Abstract
Purpose
We assessed the association of quantitative clinical and pathologic information, including serum and tissue proPSA, with outcomes among men with prostate cancer (PCa) managed expectantly.
Experimental Design
We identified 71 men enrolled in expectant management (EM) with frozen serum and tissue available from diagnosis. 39 subsequently developed unfavorable biopsies (Gleason score>=7, >=3 cores positive for cancer, >50% of any core involved with cancer), while 32 maintained favorable biopsies (median follow-up: 3.93 years). Serum tPSA, fPSA and [−2]proPSA were measured by the Beckman Coulter immunoassay. [−5/−7]proPSA was evaluated in cancer and benign adjacent tissue areas (BAA) by quantitative immunohistochemistry. Cox proportional hazards and Kaplan-Meier analyses were used to identify significant associations with unfavorable biopsy conversion.
Results
The ratio [−2]proPSA/%fPSA in serum was significantly higher at diagnosis (0.87+/−0.44pg/mL vs. 0.65+/−0.36pg/mL, p=0.02) in men developing unfavorable biopsies. [−5/−7]proPSA tissue staining was more intense (4104.09+/−3033.50 vs. 2418.06+/−1606.04, p=0.03) and comprised a greater fractional area (11.58%+/−7.08% vs. 6.88%+/−5.20%, p=0.01) in BAA of these men. Serum [−2]proPSA/%fPSA [HR:2.53(1.18–5.41), p=0.02], BAA [−5/−7]proPSA %area [HR:1.06(1.01–1.12), p=0.02] and BAA [−5/−7]proPSA stain intensity [HR:1.000213(1.000071–1.000354), p=0.003] were significantly associated with unfavorable biopsy in Kaplan-Meier and Cox analyses. Serum [−2]proPSA/%fPSA significantly correlated with BAA [−5/−7]proPSA %area (rho=0.40, p=0.002) and BAA [−5/−7]proPSA stain intensity (rho=0.33, p=0.016).
Conclusions
In a prospective cohort of men enrolled into EM for PCa, serum and tissue levels of proPSA at diagnosis are associated with need for subsequent treatment. The increase in serum proPSA/%fPSA might be driven by increased proPSA production from “pre-malignant” cells in the prostate BAA.
Keywords: Prostate cancer, expectant management, proPSA, serum, benign-adjacent, cancer, unfavorable biopsy conversion, prediction
INTRODUCTION
Prostate cancer (PCa) is the second leading cause of cancer death among men in the United States, with an anticipated 186,320 newly diagnosed cases and 28,660 deaths in 2008 (1). Over-detection and over-treatment of cases unlikely to cause morbidity represent major dilemmas for PCa management.(2). In an effort to reduce the morbidity of over-treatment, expectant management (EM), also known as active surveillance or watchful waiting, with delayed curative intervention has been proposed as a management strategy for low-grade low-stage PCa (3).
Epstein et al. (4) proposed PSA density (PSAD) <0.15 ng/ml/cm3 and favorable diagnostic needle biopsy characteristics (i.e. Gleason score <7, <3 cores involved with cancer, ≤ 50% of any core involved with cancer) as criteria to identify low-grade low-stage tumors. Men satisfying these criteria are enrolled into a prospective cohort during which they are followed with serial measurements of PSA and repeated biopsies, until tumor characteristics are discovered making them unsuitable for EM and treatment is recommended (3). To date there are very few biomarkers are associated with significant outcomes within this cohort.(3, 5–9)
Another potential candidate is proPSA.(10) Sokoll et al. (11) showed that %[−2] proPSA is the best predictor of PCa, particularly in the 2 to 10 ng/ml total PSA (tPSA) range. proPSA, the precursor of PSA, contains a 7 amino acid pro leader peptide. Additional truncated forms of proPSA with leader sequences of 5, 4 and 2 amino acids also exist in serum (12). Activational cleavage activity of the leader sequences by human kallikrein 2 and trypsin decreases with decreasing size of the propeptide leader sequence; [−2]proPSA being resistant to activation. Our group (13) has demonstrated that nuclear structure alterations and [−5/−7] proPSA staining in cancer and benign-adjacent areas (BAA) of prostate tissue can differentiate between patients of native Japanese and American Japanese origin. We sought to assess the association of proPSA staining of biopsy BAA and cancer tissue as well as quantification of serum proPSA from samples taken at diagnosis with unfavorable biopsy conversion on annual surveillance examination in our EM cohort.
MATERIAL AND METHODS
Patient Sample
Patients with signed informed consent were enrolled in our institutional review board approved EM program if they met inclusion criteria [nonpalpable tumor on digital rectal examination (DRE) (stage T1c), PSAD ≤0.15 ng/ml/cm3 (PSA before diagnosis divided by prostate volume determined by transrectal ultrasound measurement) and favorable diagnostic needle biopsy characteristics (Gleason score <7, <3 cores involved with cancer, ≤50% of any core involved with cancer)]. Patients were surveilled semiannually with serum tPSA, free PSA (fPSA) and DRE. An annual surveillance biopsy was also performed, and curative intervention was recommended if pathology were unfavorable (Gleason score ≥7, Gleason pattern 4/5, ≥3 cores involved with cancer, >50% of any core involved with cancer).
Serum PSA isoforms measurement
Serum was obtained prior to biopsy and stored at −80°C until testing. Serum specimens were analyzed in the Johns Hopkins University Clinical Chemistry Research Laboratory on the Beckman Coulter ACCESS immunoassay system for tPSA, fPSA, and [−2]proPSA. The assays are all dual monoclonal sandwich assays using Hybritech antibodies and a chemiluminescent detection system. The assays for tPSA and fPSA are commercially available, while the assay for [−2]proPSA is for research only (14). The [−2]proPSA assay is calibrated using [−2]proPSA purified from the AVA12-PSA mammalian cell line. The assay has linear ranges of less than 1 to 5,000 pg/ml for [−2]proPSA with intra- and inter-assay precision of 2.3–5.3% and 2.7–3.5% (range 9–69 pg/mL), respectively. Cross-reactivity of other PSA isoforms in the assay is minimal.
Quantitative Immunohistochemistry (QIHC)
The [−5/−7]proPSA antibody was provided by Beckman Coulter research and development (Beckman Coulter, San Diego, CA). IHC was performed using a Dako AutoStainer Universal Staining System (Dako Cytomation, Carpentaria, CA). After dewaxing and dehydration, biopsy sections were placed in a rice steamer with antigen retrieval solution (Dako # S1700) for 20 minutes. Biopsy sections were next pretreated with 0.3% hydrogen peroxide for 10 minutes to remove endogenous peroxidase activity. Subsequently, biopsy sections were incubated for 1 hour at room temperature with [−5/−7]proPSA antibody (6.31 ng/ml) at 1:320 dilution and followed by detection with the envision plus kit (Dako # K1392). Biopsy sections were incubated with horse radish peroxidase (HRP) labeled polymer-secondary antibody for 20 minutes then with freshly prepared liquid 3, 3′-diaminobenzidine (DAB) + Substrate-Chromogen solution for 5 minutes. Biopsy sections were counterstained with hematoxylin for 1 minute.
Next, [5/−7]proPSA IHC stained benign-adjacent and cancer areas of the same slide were marked as such by J.I.E for each patient. Images were individually captured using the Zeiss Axioskop microscope at 200× magnification (Supplementary Figure 1). Since marked cancer areas on biopsy cores for most patients were very small, only one 200× field of view was captured in each benign-adjacent and cancer area. We used Image-Pro Plus V6.0 (Media Cybernetics, Bethesda MD) software to quantify [−5/−7] proPSA IHC as well as to calculate proPSA (DAB) %IHC area and staining intensity.
Because the original biopsy cores from men in the EM cohort had very limited amounts of cancer (only small foci in certain instances), further cutting into those blocks sometimes produced slides without visible cancer. As we wanted to study staining differences in both the cancer and BAA, cases were excluded from IHC analysis if cancer tissue was not available for a given case.
Statistical Analysis
All data were analyzed using Stata™ v10.0 statistical analysis software (Stata Corporation, College Station, TX). The Mann-Whitney test was used to determine distribution differences across favorable and unfavorable biopsy groups. Bivariate Cox proportional hazard regression was used to identify significant prognostic factors for unfavorable biopsy conversion on annual surveillance examination. Ties were handled by the Breslow method. The proportional hazard assumption was verified by examination of residual plots and Schoenfeld residuals. We determined optimal cutoffs to dichotomize continuous variables using the classification and regression tree (CART) method. The Kaplan-Meier analysis and the Logrank test were used to test equality of survivor functions across two groups. Statistical significance in this study was set as p ≤ 0.05.
RESULTS
Of the 71 PCa patients from the Johns Hopkins EM cohort who had banked serum and tissue available from the time of diagnosis, 39 developed unfavorable biopsies and 32 maintained favorable biopsies on annual surveillance (median follow-up: 3.93 years). Demographic, clinical and pathologic information for favorable and unfavorable biopsy groups is shown in Table 1.
Table 1.
Expectant Management Cohort Patients Characteristics at Diagnosis
| Variable | Favorable (N = 32) | Unfavorable (N = 39) | p value* |
|---|---|---|---|
| Mean ± SD [Median] | Mean ± SD [Median] | ||
| Age | 65.42 ± 4.37 [65.03] | 64.82 ± 4.70 [64.97] | 0.991 |
| tPSA (ng/ml) | 4.61 ± 2.75 [4.36] | 5.35 ± 2.02 [5.53] | 0.056 |
| fPSA (ng/ml) | 0.88 ± 0.59 [0.74] | 0.97 ± 0.52 [0.89] | 0.298 |
| %fPSA | 19.15 ± 6.36 [18.98] | 18.40 ± 6.44 [18.21] | 0.587 |
| [−2] proPSA (pg/ml) | 12.21 ± 7.43 [11.36] | 15.00 ± 6.97 [14.24] | 0.051 |
| [−2] proPSA/%fPSA | 0.65 ± 0.36 [0.55] | 0.87 ± 0.44 [0.88] | 0.018 |
| PSAD (ng/ml/cm3) | 0.094 ± 0.058 [0.087] | 0.100 ± 0.0369 [0.100] | 0.173 |
| Prostate Volume | 51.86 ± 17.50 [50.00] | 57.29 ± 29.27 [50.00] | 0.737 |
| Number of Positive Core | 1.19 ± 0.40 [1.00] | 1.18 ± 0.39 [1.00] | 0.931 |
| Maximum %core involvement with Cancer | 7.39 ± 10.13 [1.00] | 8.29 ± 11.60 [1.00] | 0.741 |
| Cancer IHC† | |||
| [−5/−7] proPSA %Area | 11.87 ± 9.67 [9.51] | 13.70 ± 9.62 [12.90] | 0.371 |
| [−5/−7] proPSA Stain Intensity | 4552.54 ± 2923.61 [4055.18] | 4611.47 ± 3293.73 [3119.32] | 0.786 |
| Benign Adjacent Area IHC† | |||
| [−5/−7] proPSA %Area | 6.88 ± 5.20 [5.38] | 11.58 ± 7.08 [10.14] | 0.009 |
| [−5/−7] proPSA Stain Intensity | 2418.06 ± 1606.04 [1819.76] | 4104.09 ± 3033.50 [3196.52] | 0.025 |
For Cancer and Benign Adjacent Area IHC number of subjects (favorable/unfavorable) were 57 (27/30) and 54 (25/29) respectively
p values calculated using Mann-Whitney test.
In bivariate Cox regression analysis, age (p = 0.860), tPSA (p = 0.441), fPSA (p = 0.948), %fPSA (p = 0.433), PSAD (p = 0.816), [−2] proPSA (p = 0.280), number of cores involved with cancer (p = 0.780), maximum %core involved with cancer (p = 0.469), cancer [−5/−7] proPSA %area (p = 0.163), cancer [−5/−7] proPSA stain intensity (p = 0.301) were not significantly associated with unfavorable biopsy conversion.
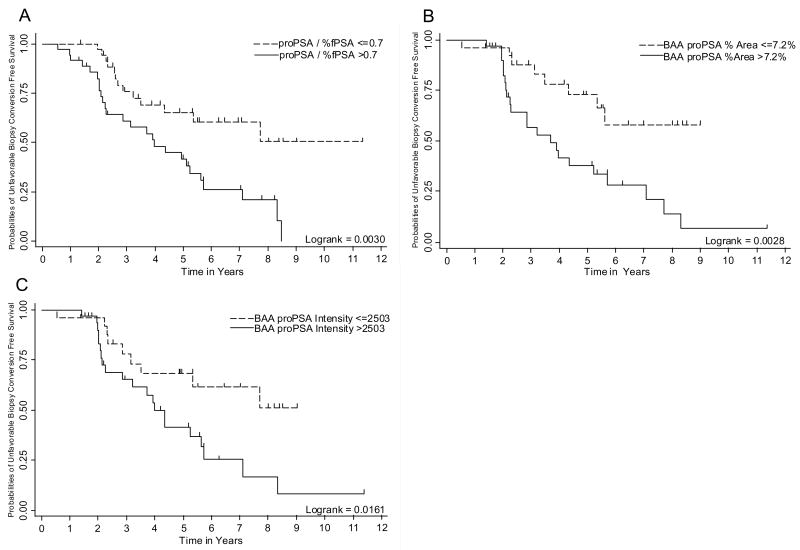
Serum [−2]proPSA/%fPSA (p = 0.017), BAA [−5/−7]proPSA %area (p = 0.019), BAA [−5/−7] proPSA stain intensity (p = 0.003) were significantly associated with unfavorable biopsy conversion (Table 2, Figure 1). Further, serum [−2] proPSA/%fPSA significantly correlated with BAA [−5/−7]proPSA %area (rho=0.403, p=0.002) and BAA [−5/−7]proPSA stain intensity (rho=0.325, p=0.016) but not with cancer [−5/−7]proPSA %area (rho=0.187, p=0.164) or cancer [−5/−7]proPSA stain intensity (rho=0.068, p=0.614). In order to avoid multicolinearity, serum [−2] proPSA/%fPSA, BAA [−5/−7] proPSA %area, BAA [−5/−7] proPSA stain intensity were not evaluated in multivariable analysis due to our small sample size and significant correlations between these variables. Standard clinical/pathological parameters were not evaluated in multivariable logistic regression because none of them were associated with unfavorable biopsy on bivariate analysis
Table 2.
Bivariate Cox Regression Analysis for Predicting Unfavorable Biopsy Conversion on Annual Surveillance Biopsy Examination in the Expectant Management Cohort
| Continuous |
Dichotomized |
|||||
|---|---|---|---|---|---|---|
| HR (95%CI) | C Index | p value | HR (95%CI) | C Index | p value | |
| Serum† | ||||||
| proPSA/%fPSA | 2.53 (1.18–5.41) | 0.610 | 0.017 | 2.65 (1.36–5.16) | 0.616 | 0.004 |
| IHC‡ | ||||||
| BAA [−5/−7] proPSA %Area | 1.06 (1.01–1.12) | 0.632 | 0.019 | 3.22 (1.43–7.24) | 0.636 | 0.005 |
| BAA [−5/−7] proPSA stain intensity | 1.000213 (1.000071–1.0000354) | 0.616 | 0.003 | 2.55 (1.16–5.61) | 0.600 | 0.020 |
Number of subjects (favorable/unfavorable) were 71 (32/39)
Number of subjects (favorable/unfavorable) were 57 (27/30)
Figure 1. Kaplan-Meier plots demonstrating unfavorable biopsy free survival: Figure 1a. Unfavorable biopsy free survival as a function of serum [−2] proPSA/%fPSA.
Figure 1b. Unfavorable biopsy free survival as a function of initial diagnostic biopsy tissue benign-adjacent area (BAA) [−5/−7] proPSA % Area
Figure 1c. Unfavorable biopsy free survival as a function of initial diagnostic biopsy tissue BAA [−5/−7] proPSA stain intensity [ Figure 1c ]
DISCUSSION
Over-detection and over-treatment of PCa is an important public health issue among older men in the United States (15). While the 12 year update of the Scandinavian Prostate Cancer Group Trial demonstrated decrease in PCa-specific mortality across the entire cohort, the absolute decrease in PCa-specific mortality in the subset of men aged 65 years or older randomized to radical prostatectomy vs. watchful waiting was only 0.1% [13.1 (8.8–19.5) vs. 13.2 (8.9 – 19.6)] (16). Further, men 65 years or older in the radical prostatectomy group had 2.7% higher overall mortality compared to those in the watchful waiting group [42 (35 – 50.5) vs. 39.3 (32.5 – 47.7)] (16). Thus, a majority of older men diagnosed with screen detected PCa may not gain survival advantage with curative intervention.
Since 1995, our department has prospectively enrolled and followed an EM cohort for men with low-grade low-stage tumors. Curative intervention is suggested by a change in clinical exam or unfavorable pathology on annual surveillance biopsy (3). While delayed surgical intervention for low-grade low-stage tumors does not appear to compromise curability compared to immediate surgical intervention (17), it would be helpful to patients, clinicians and to researchers to identify those men at increased risk of developing an unfavorable biopsy earlier in the course of their disease. Since the patients are effectively matched for tPSA, Gleason grade, and tumor volume at the time of entry into the study, we wanted to investigate the role of both tissue and serum proPSA isoforms, collected at the time of entry into EM, in determining subsequent unfavorable biopsy conversion.
There has been recent interest in the role of proPSA in PCa early detection and prognosis. Both the overall percentage of proPSA (proPSA to fPSA ratio) and levels of its truncated forms (particularly [−2]proPSA) have been able to determine the presence of PCa. Sokoll et al. (18) demonstrated %proPSA could reduce unnecessary biopsies among men with tPSA between 2.5–4.0 ng/ml. This result has been validated in a multi-institutional cohort of men with tPSA between 2–10 ng/ml (11). Men with tPSA between 4–10 ng/ml who were diagnosed with PCa have demonstrated higher fractions of proPSA than men with similar tPSA and no evidence of PCa (19). Catalona et al. (20) showed higher preoperative proPSA was associated with higher grade disease and more advanced pathology at the time of surgery. Stephan et al. demonstrated using a neural network that ratios of proPSA to %fPSA are associated with features of aggressive CaP among men undergoing CaP screening.(21)
There has been a great deal of interest in the research community to determine prognostic biomarkers for PCa managed with watchful waiting. PSA kinetics (8), p53 nuclear staining (22), Ki-67 (6), microvessel density (23), neuroendocrine differentiation (24), TMPRSS2–ERG fusions (5) apparent diffusion coefficient (ADC) on diffusion-weighted magnetic resonance imaging (DW-MRI) (25) have demonstrated prognostic value in patients managed by watchful waiting. Notably, our approach for selecting and monitoring patients (T1c, Gleason score<7, fewer than 3 cores involved with cancer, and ≤ 50% of any core involved with cancer) differs from those described by others, who may enroll men with T2 lesions and Gleason 7 tumors (5, 6, 8, 16, 22–25), and may be considered more conservative. Because of this more highly stringent entrance criteria, the patients in our cohort are better “matched” and have fewer differing characteristics between them. Our group has also demonstrated the clinical utility of several biomarkers in this cohort (7, 9).
Recently Hoshida et al. (26) showed that gene expression profiles of tumor tissue failed to show a significant association with survival in patients with hepatocellular carcinoma, while profiles of the surrounding non-tumor liver tissue were highly correlated. Alterations in DNA content are present in both benign-adjacent and cancer tissue areas of PCa and represent the up-regulation of proliferation related genes including transcription factors, signal transducer, and growth regulators (27, 28). Additionally, mitochondrial DNA alterations are known to be present in PCa and histologically normal appearing adjacent prostate glands (29).
In the current study, we sought to determine both the association of serum and tissue, cancer and BAA, proPSA levels with outcomes among men enrolled in EM. We demonstrated that serum [−2] proPSA/%fPSA, BAA [−5/−7] proPSA %area, and BAA [−5/−7] proPSA IHC stain intensity are associated with unfavorable biopsy conversion on annual surveillance biopsy in an EM cohort (Figure 1, Table 2). Further, serum [−2] proPSA/%fPSA significantly correlated with [−5/−7] proPSA %area in BAA tissue as well as BAA [−5/−7] proPSA stain intensity. However, cancer tissue [−5/−7] proPSA %area and cancer tissue [−5/−7] proPSA IHC stain intensity were not associated with unfavorable biopsy conversion and did not correlate with serum [−2] proPSA/%fPSA. Based on these results we formulated a novel hypothesis: potentially, the increase in serum proPSA/%fPSA is driven by increased proPSA production from prostate cells in BAA of the prostate.
Our study has many advantages. We have one of the largest EM cohorts of men with PCa. Our cohort is unique in its stringent entry requirements and close follow up. It also has the advantage of having banked serum and tissue samples. However, our study has limitations, the most important of which is the limited sample size of men having both serum and tissue samples available for study. Another limitation is the use of prostate biopsy status as a defined endpoint; because prostate biopsies sample only a very small fraction of the gland, our results are prone to verification bias. Our observations and interpretations can only be viewed as exploratory and hypothesis generating at this juncture because of the problem of examining multiple variables in a small cohort. Future research needs to expand the use of PSA isoforms in combination with other molecular biomarkers to assess prognosis of low-grade low-stage PCa in EM. Such biomarkers must be examined in larger, potentially multi-institutional cohorts in prospective trials with pre-specified plans of analysis.
In conclusion, measurement of serum and tissue levels of proPSA, at the time of diagnosis, are associated with future unfavorable biopsy conversation and could potentially determine which men enrolled in an EM cohort will ultimately require treatment for PCa.
Supplementary Material
Acknowledgments
Funding for this project was provided by The Johns Hopkins University Prostate Cancer SPORE (Grant number: P50CA58236), Early Detection Research Network (EDRN) NCI/NIH (Grant numbers CA086323-06 and CA115102-04), Prostate Cancer Foundation, Patana Fund, National Kidney Foundation of Maryland, Inc. Mini-Grant, The Robert Wood Johnson Foundation, and The United States Department of Veterans Affairs. Beckman Coulter provided [−5/−7] proPSA antibody for IHC and [−2] proPSA serum assay under materials transfer agreement.
Footnotes
STATEMENT OF TRANSLATIONAL RELEVENCE
The current research is highly applicable in a translational setting. Once validated, the techniques in this analysis can help to determine which patients with low risk prostate cancer are suitable for expectant management. Such an advance could prevent the unnecessary morbidity of treatment in men whose disease is unlikely to affect them during their lifetime.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Draisma G, Boer R, Otto SJ, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 3.Carter HB, Kettermann A, Warlick C, et al. Expectant Management of Prostate Cancer With Curative Intent: An Update of The Johns Hopkins Experience. J Urol. 2007;178:2359–64. doi: 10.1016/j.juro.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein JI, Walsh PC, Carmichael M, Brendler CB. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. Jama. 1994;271:368–74. [PubMed] [Google Scholar]
- 5.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 6.Jhavar S, Bartlett J, Kovacs G, et al. Biopsy tissue microarray study of Ki-67 expression in untreated, localized prostate cancer managed by active surveillance. Prostate Cancer Prostatic Dis. 2008 doi: 10.1038/pcan.2008.47. [DOI] [PubMed] [Google Scholar]
- 7.Khan MA, Carter HB, Epstein JI, et al. Can prostate specific antigen derivatives and pathological parameters predict significant change in expectant management criteria for prostate cancer? The Journal of urology. 2003;170:2274–8. doi: 10.1097/01.ju.0000097124.21878.6b. [DOI] [PubMed] [Google Scholar]
- 8.Khatami A, Aus G, Damber JE, Lilja H, Lodding P, Hugosson J. PSA doubling time predicts the outcome after active surveillance in screening-detected prostate cancer: results from the European randomized study of screening for prostate cancer, Sweden section. Int J Cancer. 2007;120:170–4. doi: 10.1002/ijc.22161. [DOI] [PubMed] [Google Scholar]
- 9.Makarov DV, Marlow C, Epstein JI, et al. Using nuclear morphometry to predict the need for treatment among men with low grade, low stage prostate cancer enrolled in a program of expectant management with curative intent. Prostate. 2008;68:183–9. doi: 10.1002/pros.20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peyromaure M, Fulla Y, Debre B, Dinh-Xuan AT. Pro PSA: a “pro cancer” form of PSA? Med Hypotheses. 2005;64:92–5. doi: 10.1016/j.mehy.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Sokoll LJ, Wang Y, Feng Z, et al. [−2]proenzyme prostate specific antigen for prostate cancer detection: a national cancer institute early detection research network validation study. The Journal of urology. 2008;180:539–43. doi: 10.1016/j.juro.2008.04.015. discussion 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peter J, Unverzagt C, Krogh TN, Vorm O, Hoesel W. Identification of precursor forms of free prostate-specific antigen in serum of prostate cancer patients by immunosorption and mass spectrometry. Cancer Res. 2001;61:957–62. [PubMed] [Google Scholar]
- 13.Veltri RW, Khan MA, Marlow C, et al. Alterations in nuclear structure and expression of proPSA predict differences between native Japanese and Japanese-American prostate cancer. Urology. 2006;68:898–904. doi: 10.1016/j.urology.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Weinzierl CF, Su SX, Pierson TB, et al. Measuring [–2]proPSA in serum: Analytical performance of the Access p2PSA assay from Beckman Coulter. Annual Meeting of the American Association for Clinical Chemistry 2007; Abstract No. C-38. [Google Scholar]
- 15.Schroder FH. Screening, early detection, and treatment of prostate cancer: a European view. Urology. 1995;46:62–70. doi: 10.1016/s0090-4295(99)80252-0. [DOI] [PubMed] [Google Scholar]
- 16.Bill-Axelson A, Holmberg L, Filen F, et al. Radical prostatectomy versus watchful waiting in localized prostate cancer: the Scandinavian prostate cancer group-4 randomized trial. J Natl Cancer Inst. 2008;100:1144–54. doi: 10.1093/jnci/djn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warlick C, Trock BJ, Landis P, Epstein JI, Carter HB. Delayed versus immediate surgical intervention and prostate cancer outcome. J Natl Cancer Inst. 2006;98:355–7. doi: 10.1093/jnci/djj072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokoll LJ, Chan DW, Mikolajczyk SD, et al. Proenzyme psa for the early detection of prostate cancer in the 2.5–4.0 ng/ml total psa range: preliminary analysis. Urology. 2003;61:274–6. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 19.Khan MA, Partin AW, Rittenhouse HG, et al. Evaluation of proprostate specific antigen for early detection of prostate cancer in men with a total prostate specific antigen range of 4.0 to 10.0 ng/ml. The Journal of urology. 2003;170:723–6. doi: 10.1097/01.ju.0000086940.10392.93. [DOI] [PubMed] [Google Scholar]
- 20.Catalona WJ, Bartsch G, Rittenhouse HG, et al. Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol. 2004;171:2239–44. doi: 10.1097/01.ju.0000127737.94221.3e. [DOI] [PubMed] [Google Scholar]
- 21.Stephan C, Kahrs AM, Cammann H, et al. A [−2]proPSA-based artificial neural network significantly improves differentiation between prostate cancer and benign prostatic diseases. Prostate. 2009;69:198–207. doi: 10.1002/pros.20872. [DOI] [PubMed] [Google Scholar]
- 22.Borre M, Stausbol-Gron B, Overgaard J. p53 accumulation associated with bcl-2, the proliferation marker MIB-1 and survival in patients with prostate cancer subjected to watchful waiting. The Journal of urology. 2000;164:716–21. doi: 10.1097/00005392-200009010-00023. [DOI] [PubMed] [Google Scholar]
- 23.Borre M, Offersen BV, Nerstrom B, Overgaard J. Microvessel density predicts survival in prostate cancer patients subjected to watchful waiting. Br J Cancer. 1998;78:940–4. doi: 10.1038/bjc.1998.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borre M, Nerstrom B, Overgaard J. Association between immunohistochemical expression of vascular endothelial growth factor (VEGF), VEGF-expressing neuroendocrine-differentiated tumor cells, and outcome in prostate cancer patients subjected to watchful waiting. Clin Cancer Res. 2000;6:1882–90. [PubMed] [Google Scholar]
- 25.van As NJ, de Souza NM, Riches SF, et al. A Study of Diffusion-Weighted Magnetic Resonance Imaging in Men with Untreated Localised Prostate Cancer on Active Surveillance. Eur Urol. 2008 doi: 10.1016/j.eururo.2008.11.051. [DOI] [PubMed] [Google Scholar]
- 26.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deitch AD, Miller GJ, deVere White RW. Significance of abnormal diploid DNA histograms in localized prostate cancer and adjacent benign prostatic tissue. Cancer. 1993;72:1692–700. doi: 10.1002/1097-0142(19930901)72:5<1692::aid-cncr2820720533>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parr RL, Dakubo GD, Crandall KA, et al. Somatic mitochondrial DNA mutations in prostate cancer and normal appearing adjacent glands in comparison to age-matched prostate samples without malignant histology. J Mol Diagn. 2006;8:312–9. doi: 10.2353/jmoldx.2006.050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.