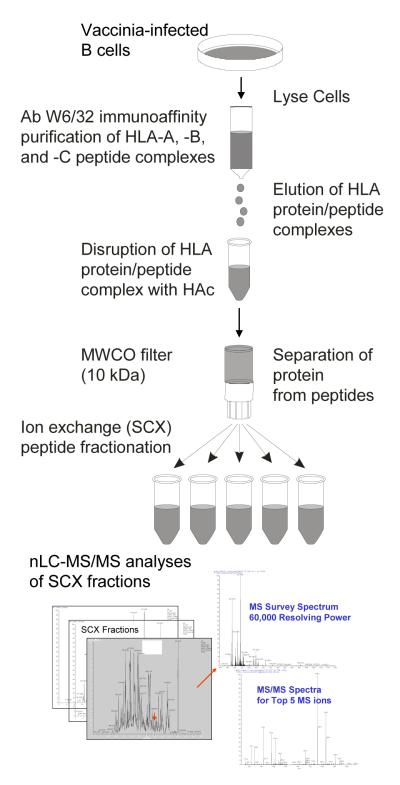
Figure 1 provides an overview of the protocol used to sequence HLA class I peptides isolated from B-cells after infection with vaccinia. Sixteen strong cation exchange fractions were analyzed by nano-scale liquid chromatography coupled with tandem mass spectrometry (nLC-MS/MS) on the LTQ-Orbitrap. Two data sets were acquired, each using multiple injections as described in the methods sections. The initial data set was acquired with external mass calibration, followed by a second data set collected with internal mass calibration using lock masses at m/z 391.2843 and m/z 445.1200 as described by Olsen et al. [35]. These analyses generated 214,800 MS/MS spectra that were searched against the human, bovine, and vaccinia subset of proteins in the SwissProt database (January 2007 version).
Figure 1.
Overview of protocol for isolating and identifying HLA class I peptides from B cells infected with vaccinia virus.
Peptide sequencing by mass spectrometry involves matching MS/MS fragmentation spectra against theoretical fragmentation spectra calculated for any peptide sequence in the database within a tolerance window of the molecular weight of the peptide as measured by the mass spectrometer. As a result, every database search will return a result, and each of these matches to a sequence must be evaluated for their validity. A variety of scoring metrics exist from which a threshold is established for accepting or rejecting the search result from any MS/MS spectrum. The goal of that threshold is to minimize the number of incorrect or random matches that are accepted (false positives) while also trying to minimize the rejection of correct sequence matches (false negatives). Most of these scoring criteria have been developed within the context of identifying peptides generated from cleavage of proteins with trypsin. Trypsin cleaves on the C-terminal side of arginine and lysine and this cleavage specificity greatly reduces the list of candidate peptide sequences that are matched against the experimental spectra. The basic C-terminus of tryptic peptides provided by the Arg and Lys side chains, favorably directs fragmentation during MS/MS, influencing the scores from the database search results.
HLA class I peptides are not constrained to a basic C-terminal amino acid. For most common alleles, hydrophobic amino acids predominate in the C-terminal position, though basic residues such as Lys, Arg, Pro, or His, are often found somewhere in the C-terminal half of the peptide. Also by not requiring Lys or Arg as the C-terminal amino acid from the database, greatly increasing the number of candidate sequences whose theoretical fragmentation spectrum must be matched against the experimental fragmentation spectrum. Because of these differences we implemented the use of decoy database entries during the search as described by Elias and Gygi [34]. For each protein in the database an additional entry is created with its amino acid sequence randomized, and labeled as such, in its accession identifier. During the database search these decoy proteins compete against authentic proteins for the best match to experimental spectra. Search results that identify peptides from a decoy protein are known to be incorrect. The rate of matches to peptides from the decoy proteins is used to estimate the false positive rate as described below.
Search results are summarized in Table 1 at the estimated 1% and 5% FPR. 5,915 MS/MS spectra were identified at the 1% FPR (30 matches against decoy peptides), representing 2,731 unique sequences. 65 of these peptides were unique to vaccinia virus, originating from 44 vaccinia virus proteins. At the 5% FPR, the number of matched spectra increased from 5,915 to 12,819 (313 matches against decoy peptides), 5,601 of which were unique sequences. Of these 5,601 unique sequences, 116 peptides originated from 61 vaccinia proteins.
Table 1.
Summary of peptides identified by two-dimensional liquid chromatography and tandem mass spectrometry.
| 1% FPRa | 5% FPRa | |
|---|---|---|
| # MS/MS spectra identifiedb | 5,915 | 12,819 |
| # Unique sequencesc | 2,731 | 5,601 |
| # Unique vaccinia sequencesd | 65 | 116 |
| # Vaccinia proteins representede | 44 | 61 |
Database search results summarized at the 1% false positive rate (FPR) and the 5% FPR.
Number of tandem mass spectrometry (MS/MS) search results surpassing scoring thresholds that characterize the 1% and 5% FPR. Includes results for peptides identified in multiple strong cation exchange fractions, multiple MS/MS spectra from the same precursor m/z, at multiple charge states, and from multiple database entries containing the identified sequence.
Number of unique peptide sequences identified, from all species represented in the database (human, bovine, vaccinia proteins).
Number of peptide sequences identified that are unique to vaccinia proteins.
Number of vaccinia proteins represented by the identified vaccinia peptides.
The naturally processed and presented vaccinia peptides we identified are listed in Table 2. The peptides are sorted by the open reading frame (ORF) they originated from and are marked whether they were identified within database search scoring criteria that characterized the 5% FPR (shaded background) or the 1% FPR (unshaded). Vaccinia peptide sequences identified at the 5% or better FPR have been selected for additional studies to characterize their immunogenic properties.
Table 2.
Class I peptides from vaccinia virus identified by two-dimensional liquid chromatography and tandem mass spectrometry after vaccinia infection of human B cells.
| Peptide Sequencea | ORF | Vaccinia Strainb |
Other Pox Virusesc | Putative Alleled |
BIMASe | CBS IC50 (nM)f |
SYFPEITHI Predictiong |
|---|---|---|---|---|---|---|---|
| oxMQKFTILEYh | A,C,V | Vr,Cm,Cw,Mn | B*1501 | 288.0 | 65 | 23 | |
| ILIRGIINV | A ORF T | C,V | None | A*0201 | 271.9 | 26 | 30 |
| AQITTDDLVKSY | A10L | C,V | Vr,Cw,Mn,Ra | B*1501 | |||
| VQAVTNAGKIVY | A12L | A,C,V | Vr,Cm,Cw,Mn,Ra | B*1501 | |||
| SQIFNIISY | A17L | C,V | Vr,Cm,Cw,Mn | B*1501 | 96.0 | 74 | 9 |
| SoxMADVSIKTNSV | A1L | A,C,V | Cm,Cw | A*0201 | |||
| ALDEKLFLI | A23R | A,C,T,V | Vr,Cm,Cw,Mn | NA | 228.2 | 6 | 27 |
| HMIDKLFYV | A23R | A,C,T,V | Vr,Cm,Cw,Mn | A*0201 | 513.8 | 2 | 26 |
| LLFEDIIQNEY | A23R | A,C,T,V | Vr,Cm,Cw,Mn | B*1501 | 114 | ||
| FTVNIFKEV | A24R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | A*0201 | 8.4 | 195 | 16 |
| GDKFTTRTSQKGTVAY | A24R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | |||
| ILYDPETDKPY | A24R | A,C,T,V | Vr,Cm,Cw,Fw,Mn,Ra | B*1501 | 81 | ||
| TTRTSQKGTVAY | A24R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | |||
| VIINSTSIF | A24R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 10.0 | 223 | 13 |
| LTREMGFLVY | A29L | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 17.4 | 132 | 15 |
| TVINEDIVSKLTF | A29L | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | |||
| TLRFLEKTSF | A31R | C,V | Vr,Cm,Cw,Mn | B*1501 | 72.0 | 507 | 11 |
| QIDVRDIKY | A35R | C,V | Cm,Cw | B*1501 | 1.8 | 12810 | 16 |
| SIMDFIGPYI | A35R | C,V | Cm,Cw,Mn,Ra | NA | 119.7 | 11 | 21 |
| VQIDVRDIKY | A35R | C,V | Cm,Cw | B*1501 | 52.8 | 131 | 23 |
| YIIGNIKTV | A35R | C,V | Cm,CwRa | A*0201 | 101.2 | 143 | 29 |
| IQYPGSEIKGNAY | A44L | A,V | Cm,Cw | B*1501 | |||
| IQYPGSKIKGNAY | A44L | C | Ra | B*1501 | |||
| IQYPGSKIKGNAYF | A44L | C | Ra | B*1501 | |||
| KISNTTFEV | A44L | A,C,V | Cm,Cw,Mn,Ra | A*0201 | 194.1 | 10 | 23 |
| LLISADDVQEIRV | A44L | A,C,V | Vr,Cm,Cw,Mn,Ra | A*0201 | |||
| TLYDISPGHVYA | A44L | A,C,V | Cm,Cw,Mn,Ra | NA | |||
| YPGSKIKGNAY | A44L | C | Ra | B*1501 | 8183 | ||
| VIRNEVNDTHY | A46R | C,V | Vr,Cm,Cw,Ra | B*1501 | 476 | ||
| FQQKVLQEY | A48R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 160.0 | 112 | 21 |
| IVIEAIHTV | A48R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | A*0201 | 97.6 | 53 | 27 |
| VAYAAAKGASM | A48R | A,C,T,V | Cm,Cw,Ra | NA | 14028 | ||
| YAAAKGASM | A48R | A,C,T,V | Cm,Cw,Ra | NA | 0.3 | 4895 | 17 |
| IoxMNNPDFKTTYh | A49R | C,V | Vr,Cw | B*1501 | 97 | ||
| ILQNRLVYV | A52R | C,V | Cw | A*0201 | 1495.7 | 13 | 28 |
| VQKQDIVKLTVY | A52R | C,V | Cw | B*1501 | |||
| ILSDENYLL | A6L | C,V | Vr,Cm,Cw,Mn,Ra | A*0201 | 148.9 | 10 | 24 |
| KLFNEDLSSKY | A7L | A,C,T,V | Vr,Cw,Mn | B*1501 | 183 | ||
| LIQEIVHEV | A7L | A,C,T,V | Vr,Cm,Cw,Mn | A*0201 | 153.3 | 18 | 29 |
| LVIENDSQF | A8R | A,C,V | Vr,Cm,Cw | B*1501 | 1.1 | 215 | 19 |
| KLYKSGNSHIDY | B12R | A,C,V | Cw,Ra | B*1501 | |||
| RVFAPKDTESVF | B12R | A,C,V | Cw,Ra | B*1501 | |||
| KVSAQNISF | B13R | C,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 2.4 | 117 | 7 |
| GQLYSTLLSF | B15R | C,V | Vr,Cm,Ra | B*1501 | 96.0 | 106 | 21 |
| IQFMHEQGY | B1R | A,C,V | Vr,Cm,Cw,Mn | B*1501 | 52.0 | 106 | 21 |
| LQYAPRELLQY | B1R | A,C,V | Vr,Cm,Cw,Mn | B*1501 | 78 | ||
| HCYLoxMNEGFES | B21R,C15L | C | Cw | NA | 39041 | ||
| TLLDHIRTA | B22R,C16L | C | Cw,Ra | NA | 34.7 | 84 | 23 |
| oxMLINYLoxMLLi | C11R | A,T | None | A*0201 | 83.5 | 4 | 27 |
| KIKDDFQTVNF | C12L | C,V | Vr,Cm,Cw,Mn | B*1501 | 731 | ||
| KIYGSDSIEF | C12L | C,V | Vr,CwMn | B*1501 | 14.4 | 229 | 12 |
| RQFYNANVL | C2L | C,T,V | Cm,Cw,Ra | A*0201 | 6960 | 11 | |
| YVoxMGGVYTTY | C2L | C,T,V | Cm,Cw,Ra | B*1501 | 6.3 | 68 | 23 |
| ILKINSVKY | D12L | A,C,V | Vr,Cm,Mn | B*1501 | 187.2 | 309 | 22 |
| KLFTHDIML | D12L | A,C,V | Vr,Cm,Mn | A*0201 | 276.6 | 15 | 22 |
| KLSDSKITV | D13L | A,C,T,V | Vr,Cm,Cw,Mn | A*0201 | 998.1 | 21 | 24 |
| VLSLELPEV | D13L | A,C,T,V | Vr,Cm,Cw,Mn | A*0201 | 271.9 | 14 | 28 |
| ILVPNINILKI | D6R | A,C,T,V | Vr,Cm,Cw,Mn | NA | 78 | ||
| IoxMSESYTLKEV | D6R | A,C,T,V | Vr,Cm,Cw,Fw,Mn,Ra | A*0201 | 46 | ||
| RLKPLDIHY | D8L | A,C,T,V | Cm,Cw,Ra | B*1501 | 172.8 | 135 | 23 |
| YAIDVSKVKPL | E10R | C | Vr,Cm,Cw,Mn | A*0201 | 1337 | ||
| KIDYYIPYV | E2L | C,V | Vr,Cm,Cw,Mn,Ra | A*0201 | 169.4 | 2 | 24 |
| GKASQNPSKoxMVY | E5R | C,D | Cw,Ra | B*1501 | |||
| IGKASQNPSKoxMVY | E5R | C,D | Cw,Ra | B*1501 | |||
| KLFSDISAI | E5R | C,D,V | Vr,Cm,Cw,Ra | NA | 310.7 | 8 | 25 |
| SQNPSKoxMVY | E5R | C,D | Cw,Ra | B*1501 | 52.8 | 88 | 22 |
| LARLGLVL | E6R | C,V | Vr,Cm,Cw,Mn,Ra | A*0201 | |||
| GSFSGRYVSY | E8R | C,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 1.2 | 204 | 15 |
| KQKFPYEGGKVF | E9L | A,C,V | Vr,Cm,Cw,Ra | B*1501 | |||
| LLLETKTILV | E9L | A,C,V | Vr,Cm,Cw,Mn,Ra | A*0201 | 1793.7 | 11 | 26 |
| IQHRQQLELAY | F11L | C,P | Ra | B*1501 | 83 | ||
| IQKDINITH | F11L | C,P | Vr,Cm,Cw,Ra | NA | 0.0 | 42108 | 4 |
| MLTEFLHYC | F11L | C,P | Vr,Cw,Mn,Ra | NA | 1664.5 | 105 | 17 |
| SLSNLDFRL | F11L | C,P | Cw,Mn,Ra | A*0201 | 123.9 | 30 | 23 |
| FLTSVINRV | F12L | C | Vr,Cm,Cw,Mn,Ra | A*0201 | 735.9 | 5 | 25 |
| LFoxMDEIDHESYh | F12L | C,P | Vr,Cm,Cw,Mn,Ra | B*1501 | 566 | ||
| NLFDIPLLTV | F12L | C,P | Vr,Cm,Cw,Mn,Ra | A*0201 | 2426.7 | 7 | 29 |
| VQILMKTANNY | F12L | C | Vr,Cm | B*1501 | 106 | ||
| KQISISTGVLY | F16L | C | Vr,Cm,Cw,Mn | B*1501 | 85 | ||
| RVKQISISTGVLY | F16L | C | Vr,Cm,Cw,Mn | B*1501 | |||
| ILoxMDNKGLGV | F1L | A,C,P,T,V | Cw | A*0201 | 1793.7 | 8 | 26 |
| ILoxMDNKGLGVRL | F1L | A,C,P,T,V | Cw | A*0201 | |||
| RQLPTKTRSY | F1L | A,C,P,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 96.0 | 80 | 25 |
| QLIYQRIYY | F2L | C,P,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 26.4 | 250 | 21 |
| ILKSEIEKATY | G4L | C,V | Cw | B*1501 | 374 | ||
| SLKDVLVSV | G5.5E | A,C,T,V | Vr,Mn | A*0201 | 23.0 | 18 | 30 |
| ILDDNLYKV | G5R | C | Vr,Cm,Cw,Mn,Ra | A*0201 | 446.0 | 4 | 30 |
| VPLPCQLoxMY | G5R | C | Vr,Cm,Cw,Mn,Ra | B*1501 | 2.8 | 18814 | 12 |
| ILIEIIPKI | H4L | A,C,V | Vr,Cm,Cw,Mn,Ra | NA | 167.2 | 4 | 31 |
| ITNKADTSSF | H5R | C,V | Vr | B*1501 | 2.6 | 276 | 10 |
| IIKEDISEY | H7R | A,C,T,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 42.9 | 171 | 19 |
| NTIDKSSPL | I1L | A,C,T,V | Mn | A*0201 | 1.2 | 3795 | 18 |
| TQFNFNGHTY | I1L | A,C,P,T,V | Cm,Cw,Mn | B*1501 | 88.0 | 70 | 22 |
| YSKKFQESF | I1L | A,C,P,T,V | Cm,Cw,Mn | B*1501 | 6.0 | 185 | 11 |
| KLLLGELFFL | J3R | A,C,V | Vr,Cm,Cw,Mn | A*0201 | 20297.3 | 7 | 27 |
| LQKGHNKFPVNF | J4R | C,V | Vr,Cm,Cw,Mn | B*1501 | |||
| VVIGNTLIKY | J6R | A,C,V | Vr,Cw,Mn,Ra | B*1501 | 2.9 | 242 | 22 |
| YoxMIERFISFh | J6R | A,C,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 2.6 | 49 | 11 |
| KoxMIIEKHVEY | K1L | C,V | None | B*1501 | 2.4 | 103 | 16 |
| oxMIIEKHVEY | K1L | C,V | None | B*1501 | 13.2 | 154 | 18 |
| SLLFIPDIKL | K1L | C,V | Vr,Cw,Mn,Ra | A*0201 | 79.0 | 61 | 25 |
| SQFDDKGNTALY | K1L | C,V | Cm,CwMn,Ra | B*1501 | |||
| VLLDDAEIAKoxM | K1L | V | Cm,Cw,Mn,Ra | NA | 55 | ||
| VLLDDAEIAKoxMIIh | K1L | V | Cm,Cw,Mn,Ra | NA | |||
| KLVGKTVKV | K3L | C,V | Cm,Cw,Mn | A*0201 | 243.3 | 54 | 30 |
| YLFDYPHFEA | K3L | V | None | NA | 2010.7 | 8 | 20 |
| ITYPKALVF | K6L | C,V | Cw,Mn | B*1501 | 4.1 | 168 | 11 |
| MMIDDFGTARGNY | K6L | C,V | None | B*1501 | |||
| LoxMKFDDVAIRY | K7R | A,C,V | Cw | B*1501 | 147 | ||
| RLYKELoxMKFh | K7R | A,C,V | Cm,Cw,Ra | B*1501 | 40.0 | 763 | 20 |
| IINDKGKQY | M1L | C,V | Vr,Cw,Mn,Ra | B*1501 | 14.3 | 423 | 18 |
| QAIEPSGNNY | M1L | C,V | Cw,Ra | B*1501 | 2.2 | 146 | 12 |
| ILFRMIETY | N1L | C,V | Vr,Cw | B*1501 | 57.2 | 208 | 24 |
| HIIKEFMTY | N2L | C,V | Vr,Cm,Cw,Mn,Ra | B*1501 | 11.0 | 797 | 18 |
| SIIAILDRF | N2L | V | Vr,Cm,Cw,Ra | B*1501 | 20.0 | 3775 | 16 |
| GLLDRLYDL | O1L | C,V | Cm,Cw,Ra | A*0201 | 745.4 | 10 | 29 |
Peptides without shading were identified at a 1% false positive identification rate (1% FPR); shaded peptides were identified within scoring criteria characteristic of a 5 % FPR. Peptide sequences in boldface are predicted to be strong binders (IC50 < 50 nanoM) by the epitope prediction algorithm at: http://www.cbs.dtu.dk/services/NetMHC/ (accessed Jan.4, 2008). oxM denotes that the peptide contains an oxidized methionine (not considered in calculation of IC50 values).
Epitope is common to the following vaccinia strains: A=Ankara, C=Copenhagen, D=Dairen I, P=L-IVP, T=Tian Tan, V= Western Reserve as listed in the SwissProt database.
Epitope also found in the following common poxviruses: Vr=variola, Cm=camelpox, Cw=cowpox, Fw=fowlpox, Mn=monkeypox, Ra=rabbitpox, None=not found in other poxviruses as reported in the NCBI nr database (http://www.ncbi.nlm.nih.gov/ ).
Putative association of peptide with A*0201 or B*1501 alleles based upon the C-terminal amino acid: amino acids V and L classified as A2; amino acids F and Y classified as B15; NA = not assigned.
Score is proportional to predicted affinity. Bioinformatics and Molecular Analysis Section, National Institute of Health, http://www-bimas.cit.nih.gov/molbio/hla_bind/
Center for Biological Sequence Analysis, Technical University of Denmark, http://www.cbs.dtu.dk/services/NetMHC/
Score is proportional to predicted affinity. Department of Immunology, University of Tübingen, http://www.syfpeithi.de/
Peptide was identified with oxidized and non-oxidized forms of methionine
Peptide was identified in four oxidative forms: without methionine oxidation, oxidized at the N-terminal methionine, oxidized at the position-7 methionine, and oxidized at both methionines.
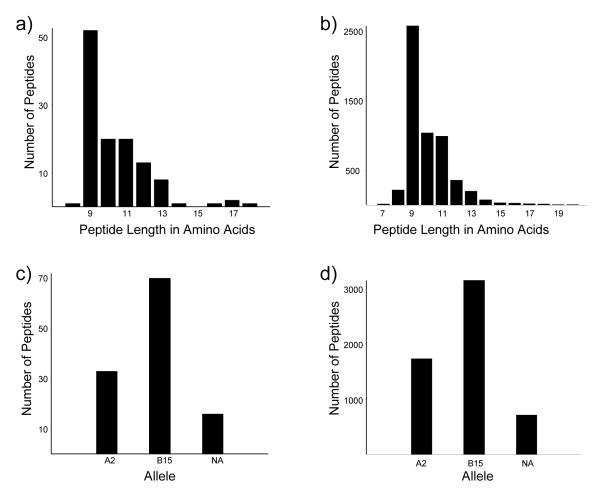
Figure 2 shows the distribution of peptide lengths and putative allele from the vaccinia subset (Figure 2a) as well as all class I peptides identified at the 5% FPR rate (Figure 2b). As expected, the majority of the peptides are 9-mers in both the full set of peptides as well as the vaccinia subset. However, there are a significant number of peptides longer than 11 amino acids in the full set of identified peptides as well as in the vaccinia subset.
Figure 2.
Distribution of HLA peptide amino acid length for a) vaccinia peptides and b) all identified peptides; putative sorting of peptides by binding allele for c) vaccinia peptides, and d) all identified peptides. Peptides classified by allele using their C-terminal amino acid: L or V assigned to A*0201, F or Y assigned to B*1501, and all other peptides marked as NA (not assigned).
Since the W6/32 antibody used to isolate the HLA-peptide complexes has affinity for each of the major HLA class I alleles, the list of identified peptides reflects the allotypes of the host cell line; in this case HLA-A*0201, B*1501, and C*03. As a first approximation, we associated identified peptides with an allele by comparing the C-terminal amino acid to the reported binding motifs at the P9 position [36]. We assigned peptides terminating in L or V to A*0201, and peptides terminating in F or Y to B*1501, while designating the remaining peptides as “not assigned”. Figure 2c and 2d shows an estimate of the distribution of peptides among the A and B alleles, and while our method of associating a peptide with its putative allele is rudimentary, it clearly shows a significant subset of the peptides being presented by the B allele. There is also an unknown contribution of peptides from the HLA-C allele, although the density of HLA-C molecules on the surface of cells has been reported as being 6-fold less than that of the A and B alleles [37, 38].
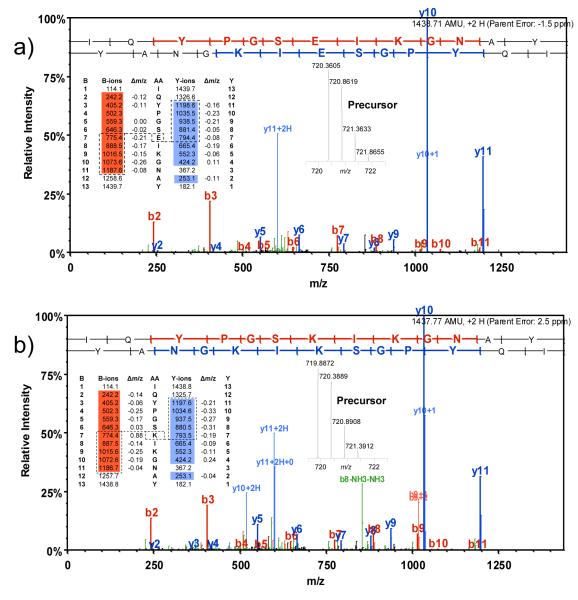
The vaccinia peptides identified also bear evidence of the genetic heterogeneity of the Dryvax strain [39]. For example, two forms of the same peptide from ORF A44L were identified: IQYPGSKIKGNAY from the Copenhagen strain, and IQYPGSEIKGNAY from the Western Reserve and Ankara strains of vaccinia. At first glance these two sequences were flagged as redundant identities of the same peptide, since they vary only in one amino acid residue resulting in a mass difference of one dalton. However, these sequences were assigned from two distinctly different peptides with IQYPGSEIKGNAY identified from SCX fractions 4 and 5, and IQYPGSKIKGNAY identified from SCX fractions 14-16. Figure 3a and 3b shows the annotated MS/MS spectra for the two peptides. Orbitrap spectra for each precursor mass (inset) illustrate the difference in mass for the two peptides (0.5 on the m/z axis, where z=2 as shown by the isotope spacing). Fragment ions are observed from the N-terminal end of the peptide (B-ions, highlighted in red) and from the C-terminal end of the peptide (Y-ions, highlighted in blue), while the y- and b-ions highlighted within the dashed box delineate the sequence difference between the two peptides. Table 2 contains other instances where peptides were identified that were unique to specific vaccinia strains.
Figure 3.
Figure 3a shows the MS/MS spectrum of the vaccinia peptide IQYPGSEIKGNAY found in SCX fractions 4 and 5, as annotated by Scaffold, including mass accuracy of the precursor mass in parts-per-million (ppm). A table of the fragment ions matched and the experimental error of the fragment ions is included. The Orbitrap survey spectrum of the precursor ion is shown. Figure 3b shows the same information for the vaccinia peptide IQYPGSKIKGNAY as identified from SCX fractions 14-16. Note the different precursor mass, as well as concomitant changes to fragment ion masses consistent with the change in amino acid E (Glu) to K (Lys).
Additionally, annotated MS/MS spectra of all vaccinia peptide sequences that were identified at the 5% FPR criteria, but did not meet the criteria for a 1% FPR, are included in the supplemental information. An Excel spreadsheet is also included as supplemental information that contains the entire search dataset used for calculating false positive identification rates.
3.2. Comparison of directly identified vaccinia class I peptides to predictive algorithms
The vaccinia protein sequences represented by this set of peptides were submitted to three algorithms for predicting potential epitopes according to their predicted alleles. Results from the NetMHC, [40, 41] BIMAS, [42] and SYFPEITHI [43] algorithms are shown in Table 2. The NetMHC algorithm uses a neural network approach to determine likely epitopes from protein sequence, calculates a predicted binding affinity to HLA molecules, and classifies this binding as being strong (IC50 < 50 nM), weak (IC50 > 50 nM and < 500 nM), or less than weak (IC50 > 500 nM), (http://www.cbs.dtu.dk/services/NetMHC/, accessed Jan. 4, 2008). Figure 4 summarizes how the peptides we directly identified by MS are predicted by the NetMHC algorithm to bind with HLA molecules. Twenty seven (22%) of the vaccinia peptides identified by MS were predicted by the algorithm to be strong binders with another 49 (41%) predicted to be weak binding. Twenty four peptides identified by MS/MS were not predicted by the algorithm, 23 because of excess length.
Figure 4.
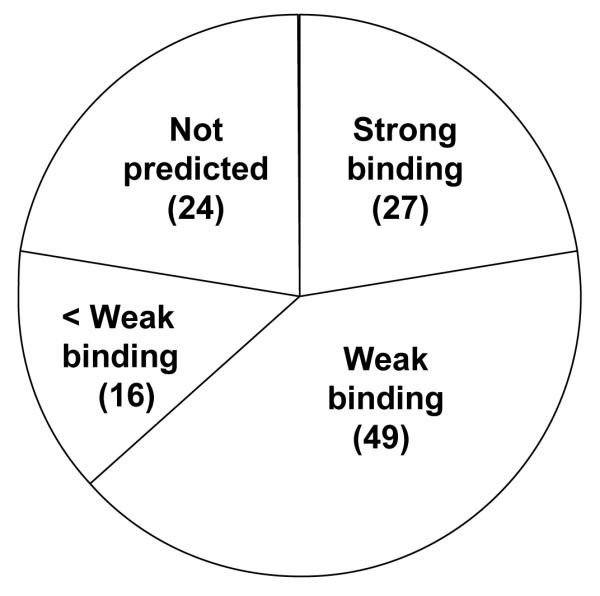
Vaccinia epitopes directly identified by MS/MS are classified by predicted HLA-binding strength as determined by the netMHC algorithm at the Center for Biological Sequence Analysis, Technical University of Denmark, (http://www.cbs.dtu.dk/services/NetMHC/. Peptide sequences with calculated IC50 values < 50 nM were classified as strong binding, IC50 values between 50 nM and 500 nM were classified as weak binding, and IC50 > 500 nM were classified as non-binding peptides.
Additionally, we compared our list of identified vaccinia peptides to vaccinia epitopes, from all alleles, contained in the Immune Epitope Database and Analysis Resource (IEDB, http://www.immuneepitope.org/home.do, accessed Jan. 4, 2008) [44]. From our list of 116 directly identified peptides, 23 were an exact match to an IEDB database record, while another 7 peptides were contained within a vaccinia epitope from the IEDB database. These results demonstrate both the complementary information provided by direct identification of epitopes by MS, and the limitations inherent in relying solely on computer algorithms for understanding the spectrum of peptides presented by natural infection.
4. Discussion
While very effective, the Dryvax vaccine has the highest rate of serious adverse events of any Food and Drug Administration (FDA)-approved vaccine [45-47]. Because of the risks associated with administration of the current live viral smallpox vaccine, and the large percentage of the population for whom this live virus vaccine is contraindicated, public health interests would best be served by the development of an efficacious recombinant or peptide-based vaccine. One approach for developing a safer smallpox vaccine is to utilize naturally processed immunogenic vaccinia-derived peptides rather than live whole vaccinia virus.
Recombinant vaccinia viruses have been created from several strains of vaccinia virus. In the U.S. most recombinant viruses have been made from either the NYCBOH strain or a mouse neuroadapted derivative, the Western Reserve (WR) vaccinia strain. Several recombinants have been made from the Copenhagen and Lister vaccinia strains, which are more pathogenic in animals than the NYCBOH strain [48]. In general, genes located in the center of the virus genome are conserved between orthopoxviruses and are important for virus replication [49].
In this study we identified a set of naturally processed and HLA (A*0201, B*1501, C*03)-presented peptides, derived from the Dryvax virus strain (NYCBOH), which are encoded by 61 vaccinia virus genes. Since there are approximately 230 ORFs in vaccinia, multiple viral proteins can be potentially targeted by human cellular (CTL) immune responses. In fact, some of these gene products have been demonstrated to be immunogenic in both humans and a mouse model, including A6L, A8R, A10L, A23R, D12L, E9L, J6R, and M1L [4, 6, 12, 14, 50].
Seven of the peptides we have identified after infection with vaccinia virus have previously been reported to have immunogenic properties, including ILDDNLYKV (ORF G5R) which has been classified as an immunodominant epitope [14, 51]. The other peptides are: KLFTHDIML (D12L), ILSDENYLL (A6L), KIDYYIPYV (E2L), FLTSVINRV (F12L), NLFDIPLLTV (F12L), and GLLDRLYDL (O1L). All seven of these peptides were predicted to be strong binding peptides by the NetMHC algorithm. We also identified the naturally processed peptide SQIFNIISY (ORF A17L) that is homologous to QIFNIISYI (A17L). QIFNIISYI was reported by Assersson at al., to be one of 15 immunodominant A*0201-restricted epitopes [14]. SQIFNIISY is predicted by NetMHC to have an IC50 value of 74nM for the B*1501 allele, making some form of this sequence a potentially useful epitope across multiple alleles. As shown in Table 2, 30 of the 61 vaccinia proteins were represented by a single class I peptide, while the remainder of the vaccinia proteins were represented by two to seven peptides. These data provide the first direct evidence of multiple naturally processed vaccinia peptides, resulting from natural viral infection, in the context of A2 and B62 HLA supertypes.
A goal of future work will be to study cellular (CTL) class I-restricted immune responses in vitro to identified peptides in unvaccinated and previously smallpox vaccinated subjects for evidence of peptide recognition by T cells. Ostrout et al. [4] recently described T cell memory IFN-γ responses to theoretical HLA-A2 epitopes in humans vaccinated against smallpox. This study revealed six peptides encoded by 14L, G1L, A8R, 18R, D12L, and H3L ORFs that were identical for vaccinia (Copenhagen), variola major (Bangledesh 1975) and modified vaccinia Ankara (MVA) strains [4]. In addition, Oseroff et al. [12] identified a total of 48 computational epitopes derived from 35 vaccinia proteins, some of which (B8R, D1R, D5R, C10L, C19L, C7L, F12, and O1L) were recognized in an ex vivo Elispot assay in multiple donors of different HLA types. Immunogenic peptide identification for poxviruses is just beginning, and it is likely that as more peptides are identified, a growing number of new vaccinia antigens that contain multiple epitopes will be identified [20]. In this regard, MS techniques, such as those described here, will further facilitate the search for unknown naturally processed peptides from poxvirus proteins. Sequence analysis of poxvirus genomes has improved our knowledge of the structure and function of poxviral genes and increased our understanding of host-virus interactions [49].
Several lines of evidence indicate that in humans a class I-mediated T cell response is sufficient to provide protection against a poxviral challenge [52-54], justifying initial research that focused on class I-related activation. However, recent studies indicate an important role for humoral immunity (neutralizing antibodies) as well as CD4+ T cells in protection against poxvirus infection [55, 56]. Recently two HLA-DR1-restricted computer-predicted epitopes on vaccinia virus A24R and D1R enzyme proteins were described, that are conserved among poxviruses [5]. This study identified five new peptides from the A24R enzyme protein, which is involved in mRNA synthesis. Although the involvement of CD8+ T lymphocytes in the immune response to smallpox virus has been demonstrated, naturally processed vaccinia virus-derived peptides presented in the context of several HLA class I molecules, have not yet been identified. Our results show that approximately 60% of the peptides identified in this study are conserved from vaccinia to variola. If found to be immunodominant, these peptides may allow for monitoring of cellular immune responses induced by licensed smallpox immunization and the development of new poxvirus multiepitope-based vaccines. Future studies are warranted to investigate the value of these naturally processed and HLA-presented class I vaccinia-derived peptides for immunostimulatory responses, T cell recognition, and HLA restriction. Finally, our study demonstrates that computer-based algorithms are insufficiently robust to predict the full repertoire of naturally presented pathogen-derived peptides, in this case failing to predict 20% of the peptides identified by MS.
The technical challenges of comprehensively identifying the HLA ligandome [57] by mass spectrometric methods are the same challenges being faced in proteomics in terms of number of components, their range of relative abundance, and how the abundance of components change as a function of disease. Proteomics continues to drive significant improvements in sensitivity and throughput of MS-based methods that are immediately applicable to immunology. Recent reviews have speculated on what future role enhanced mass spectrometric capabilities will play in immunology [58, 59]. We believe that as larger data sets become available, predictive algorithms will be used to rank the potential immunological significance of directly identified HLA ligands for further study, as well as provide additional insight into ligands that are exceptions to known binding motifs [19, 58, 59]. As stated by Kessler and Melief, “It is expected that the rapidly increasing power of mass spectrometric techniques will have a tremendous impact on the unraveling of the cancer-specific HLA-bound ‘ligandome’. The identification of HLA class I ligands by reverse immunology-based predictions may eventually be bypassed by direct identification of cell surface-presented peptides with mass spectrometry”[59].
Supplementary Material
Acknowledgements
The authors thank Cheryl A. Hart and Diana L. Ayerhart for their help in preparing this manuscript. Funding for this work was provided by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program (NIH award 1-U54-AI-057153), NIH/NIAID N01 AI40065, and Mayo Clinic.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented in part: Eleventh Annual Conference on Vaccine Research, Baltimore, MD, 5-7 May 2008 (abstract S10).
References
- [1].Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: The role of immunogenetics and the emerging field of vaccinomics. Clin.Pharmacol.Ther. 2007;82(6):653–64. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- [2].Mitaksov V, Fremont DH. Structural definition of the H-2Kd peptide-binding motif. J Biol.Chem. 2006;281(15):10618–25. doi: 10.1074/jbc.M510511200. [DOI] [PubMed] [Google Scholar]
- [3].Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–12. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- [4].Ostrout ND, McHugh MM, Tisch DJ, Moormann AM, Brusic V, Kazura JW. Long-term T cell memory to human leukocyte antigen-A2 supertype epitopes in humans vaccinated against smallpox. Clin Exp.Immunol. 2007;149(2):265–73. doi: 10.1111/j.1365-2249.2007.03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mitra-Kaushik S, Cruz J, Stern LJ, Ennis FA, Terajima M. Human cytotoxic CD4+ T cells recognize HLA-DR1-restricted epitopes on vaccinia virus proteins A24R and D1R conserved among poxviruses. J Immunol. 2007;179(2):1303–12. doi: 10.4049/jimmunol.179.2.1303. [DOI] [PubMed] [Google Scholar]
- [6].Pasquetto V, Bui HH, Giannino R, Banh C, Mirza F, Sidney J, et al. HLA-A*0201, HLA-A*1101, and HLA-B*0702 transgenic mice recognize numerous poxvirus determinants from a wide variety of viral gene products. J Immunol. 2005;175(8):5504–15. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- [7].Stanley SL, Jr., Frey SE, Taillon-Miller P, Guo J, Miller RD, Koboldt DC, et al. The immunogenetics of smallpox vaccination. J Infect Dis. 2007;196(2):212–9. doi: 10.1086/518794. [DOI] [PubMed] [Google Scholar]
- [8].Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum.Immunol. 2001;62(11):1200–16. doi: 10.1016/s0198-8859(01)00319-6. [DOI] [PubMed] [Google Scholar]
- [9].Sidney J, Southwood S, del Guercio MF, Grey HM, Chesnut RW, Kubo RT, et al. Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J Immunol. 1996;157(8):3480–90. [PubMed] [Google Scholar]
- [10].Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50(34):201–12. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- [11].Drexler I, Staib C, Kastenmüller W, Stevanović S, Schmidt B, Lemonnier FA, et al. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc.Natl.Acad.Sci.USA. 2003;100:217–22. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl.Acad.Sci U.S.A. 2005;102(39):13980–5. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat.Biotechnol. 2006;24(7):817–9. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- [14].Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178(12):7890–901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- [15].Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–3. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- [16].van der Heeft E, ten Hove G Jan, Herberts CA, Meiring HD. A microcapillary column switching HPLC-electrospray ionization MS system for the direct identification of peptides presented by major histocompatibility complex class I molecules. Anal.Chem. 1998;70:3742–51. doi: 10.1021/ac9801014. [DOI] [PubMed] [Google Scholar]
- [17].Poland GA, Ovsyannikova IG, Johnson KL, Naylor S. The role of mass spectrometry in vaccine development. Vaccine. 2001;19:2692–700. doi: 10.1016/s0264-410x(00)00505-3. [DOI] [PubMed] [Google Scholar]
- [18].Meiring HD, Kuipers B, van Gaans-van den Brink JA, Poelen MC, Timmermans H, Baart G, et al. Mass tag-assisted identification of naturally processed HLA class II-presented meningococcal peptides recognized by CD4+ lymphocytes. J.Immunol. 2005;174:5636–43. doi: 10.4049/jimmunol.174.9.5636. [DOI] [PubMed] [Google Scholar]
- [19].Engelhard VH. The contributions of mass spectrometry to understanding of immune recognition by T lymphocytes. International Journal of Mass Spectrometry. 2007;259(13):32–9. doi: 10.1016/j.ijms.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ovsyannikova IG, Johnson KL, Bergen HR, III, Poland GA. Mass spectrometry and peptide-based vaccine development. Clin.Pharmacol.Ther. 2007;82(6):644–52. doi: 10.1038/sj.clpt.6100389. [DOI] [PubMed] [Google Scholar]
- [21].Schirle M, Keilholz W, Weber B, Gouttefangeas C, Dumrese T, Becker HD, et al. Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur.J.Immunol. 2000;30:2216–25. doi: 10.1002/1521-4141(2000)30:8<2216::AID-IMMU2216>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- [22].Pascolo S, Schirle M, Guckel B, Dumrese T, Stumm S, Kayser S, et al. A MAGE-A1 HLA-A A*0201 epitope identified by mass spectrometry. Cancer Res. 2001;61(10):4072–7. [PubMed] [Google Scholar]
- [23].Carralot JP, Lemmel C, Stevanovic S, Pascolo S. Mass spectrometric identification of an HLA-A*0201 epitope from Plasmodium falciparum MSP-1. Int Immunol. 2008;20(11):1451–6. doi: 10.1093/intimm/dxn102. [DOI] [PubMed] [Google Scholar]
- [24].Ovsyannikova IG, Johnson KL, Muddiman DC, Vierkant RA, Poland GA. Identification and characterization of novel, naturally processed measles virus class II HLA-DRB1 peptides. J.Virol. 2004;78(1):42–51. doi: 10.1128/JVI.78.1.42-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Johnson KL, Ovsyannikova IG, Poland G, Muddiman DC. Identification of class II HLA-DRB1*03-bound measles virus peptides by 2D-liquid chromatography tandem mass spectrometry. Journal of Proteome Research. 2005;4:2243–9. doi: 10.1021/pr0501416. [DOI] [PubMed] [Google Scholar]
- [26].Tsai SL, Chen MH, Yeh CT, Chu CM, Lin AN, Chiou FH, et al. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD8+ cytotoxic T lymphocytes. J.Clin.Invest. 1996;97:577–84. doi: 10.1172/JCI118450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meiring HD, Soethout EC, Poelen MC, Mooibroek D, Hoogerbrugge R, Timmermans H, et al. Stable isotope tagging of epitopes: a highly selective strategy for the identification of major histocompatibility complex class I-associated peptides induced upon viral infection. Mol.Cell Proteomics. 2006;5(5):902–13. doi: 10.1074/mcp.T500014-MCP200. [DOI] [PubMed] [Google Scholar]
- [28].Slingluff CL, Cox AL, Henderson RA, Hunt DF, Engelhard VH. Recognition of human melanoma cells by HLA-A2.1-restricted cytotoxic T lymphocytes is mediated by at least six shared peptide epitopes. J.Immunol. 1993;150:2955–63. [PubMed] [Google Scholar]
- [29].Johnson KL, Ovsyannikova IG, Madden BJ, Poland GA, Muddiman DC. Accurate mass precursor ion data and tandem mass spectrometry identify a class I Human Leukocyte Antigen A*0201-presented peptide originating from vaccinia virus. Journal of American Society for Mass Spectrometry. 2005;16(11):1812–7. doi: 10.1016/j.jasms.2005.07.015. [DOI] [PubMed] [Google Scholar]
- [30].Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123(1):342–9. [PubMed] [Google Scholar]
- [31].Hogan KT, Sutton JN, Chu KU, Busby JA, Shabanowitz J, Hunt DF, et al. Use of selected reaction monitoring mass spectrometry for the detection of specific MHC class I peptide antigens on A3 supertype family members. Cancer Immunol.Immunother. 2005;54(4):359–71. doi: 10.1007/s00262-004-0592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol.Cell Proteomics. 2005;4(12):2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- [33].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat.Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [34].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal.Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [35].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal.Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [36].Rammensee H-G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- [37].Snary D, Barnstable CJ, Bodmer WF, Crumpton MJ. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur.J Immunol. 1977;7(8):580–5. doi: 10.1002/eji.1830070816. [DOI] [PubMed] [Google Scholar]
- [38].Neisig A, Melief CJ, Neefjes J. Reduced cell surface expression of HLA-C molecules correlates with restricted peptide binding and stable TAP interaction. J Immunol. 1998;160(1):171–9. [PubMed] [Google Scholar]
- [39].Osborne JD, Da Silva M, Frace AM, Sammons SA, Olsen-Rasmussen M, Upton C, et al. Genomic differences of Vaccinia virus clones from Dryvax smallpox vaccine: the Dryvax-like ACAM2000 and the mouse neurovirulent Clone-3. Vaccine. 2007;25(52):8807–32. doi: 10.1016/j.vaccine.2007.10.040. [DOI] [PubMed] [Google Scholar]
- [40].Buus S, Lauemøller SL, Worning P, Kesmir C, Frimurer T, Corbet S, et al. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens. 2003;62(5):378–84. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- [41].Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–17. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J.Immunol. 1994;152:163–75. [PubMed] [Google Scholar]
- [43].Rammensee H-G, Bachmann J, Emmerich NPN, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- [44].Peters B, Sette A. Integrating epitope data into the emerging web of biomedical knowledge resources. Nat.Rev.Immunol. 2007;7(6):485–90. doi: 10.1038/nri2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Goldstein JA, Neff JM, Lane JM, Koplan JP. Smallpox vaccination reactions, prophylaxis, and therapy of complications. Pediatrics. 1975;55:342–7. [PubMed] [Google Scholar]
- [46].Halsell JS, Riddle JR, Atwood JE, Gardner P, Shope R, Poland GA, et al. Myopericarditis following smallpox vaccination among vaccinia-naive US military personnel. JAMA. 2003;289:3283–9. doi: 10.1001/jama.289.24.3283. [DOI] [PubMed] [Google Scholar]
- [47].Poland GA, Neff JM. Smallpox vaccines: problems and prospects. In: Poland GA, editor. Immunology and Allergy Clinics of North America: Vaccines in the 21st Century. WB Saunders Company; Philadelphia: 2003. pp. 731–43. [DOI] [PubMed] [Google Scholar]
- [48].Vaccinia (smallpox) vaccine. Recommendations of the Immunization Practices Advisory Committee (ACIP) MMWR Recomm.Rep. 1991;40(RR14):1–10. [PubMed] [Google Scholar]
- [49].Antoine G, Scheiflinger F, Dorner F, Falkner FG. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244(2):365–96. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- [50].Kennedy R, Poland GA. T-Cell epitope discovery for variola and vaccinia viruses. Rev.Med.Virol. 2006;17(2):93–113. doi: 10.1002/rmv.527. [DOI] [PubMed] [Google Scholar]
- [51].Smith CL, Mirza F, Pasquetto V, Tscharke DC, Palmowski MJ, Dunbar PR, et al. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J Immunol. 2005;175(12):8431–7. doi: 10.4049/jimmunol.175.12.8431. [DOI] [PubMed] [Google Scholar]
- [52].Erickson AL, Walker CM. Class I major histocompatibility complex-restricted cytotoxic T cell responses to vaccinia virus in humans. J.Gen.Virol. 1993;74:751–4. doi: 10.1099/0022-1317-74-4-751. [DOI] [PubMed] [Google Scholar]
- [53].McClain DJ, Harrison S, Yeager CL, Cruz J, Ennis FA, Gibbs P, et al. Immunologic responses to vaccinia vaccines administered by different parenteral routes. J.Infect.Dis. 1997;175:756–63. doi: 10.1086/513968. [DOI] [PubMed] [Google Scholar]
- [54].Demkowicz WE, Jr., Littaua RA, Wang J, Ennis FA. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J.Virol. 1996;70:2627–31. doi: 10.1128/jvi.70.4.2627-2631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Belshe RB, Newman FK, Frey SE, Couch RB, Treanor JJ, Tacket CO, et al. Dose-dependent neutralizing-antibody responses to vaccinia. J.Infect.Dis. 2004;189:493–7. doi: 10.1086/380906. [DOI] [PubMed] [Google Scholar]
- [56].Belyakov IM, Earl P, Dzutsev A, Kuznetsov VA, Lemon M, Wyatt LS, et al. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc.Natl.Acad.Sci.U.S.A. 2003;100(16):9458–63. doi: 10.1073/pnas.1233578100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rammensee HG, Weinschenk T, Gouttefangeas C, Stevanovic S. Towards patient-specific tumor antigen selection for vaccination. Immunol.Rev. 2002;188:164–76. doi: 10.1034/j.1600-065x.2002.18815.x. [DOI] [PubMed] [Google Scholar]
- [58].Hillen N, Stevanovic S. Contribution of mass spectrometry-based proteomics to immunology. Expert Rev.Proteomics. 2006;3(6):653–64. doi: 10.1586/14789450.3.6.653. [DOI] [PubMed] [Google Scholar]
- [59].Kessler JH, Melief CJ. Identification of T-cell epitopes for cancer immunotherapy. Leukemia. 2007;21(9):1859–74. doi: 10.1038/sj.leu.2404787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.