Abstract
microRNAs (miRNAs) are small, non-coding RNAs that modulate diverse biological functions through the repression of target genes. miRNA profiling studies have indicated that the levels of miRNAs are altered during normal development and pathogenesis of various diseases, including cancer and cardiovascular disorders. The signaling pathways which control miRNA biogenesis and the mechanisms of regulation, however, are not well understood. Following transcription, mature miRNAs are generated through a series of coordinated processing events mediated by large protein complexes. We recently found that signal transducers of the Transforming Growth Factor β (TGFβ) signaling pathway, the Smads, play a regulatory role in the processing of miRNA in the nucleus. In this review, we summarize the current understanding of the regulation of miRNA biogenesis mediated by the TGFβ signaling pathway.
Introduction
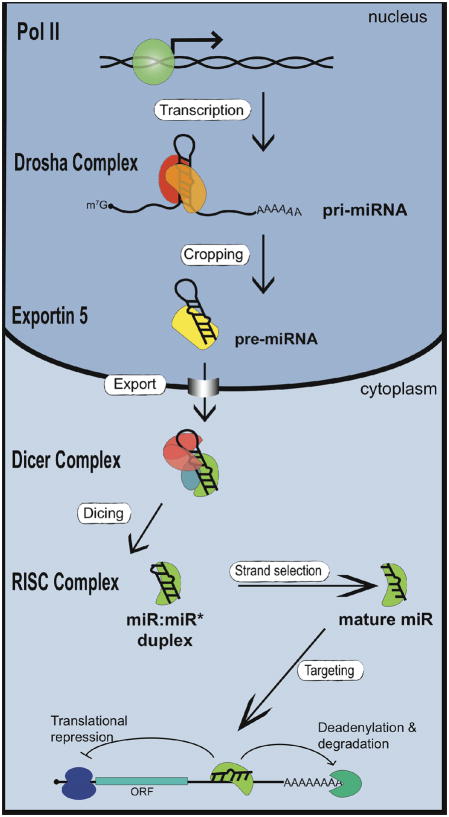
miRNAs have been reported to control diverse aspects of biology, including developmental timing, differentiation, proliferation, cell death, and metabolism. At least 30% of human genes are thought to be regulated by miRNAs. Approximately 30–50% of miRNAs are encoded within the introns of protein coding genes while the remaining miRNAs are located in intergenic sites (1). The majority of miRNAs are transcribed by RNA polymerase (RNA pol) II and bear the 5′ 7-methyl guanylate cap and 3′ poly (A) tail, characteristic of mRNAs (2, 3) (Fig. 1). The evolutionarily-conserved mechanism which gives rise to mature miRNA involves two ordered endonucleolytic cleavages by the RNase III enzymes Drosha and Dicer (Fig. 1). Following transcription by RNA polymerase II (RNA pol II), Drosha processes the primary miRNA transcript (pri-miRNA) into a ~65–80nt nucleotides (nt) hairpin structure termed the precursor-miRNA (pre-miRNA). Through the interaction with exportin-5 and Ran-GTP, the pre-miRNA is transported into the cytoplasm, where it undergoes a second round of processing catalyzed by Dicer (Fig. 1). This cleavage event gives rise to a double-stranded ~22 nt product comprised of the mature miRNA guide strand and the miRNA* passenger strand. The miRNA guide strand is then loaded into the RNA Induced Silencing Complex (RISC) while the passenger strand is degraded (Fig. 1). The RISC complex loaded with miRNA associates with target mRNAs which then leads, in majority of cases, to negative regulation of protein synthesis or mRNA degradation. The association of miRNAs with target mRNAs requires the presence of binding sites which are partially complimentary to the miRNA sequences. In particular, the 5′ region of the miRNA (nt 2–7), termed the ‘seed sequence’ seems particularly important for mRNA repression by miRNA. Functional miRNA binding sequences are often located in the 3′-untranslated region (UTR) of the target mRNA, but can also occur within the 5′UTR (4) or coding region (5). Because miRNAs exert variety of physiological functions primarily through the repression of target genes, the determination of miRNA targets has been an area of intense research. Computational and experimental approaches indicate that a single miRNA may target several dozen or even hundreds of mRNA (6). Although major progress has been made in understanding the fundamental mechanism of miRNA biogenesis, little is known about the mechanisms that regulate miRNA biogenesis.
Fig. 1. Biogenesis of miRNAs and assembly into RISC complex.
Transcription by RNA pol II leads to capped and polyadenylated pri-miRNAs which are processed by Drosha in the nucleus to generate pre-miRNAs. After translocation to the cytoplasm by exportin 5, pre-miRNAs are processed by Dicer to form the mature miRNA/miRNA* duplex. Following processing, miRNAs are assembled into the RISC complex. Only one strand of the duplex is stably associated with the RISC complex. The mature miRNA directs repression of mRNA containing partially complementary miRNA binding sites within the 3′UTR.
Spatiotemporal regulation of miRNA expression
lin-4 and let-7; the founding members of the miRNAs in C. elegans were identified and originally named as small temporal RNAs (stRNA) because of their specific pattern of expression during development (7, 8). The temporal regulation of let-7 miRNA was conserved during evolution, suggesting that miRNAs could be expressed in defined spatiotemporal patterns (9). miRNA profiling in mouse and human adult organs indicated about one half of miRNAs are expressed in a tissue-specific manner (10, 11). For example, cloning of small RNAs from different mouse tissues identified miR-1, miR-122a, and miR-124 as highly enriched in the heart, liver, and brain, respectively(12). In situ hybridization of zebrafish embryos identified a large percentage of tissue-specifically expressed miRNAs which are also temporally regulated during development (13). Furthermore, increased expression of tissue-specific miRNAs represses progenitor cell-specific mRNA which increases the fidelity of cell fate transition during differentiation of progenitor cells (14, 15).
Given the importance of miRNAs in development, it is not surprising that deregulation of miRNA expression is observed in a variety of human diseases, including cancer, neurological, and cardiovascular disorders. A majority of miRNAs are located at sites of genomic instability, including duplications and fragile sites (16). Global miRNA expression is frequently reduced in tumor samples relative to normal tissues, suggesting a role for miRNA in maintaining the differentiated state. Indeed, let-7 expression is often dramatically reduced in lung cancer and exogenous expression of let-7 can dramatically inhibit tumor growth in vivo (17). Conversely, a subset of miRNAs, including miR-21 and miR-155, have been identified as highly expressed in a variety of tumors and may serve to promote tumor growth through the inhibition of pro-apoptotic pathways. miRNA expression profiling serves as a better predictor of tumor origin and prognosis than conventional gene arrays, further emphasizing the importance of miRNAs in oncogenesis (18, 19). Understanding the mechanisms that control both normal and deregulated miRNA expression may lead to new avenues for the treatment of a variety of disorders.
Control of miRNA processing by Smad
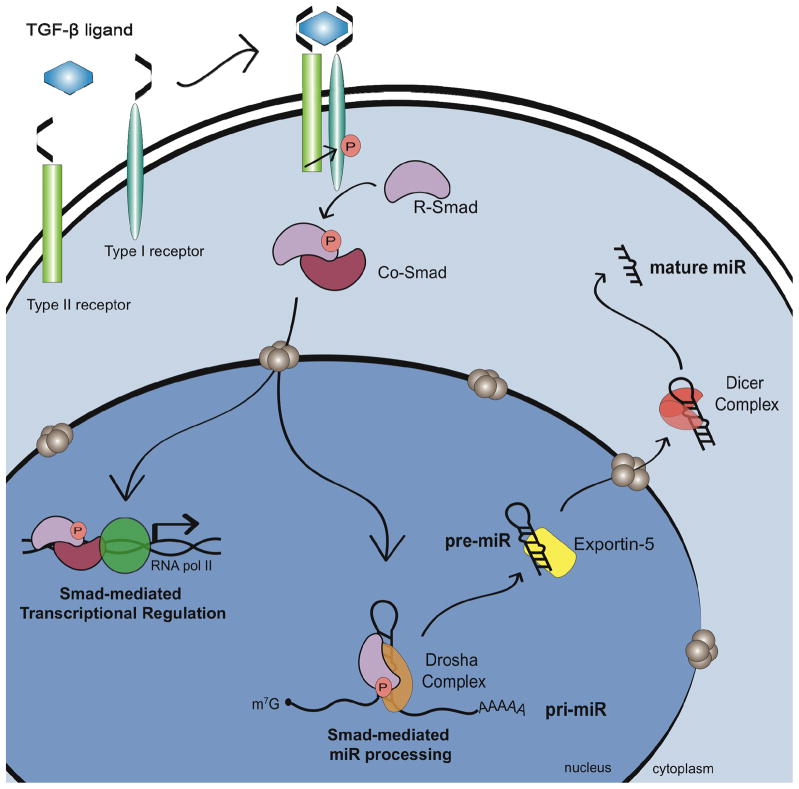
Following transcription of the miRNA gene, the first cleavage is catalyzed in the nucleus by the RNase III enzyme Drosha which generates pre-miRNA from pri-miRNA (Fig. 1). The precise position and orientation of Drosha cleavage serves a critical role in the generation of miRNAs as it determines the identity of both the 5′ and 3′ nt of the mature miRNA. An error in the Drosha cleavage site may result in the alteration of the miRNAseed sequence and cause redirection of miRNA targets. Additionally, in some cases, altered cleavage could invert the relative stability of the two miRNA strands, leading to the incorporation of the improper miRNA strand into the RISC complex. In vivo, Drosha is present in a large protein complex, and the association with co-factors present within this complex promotes the fidelity and activity of Drosha cleavage. In particular, DGCR8 is required for Drosha-mediated cleavage of pri-miRNAs (20, 21). The pri-miRNA is composed of a ~33 nt stem connected by a terminal loop and flanked by single-stranded segments. DGCR8 is thought to recognize the region between the single-stranded RNA and the stem in order to direct Drosha cleavage one helical (~11 bp) turn away from this junction (22). While the cropping of many miRNAs can be mediated in vitro by purified DGCR8 and Drosha, nuclear run-on and in vitro processing assays indicate that the pri-miRNA to pre-miRNA cleavage of some miRNA is relatively slow and inefficient (23). Therefore, the efficient processing of miRNAs by the Drosha/DGCR8 complex may require the involvement of accessory factors. The DEAD-box RNA helicases p68 (DDX5) and p72 (DDX17) were identified as components of the large Drosha processing complex by immunopreciptiation-mass spec analysis and subsequently shown to also associate with DGCR8 (24, 25). Analysis of mature miRNA levels indicated reduced steady state levels of miRNA in p68-null or p72-null mouse embryonic fibroblasts (MEFs) in comparison with control MEFs, suggesting an important role for p68 and p72 in miRNA biogenesis. Recently, the role of p68/p72 in miRNA processing is further supported by the positive regulation of Drosha processing mediated by the p68-interacting Smad proteins. The Smads are the signal transducers of the TGFβ family signaling cascade. In the canonical pathway, ligand binding to the type I and type II TGFβ receptors promotes the nuclear accumulation of receptor-Smads (R-Smads) in association with the common-Smad (co-Smad), Smad4 (Fig. 2). The complex of R-Smad and co-Smad bind to sequence elements within the promoter of target genes to positively or negatively regulate gene transcription. TGFβ and its family member, Bone Morphogenetic Protein 4 (BMP4) are particularly important for the differentiation of vascular smooth muscle cells (VSMCs). Treatment with either BMP4 or TGFβ increases expression of contractile smooth muscle genes. This process is due, at least in part, to the miR-21-mediated repression of programmed cell death protein-4 (PDCD4). miR-21 is rapidly induced by BMP4 and TGFβ in VSMC which results in a subsequent decrease in PDCD4 and increased VSMC gene expression (26). Interestingly, although knockdown of the R-Smads prevents upregulation of mature and pre-miR-21 in response to BMP4 or TGFβ, no alteration in pri-miR-21 transcription is detected (26). Furthermore, BMP4 or TGFβ could increase the expression of pre- and mature miR-21 by facilitating the Drosha processing step.. The identification of R-Smads as binding partners of p68 by yeast-two-hybrid suggested that R-Smads could associate with the Drosha complex (27). Consistently, co-immunoprecipitation (co-IP) and RNA-IP studies confirmed that Smad is present in a complex with Drosha and p68 on the pri-miR-21 hairpin following BMP4 or TGFβ stimulation (26) (Fig. 2). Drosha binding to pri-miR-21 was also elevated following ligand treatment, suggesting that Smads may promote the association of Drosha with miRNA hairpins. These results indicate that TGFβ can regulate gene expression not only through direct transcriptional regulation but also through miRNA processing (Fig. 2). Nucleo-cytoplasmic shuttling of Smads is tightly controlled by phosphorylation of serine residues at the C-terminus by the TGFβ type I receptor kinases. Interestingly, mitogen-activated protein kinase (MAPK) and glycogen synthase kinase 3 (GSK3) can also alter the subcellular localization of Smads through phosphorylation in the linker region (28, 29). Thus, it is possible that Smad-dependent regulation of miRNA biosynthesis could be modulated independently of TGF! and BMPs by signals that alter the nuclear localization of Smads, such as the ERK-MAPK and the Wnt pathways. Recently, p53 (TP53) has been shown to interact with the Drosha complex through p68 and facilitates the Drosha processing similarly to Smads (30). It is still unknown what triggers the association of p53 with the Drosha/p68 complex. Interestingly, the association of R-Smads with the Drosha processing machinery does not require co-Smad Smad4. Knockdown of Smad4 in VSMC did not affect induction of miR-21; furthermore, miR-21 is strongly induced by TGFβ in the Smad4-null MDA-MB-468 breast cancer cell line (26). It was previously reported that R-Smads and Smad4 translocate into the nucleus as a complex (31). A more recent study, however, demonstrates that R-Smads and Smad4 can be independently transported into the nucleus through different nuclear import machineries (32, 33). Thus, R-Smads that are not locked into a complex with Smad4 might preferentially participate in microRNA processing through association with the Drosha/DGCR8 complex. In contrast, the R-Smad/Smad4 heteromeric complex may preferentially associate with the Smad binding element (SBE) in promoter regions of the TGFβ target genes and act as a transcription factor.
Fig. 2. Regulation of miRNA maturation by the TGFβ superfamily signaling.
TGFβ and BMP signaling stimulates the production of pre-miR-21 by promoting the Drosha-mediated processing by controlling nuclear localization of R-Smad proteins. Thus, Smads regulate gene expression in two distinct manners; (i) transcriptional regulation by DNA binding and (ii) regulation of miRNA maturation by associating with the Drosha/DGCR8 complex.
Several other miRNAs are post-transcriptionally induced by BMP and TGFβ, suggesting that rapid modulation of miRNA levels may play an important role in cellular response to cytokine signaling (26)[B. D and A.H., unpublished observation]. More recently, the role of Smads in the regulation of the Drosha complex has been suggested by the study of a nuclear factor called Smad nuclear interacting protein 1 (SNIP1). SNIP1, which was originally identified as a nuclear protein partner of Smads, is found in complex with Drosha (34). The Arabidopsis homologue of SNIP1, DAWDLE (DDL), is required for efficient pri-miRNA to pre-miRNA processing and is thought to promote the access or recognition of pri-miRNA by DCL1 (34). Furthermore, downregulation of SNIP1 in mammalian cells reduces the expression of subset of miRNAs, including miR-21 (34). These results suggest that SNIP1 participates in miRNA biogenesis by facilitating the Drosha function possibly through interaction with Smad proteins.
In contrast with the positive regulation of Droshaprocessing mediated by the TGFβsignaling pathway, the processing of the let-7 family of miRNAs is negatively regulated by lin-28. Lin-28 binds to pri-let-7 and prevents its cropping by the Droshacomplex (35, 36). This mechanism fits with the observation that (i) although the pri-let-7 transcript is highly expressed in most cell types, mature let-7 is not detectable in undifferentiated cells which express high levels of Lin-28, and (ii) mature let-7 is detectable highly differentiated cells which do not express Lin-28 (37). In addition to inhibition of the Drosha processing step in the nucleus, Lin-28 also inhibits the Dicer processing of pre-let-7 and promotes its degradation in the cytoplasm (38, 39). It is unknown if Smad proteins have an additional role in the regulation of the second processing step by Dicer in the cytoplasm. Altogether, these observations suggest that Smads may be present in a large, multi-protein complex, and through the interaction with different RNA binding proteins regulate mature miRNA levels in a context-dependent manner.
Transcriptional regulation of miRNA genes by Smad
RNA pol II mediated transcription provides a major regulatory step for the biosynthesis of miRNAs. A large scale nucleosome positioning and chromatin immunoprecipitation-on-genomic DNA microarray chip (or ChIP-on-chip) analysis of the promoters of miRNA genes suggests that the promoter structure of miRNA genes, including the relative frequencies of CpG islands, transcription initiator elements, and histone modifications, is indistinguishable between the promoters of miRNA genes and protein coding genes (40, 41). Furthermore, DNA binding factors that regulate miRNA transcription largely overlap with those that control protein coding genes. Therefore, it is likely that the Smad proteins might modulate expression of miRNAs by regulating the transcription of miRNA genes (Fig. 2). Alternatively, it is likely that the TGFβ signaling pathways might modulate transcription of genes encoding critical enzymes or regulators of the miRNA biogenesis, such as Drosha, DGCR8, Dicer or Ago. Recently, the protein stability of Ago proteins was found to be regulated by post-translational modification. The relative level of Ago proteins may critically regulate the stability of miRNA as knockdown or overexpression of Agos markedly decrease or increase miRNA levels, respectively. (cite Diederichs Cell 2007). Ago protein stability is enhanced when a specific proline residue is hydroxylated by the type I collagen prolyl-4-hydroxylase (C-P4H) (42). Knockdown of C-P4H reduced Ago protein level (42). The alpha subunit of C-P4H is rate-limiting for the formation of active C-P4H and has been reported to be transcriptionally induced by different stimuli, including TGFβ. Thus, it is plausible that upregulation of C-P4H by TGFβ may lead to the stabilization of Ago, which may result in global increase in miRNAs.
Epigenetic control of miRNA gene loci by Smad
As the majority of miRNA genes are transcribed by the same RNA pol II as protein coding genes, the mechanisms of epigenetic control known for protein coding genes are likely to be applied to miRNA gene loci. Indeed, several miRNA loci including miR-9-1, -193a, -137, -342, -203 and -34b/c, are found to be hypermethylated in multiple human cancers (43, 44). Conversely, the let-7a-3 locus was found to be hypomethylated in lung adenocarcinoma and elevated expression of this locus resulted in enhanced oncogenic gene transcription (45). miRNA promoters are also regulated by histone modifications during development and pathogenesis. Inhibition of histone deacetylase (HDAC) activities by inhibitors has been reported to upregulate a subset of miRNAs in cancer cells (46, 47). Recruitment of HDAC or histone acetylase (HAT) activities to various gene promoters by Smads has been shown previously (48). We therefore speculate that some of the changes in mRNA expression previously linked to the Smad-mediated HAT/HDAC activity may be indirect effects due to altered miRNA expression by epigenetic control of miRNA gene loci.
Future prospects
miRNAs are generated through the concerted action of multi-subunit complexes which promote the sequential cleavage, export, and loading of miRNAs into RISC complexes. An increasing number of reports suggest that each of these steps serves as a potential point of regulation, and therefore provides additional complexity to miRNA-dependent gene regulation. So far there are only a few reports demonstrating that the TGFβ-Smad pathway modulates microRNA levels either at the transcription of miRNA gene or at the Drosha processing step from pri-miRNA to pre-miRNA. Unlike protein coding genes which must be translated following transcription, microRNAs could more rapidly modify gene expressionin response to environmental cues. Therefore, regulation of miRNA biogenesis may serve as the first line of response following TGFβ/BMP stimulation. As a single miRNA modulates the expression of hundreds of targets simultaneously, the regulation of even a handful of miRNAs by the TGFβ/BMP signaling pathway could have a dramatic impact on gene expression and cellular physiology. In the future it will be important to determine the mechanisms of miRNA coregulation, as well as the influence of such coregulated miRNAs on cellular processes. As more mechanisms of miRNA regulation are uncovered, one major challenge will be to elucidate how multiple mechanisms of miRNA regulation cooperate to orchestrate the modulation of microRNA expression in a context-dependent manner. Finally, as for Smad function in the regulation of miRNA biogenesis, it is left for future studies to uncover the mechanism that limits the Smad-mediated processing to a selective group of miRNAs and the exact role of Smad proteins in the Drosha microprocessor complex.
Acknowledgments
We are grateful to members of the Lagna and Hata laboratory for stimulating discussions. This work was supported by National Institute of Health grants HL082854 and HL093154 to A.H.
Biographies
 Akiko Hata earned her Ph.D, at the Rockefeller University under Dr. Hidesaburo Hanafusa, Akiko did postdoctoral research at the Memorial Sloan-Kettering Cancer Center under Dr. Joan Massague from 1995 to 2000. Since 2008, Akiko is an Associate Professor in the Department of Biochemistry in the Tufts University School of Medicine and MCRI at Tufts Medical Center.
Akiko Hata earned her Ph.D, at the Rockefeller University under Dr. Hidesaburo Hanafusa, Akiko did postdoctoral research at the Memorial Sloan-Kettering Cancer Center under Dr. Joan Massague from 1995 to 2000. Since 2008, Akiko is an Associate Professor in the Department of Biochemistry in the Tufts University School of Medicine and MCRI at Tufts Medical Center.
 Brandi Davis is a graduate student of Sackler Graduate Program at Tufts University School of Medicine. After receiving B.S. from University of Rochester, Brandi joined Akiko's laboratory in 2004. Brandi and Akiko are interested in understanding a mechanism of regulation of miRNA biosythesis by growth factor signaling pathways.
Brandi Davis is a graduate student of Sackler Graduate Program at Tufts University School of Medicine. After receiving B.S. from University of Rochester, Brandi joined Akiko's laboratory in 2004. Brandi and Akiko are interested in understanding a mechanism of regulation of miRNA biosythesis by growth factor signaling pathways.
Footnotes
Competing interests
The authors declare that they have no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Kim M, Han JJ, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 13.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 14.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 15.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 17.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Getz G, Miska E, Alvarez-Saavedra E, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Han JJ, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D-melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 23.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 25.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warner DR, Bhattacherjee V, Yin X, et al. Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochem Biophys Res Commun. 2004;324:70–76. doi: 10.1016/j.bbrc.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Fuentealba LC, Eivers E, Ikeda A, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 31.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 32.Xu L, Yao X, Chen X, Lu P, Zhang B, Ip YT. Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads. J Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao X, Chen X, Cottonham C, Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem. 2008;283:22867–22874. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu B, Bi L, Zheng B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 37.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Wulczyn FG, Smirnova L, Rybak A, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 39.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 40.Ozsolak F, Poling LL, Wang Z, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS ONE. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi HH, Ongusaha PP, Myllyharju J, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 44.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 46.Nasser MW, Datta J, Nuovo G, et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 48.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]