Abstract
In fungal species, lysine 56 of newly synthesized histone H3 molecules is modified by the acetyltransferase Rtt109, which promotes resistance to genotoxic agents. To further explore how H3 K56ac contributes to genome stability, we conducted screens for suppressors of the DNA damage sensitivity of budding yeast rtt109Δ mutants. We recovered a single extragenic suppressor mutation that efficiently restored damage resistance. The suppressor is a point mutation in the histone H3 gene HHT2, and converts lysine 56 to glutamic acid. In some ways, K56E mimics K56ac, because it suppresses other mutations that interfere with the production of H3 K56ac and restores histone binding to chromatin assembly proteins CAF-1 and Rtt106. Therefore, we demonstrate that enhanced association with chromatin assembly factors can be accomplished not only by acetylation-mediated charge neutralization of H3K56 but also by replacement of the positively charged lysine with an acidic residue. These data suggest that removal of the positive charge on lysine 56 is the functionally important consequence of H3 K56 acetylation. Additionally, the suppressive function of K56E requires the presence of a second H3 allele, because K56E impairs growth when it is the sole source of histones, even more so than does constitutive H3K56 acetylation. Our studies therefore emphasize how H3 K56ac not only promotes chromatin assembly but also leads to chromosomal malfunction if not removed following histone deposition.
Keywords: histone, acetylation, budding yeast, chromatin assembly
1. INTRODUCTION
Histone proteins are subject to a variety of post-translational modifications (PTMs), which modulate all chromatin-based events, including epigenetic gene silencing and DNA repair. The majority of histone modification sites reside in the flexible N-terminal tails of histones [1], which, as opposed to their globular core domains, are exposed in the context of the nucleosome [2]. However, several PTMs within the core domains have also been reported, including H3 lysine 56 and H4 lysine 91 acetylation (K56ac and K91ac), and H3 lysine 79 methylation (K79me) [3–13]. Importantly, mutagenesis studies in Saccharomyces cerevisiae have revealed that these core domain modifications, unlike those of the individual tail residues, play key roles in promoting DNA damage tolerance.
The enzyme that catalyzes H3 K56 acetylation is Rtt109 [14–17], a fungal-specific histone acetyltransferase [18] that shares structural homology but limited primary sequence similarity with metazoan p300/CBP [19–21]. In vitro, the inherent low-level HAT activity of Rtt109 is strongly stimulated by either of two histone chaperones, Asf1 or Vps75 [16,17,22,23]. However, the accumulation of H3K56ac in vivo is dependent on Rtt109 and Asf1 but not Vps75 [17,24,25]. Accordingly, asf1Δ and rtt109Δ mutants are both highly sensitive to DNA damaging agents [15,16,26–28]. Moreover, both asf1Δ and rtt109Δ deletions are epistatic with non-acetylatable H3 K56 alleles [15,24] and display similar DNA replisome instability in the presence of genotoxic agents [17,27,29]. Therefore, the damage sensitivity of asf1Δ and rtt109Δ mutant cells is caused by the failure to acetylate H3 K56.
H3 K56 acetylation occurs on nascent, soluble histones [17,23] and accumulates maximally in S phase when the synthesis of new histones is at its peak [12]. The H3K56 acetylation levels are dictated by the regulated accumulation of Rtt109 [16] and also the H3 K56 deacetylases Hst3 and Hst4 [30,31]. Whereas Rtt109 peaks just prior to H3 K56ac in S phase, Hst3/4 levels rise in late S and G2/M phases, coinciding with when H3 K56ac levels begin to drop. In addition, when cells are exposed to DNA damaging agents, DNA damage checkpoint-dependent downregulation of Hst3 and 4 [30,32] lengthens the persistence of H3 K56ac in chromatin [12]. The maintenance of H3 K56ac during DNA damage signaling illustrates the importance of this modification for DNA damage tolerance in yeast.
Recent reports suggest that re-packaging repaired DNA with H3 K56ac may be vital for checkpoint de-activation [33,34], an event that is required to survive even transient DNA damage [35]. The data show that although competent to repair DNA double-strand breaks, asf1Δ and rtt109Δ mutants fail to subsequently inactivate the DNA damage checkpoint [33]. These defects correlate with delayed chromatin reassembly after repair, suggesting that part of the essential function of H3 K56ac in damage tolerance is to facilitate the deposition of histones onto repaired DNA. This model is supported by recent findings which show that the association of histone chaperones CAF-1 and Rtt106 with histones is enhanced when H3 K56 is acetylated [36]. Combined, the current data support a model whereby efficient chromatin reassembly following damage repair is critical for subsequent checkpoint inactivation and recovery [33,34].
Epistasis miniarray profiling has confirmed the close functional relationship of ASF1 and RTT109 and has also highlighted genetic interactions with subunits of the Rtt101Mms1/Mms22 complex, a CUL4DDB1–like ubiquitin ligase [22,37]. The RTT101, MMS1 and MMS22 genes are epistatic for DNA damage tolerance [38–40] and play an essential role in the re-start of replication forks transiently stalled by DNA damage [37,41], possibly by a mechanism involving homologous recombination [42]. In addition, a direct role for Rtt101 in damage recovery is suggested by the damage-dependent recruitment of Rtt101 to chromatin [43].
To further elucidate the mechanism by which Rtt109 contributes to damage tolerance, we conducted a screen for dosage suppressors of the DNA damage sensitivity of rtt109Δ mutants. Here, we describe an extragenic suppressor mutation we isolated during the course of this screen.
2. MATERIALS AND METHODS
2.1 Yeast strains and plasmids
All the strains used in this study (Table I) are isogenic with W303 [44]. rtt109Δ sup− (PKY4342) was a spontaneous mutant isolated in an overexpression screen designed to identify plasmid-bourne genes that suppressed the MMS sensitivity of rtt109Δ mutants. Characterization of the rtt109Δ suppressor mutation was carried out using strains in which sup− had been backcrossed to non-mutagenized strains at least twice, e.g. PKY4345 and 4347. The strains shown in Figure 4, Figure 5 and Figure 8 were obtained by crossing PKY4345 (rtt109Δ sup−) to PKY4273 ((hht1-hhf1)Δ (hht2-hhf2)Δ + pPK589). To either expel or shuffle plasmids, strains were propagated in non-selective media, plated and then screened for stochastic loss of the unwanted plasmid. In most crosses sup− was traceable by phenotypic suppression. This was the case however only if H3 K56ac deficiency was the reason for the damage sensitivity caused by the other mutations in the cross. When this phenotypic screening method could not be employed, e.g. in the construction of hht1Δ hst3Δ hst4Δ hht2-K56E mutants, the mutation was instead identified by sequencing. Rtt106-TAP strains (Figure 6) were made by replacing the RTT106 locus of PKY4428, 4170 and 4345 with the RTT106-TAP allele from the corresponding Open Biosystems TAP-tagged strain. RTT101 was deleted using the rtt101Δ::kanMX allele from the Open Biosystems deletion collection knock-out strain. The HHT2-HHF2/TRP1 (pPK587), hht2(K56R)-HHF2/TRP1 (pPK589) and hht2(K56Q)-HHF2/TRP1 (pPK588) plasmids we used were isolated from yeast strains kindly provided by D. Allis. To make URA3-marked HHT2-HHF2 and hht2(K56Q)-HHF2 plasmids, the Not I/Sal I restriction fragments from pPK587 and pPK588 were sub-cloned into the corresponding sites of pRS416, creating the plasmids pPK604 and pPK605. TRP1- and URA3- K56E plasmids were made as follows: The HHT2-HHF2 locus from PKY4345 was PCR amplified, digested with Bsp EI/Bam HI and cloned into the corresponding sites of pPK587 and pPK604, creating the respective plasmids, pPK607 and pPK606.
Table 1.
Yeast strains.
| Strain No. | Relevant Genotype |
|---|---|
| PKY4342 | MATa rtt109Δ::kanMX hht2-K56E bar1-1 |
| PKY4176 | MATa rtt109Δ::kanMX bar1-1 URA3-VIIL |
| PKY3310 | MATa bar1Δ::his5+ |
| PKY4170 | MATa rtt109Δ::kanMX bar1-1 |
| PKY4345 | MATα rtt109Δ::kanMX hht2-K56E bar1-1 |
| PKY4347 | MATα hht2-K56E bar1Δ::his5+ |
| PKY3328 | MATa asf1Δ::TRP1 bar1Δ::his5+ |
| PKY4355 | MATa asf1Δ::TRP1 hht2-K56E bar1-1 |
| PKY4356 | MATa asf1Δ::TRP1 rtt109Δ::kanMX bar1Δ::his5 + |
| PKY4353 | MATa asf1Δ::TRP1 rtt109Δ::kanMX hht2-K56E bar1Δ::his5+ |
| PKY4273 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 URA3-VIIL + pPK589 |
| PKY4399 | MATa bar1Δ::his5+ + pRS414 |
| PKY4401 | MATα hht2-K56E bar1Δ::his5+ + pRS414 |
| PKY4375 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E URA3-VIIL + pRS414 |
| PKY4376 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E URA3-VIIL + pPK587 |
| PKY4366 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E URA3-VIIL + pPK589 |
| PKY4400 | MATα rtt109Δ::kanMX hht2-K56E bar1-1 + pRS414 |
| PKY4377 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E rtt109Δ::kanMX URA3-VIIL + pRS414 |
| PKY4378 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E rtt109Δ::kanMX URA3-VIIL + pPK587 |
| PKY4369 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E rtt109Δ::kanMX URA3-VIIL + pPK589 |
| PKY4387 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK604 and pPK587 |
| PKY4388 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK604 and pPK588 |
| PKY4390 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK605 and pPK588 |
| PKY4389 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK604 and pPK607 |
| PKY4392 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK606 and pPK607 |
| PKY4391 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK605 and pPK607 |
| PKY4393 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK604 and pPK587 |
| PKY4394 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK604 and pPK588 |
| PKY4396 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK605 and pPK588 |
| PKY4395 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK604 and pPK607 |
| PKY4398 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK606 and pPK607 |
| PKY4397 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK605 and pPK607 |
| PKY1252 | MATa cac1Δ::hisG trp1Δ::CAC1-FLAG-TRP1 URA3-VIIL |
| PKY4426 | MATa rtt109Δ::kanMX cac1Δ::hisG trp1Δ::CAC1-FLAG-TRP1 bar1-1(?) |
| PKY4427 | MATα rtt109Δ::kanMX hht2-K56E cac1Δ::hisG trp1Δ::CAC1-FLAG-TRP1 URA3-VIIL bar1-1(?) |
| PKY4428 | MATa bar1-1(?) |
| PKY4429 | MATa RTT106-TAP-HIS3 bar1-1(?) |
| PKY4430 | MATa rtt109Δ::kanMX RTT106-TAP-HIS3 bar1-1 |
| PKY4431 | MATα rtt109Δ::kanMX hht2-K56E RTT106-TAP-HIS3 bar1-1 |
| PKY4220 | MATa hst3Δ::natMX hst4Δ::kanMX bar1Δ::his5+ |
| PKY388 | Mata (hht1-hhf1)Δ::LEU2 |
| PKY4415 | MATa (hht1-hhf1)Δ::LEU2 hht2-K56E hst3Δ::natMX hst4Δ::kanMX URA3-VIIL |
| PKY4420 | MATa (hht1-hhf1)Δ::LEU2 hst3Δ::natMX hst4Δ::kanMX URA3-VIIL |
| PKY4402 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK587 |
| PKY4404 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 + pPK607 |
| PKY4409 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt101Δ::kanMX + pPK587 |
| PKY4410 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt101Δ::kanMX + pPK607 |
| PKY4405 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK587 |
| PKY4407 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::kanMX + pPK607 |
| PKY4413 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::natMX rtt101Δ::kanMX + pPK587 |
| PKY4414 | MATa (hht1-hhf1)Δ::LEU2 (hht2-hhf2)Δ::HIS3 rtt109Δ::natMX rtt101Δ::kanMX + pPK607 |
Figure 4.
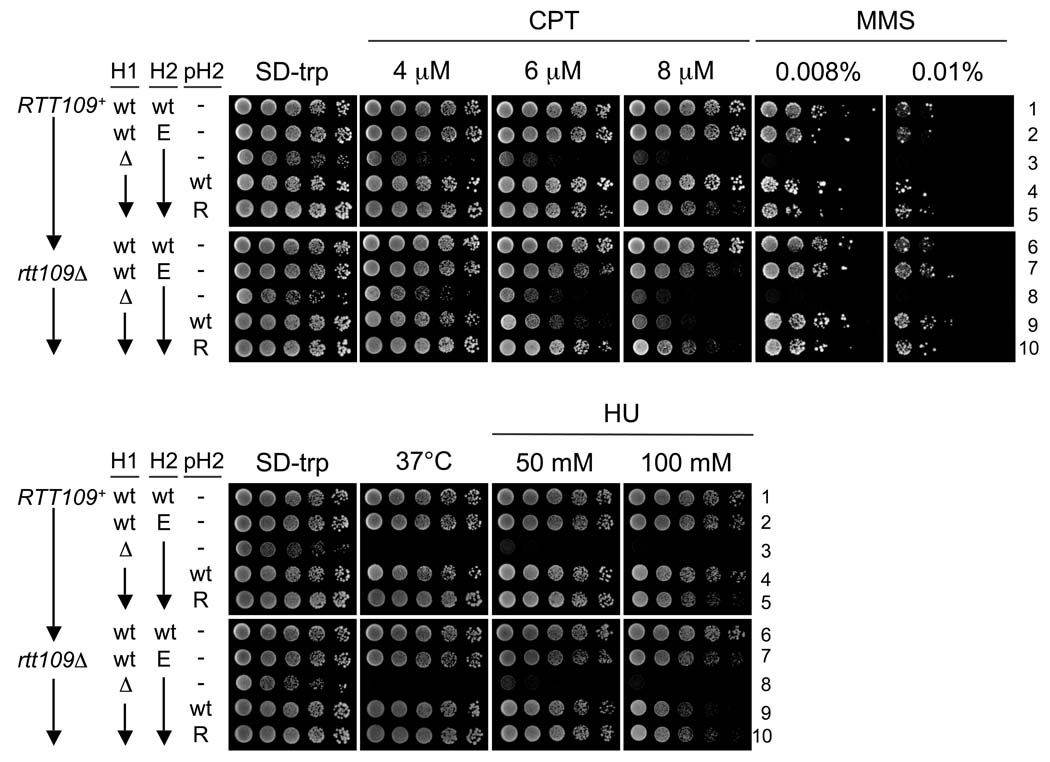
Damage sensitivity caused by the H3-K56E allele. Five-fold serial dilutions were spotted onto SD-trp plates containing the indicated compounds. Strains one through six are RTT109+, and seven through ten are rtt109Δ. wt (1 & 6: PKY4399); hht2-K56E (2, 7: PKY4401, PKY4400); hht1Δ hht2-K56E (3, 8: PKY4375, PKY4377); hht1Δ hht2-K56E + p[HHT2-HHF2] (4, 9: PKY4376, PKY4378); hht1Δ hht2-K56E + p[hht2(K56R)-HHF2]; (5, 10: PKY4366, PKY4369). p[HHT2-HHF2] and p[hht2(K56R)-HHF2], referred to in the figure as pH2 “wt” and “R”, are plasmids containing the HHT2-HHF2 genes and encode either wild-type (pPK587) or K56R (pPK589) H3. “H1” and “H2” refer to the HHT1-HHF1 and HHT2-HHF2 gene pairs. Unless otherwise noted, the strains were incubated at 30°C.
Figure 5.
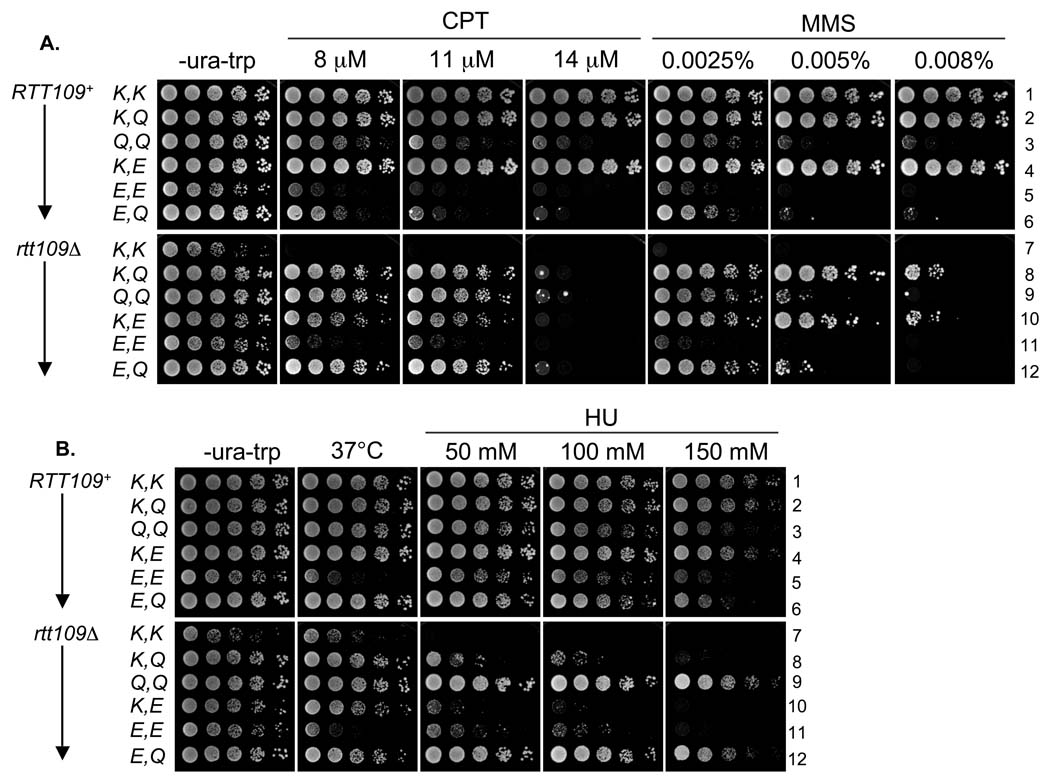
Comparison of K56E and K56Q HHT2 alleles. As in Figure 4, except the drop-out media additionally lacked uracil. The strains analyzed are deleted for both chromosomal histone H3/H4 gene pairs and are either RTT109+ (strains 1–6) or rtt109Δ (strains 7–12). The different HHT2 allele combinations were generated using parallel plasmid sets marked with either URA3 or TRP1. The strains are as follows: 1: PKY4387, 2: PKY4388, 3: PKY4390, 4: PKY4389, 5: PKY4392, 6: PKY4391, 7: PKY4393, 8: PKY4394, 9: PKY4396, 10: PKY4395, 11: PKY4398 and 12: PKY4397.
Figure 8.
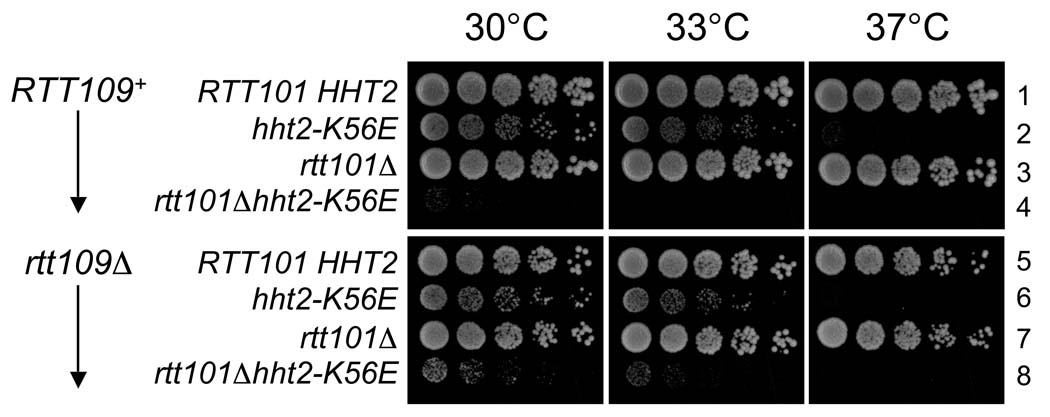
Relationship between H3 K56E and RTT101. Five-fold serial dilutions of the indicated strains were spotted onto YPD plates and incubated at 30, 33 or 37°C. 1: PKY4402, 2: PKY4404, 3: PKY4409, 4: PKY4410, 5: PKY4405, 6: PKY4407, 7: PKY4413 and 8: PKY4414.
Figure 6.
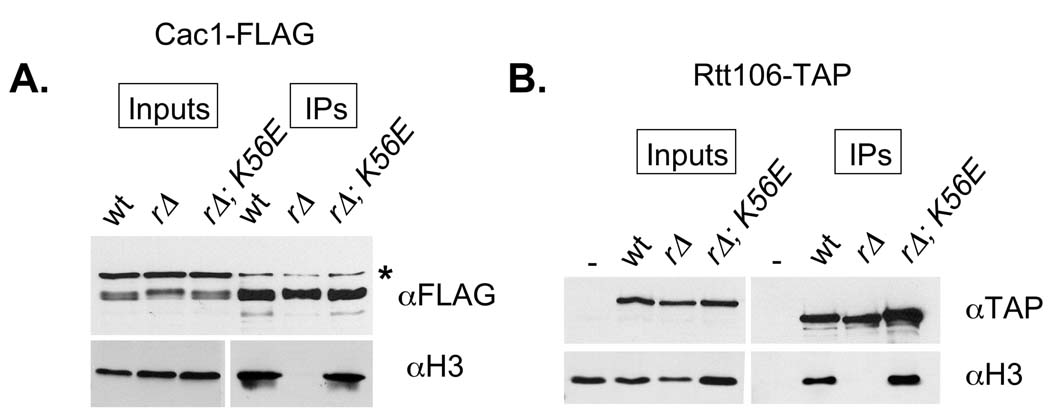
H3-K56E restores histone binding to CAF-1 and Rtt106. A. Cac1-FLAG was immunoprecipitated from the indicated strains. Relative Cac1-FLAG and H3 levels in each extract were determined by blotting with anti-FLAG (Sigma) and anti-H3 antibodies (Abcam), respectively. These lanes are labeled “inputs”. The amount of Cac1-FLAG and H3 recovered in the anti-FLAG immunoprecipitates is shown in the lanes labeled “IPs”. B. Rtt106-TAP complexes were recovered from the indicated strains, and analyzed for Rtt106-TAP and histone H3 content. Cac1-FLAG strains: wt (PKY1252), rtt109Δ (PKY4426) and rtt109Δ hht2-K56E (PKY4427). Rtt106-TAP strains: wt (PKY4429), rtt109Δ (PKY4430) and rtt109Δ hht2-K56E (PKY4431).
2.2 CAF-1 and Rtt106 purification
With slight revision, CAF-1 and Rtt106 were purified according to previously published protocols [36]. Three liters of each strain were cultured to an OD600 of approximately 2.0 and harvested by centrifugation. The cells were then washed once with water, and once with Buffer A containing 10% glycerol and PMSF (1 mM). In liquid nitrogen, the cells were drawn into flash-frozen “noodles”, and subsequently ground to a fine powder in a frozen mortar. The resulting yeast powder was resuspended in 9 ml of Buffer A containing 15 KU/ml DNase I, 1 mM PMSF, 1 mM benzamidine, 1 µg/ml E64, 1.1 µg/ml phosphoramidon, 0.5 µg/ml leupeptin, 0.7 µg/ml pepstatin and 1 µg/ml aprotinin and extracted for 30 minutes at 4°C. The extracts were then supplemented with ethidium bromide (75 µg/ml), and incubated for an additional 30 minutes. Extracts were centrifuged for 15 min at approximately 2000 ×g, and then further clarified by centrifugation for 1 hr at 100,000 × g. Cac1-FLAG complexes were retrieved from extracts (100 mg of total protein) by overnight co-incubation with 60 µl of anti-FLAG resin. The following day, the immunoprecipitates were washed extensively with Buffer A containing 200 mM NaCl, and the Cac1-FLAG complexes were eluted with FLAG peptide (0.4 mg/ml) for 30 min at 4°C. The eluates were TCA precipitated overnight and then analyzed for Cac1 and H3 content by blotting with anti-FLAG (Sigma) and anti-H3 (Abcam) antibodies. The Rtt106-TAP purifications were conducted in a similar manner, with the addition of a CL6B pre-clearing step following the 100,000 × g high-speed spin. The TAP-tagged complexes were recovered on IgG Sepharose beads and subsequently released with TEV protease (100 µg of enzyme total, added in 50 µg aliquots over the course of 16 hrs). The eluates were then TCA precipitated and analyzed for Rtt106 (Open Biosystems anti-TAP antibody) and H3 content by Western blotting.
3. RESULTS
3.1 An extragenic suppressor of rtt109Δ
To gain insight into the molecular function of Rtt109 in promoting genome stability, we screened a GAL1 overexpression library for genes that could suppress the sensitivity of rtt109Δ mutants to the DNA alkylating agent, methanesulfonate (MMS). Out of the 3.6 ×106 transformants screened, we isolated a single candidate suppressed strain. However, the MMS-resistance of this isolate proved to be plasmid-independent (data not shown), suggesting that the suppressed phenotype was caused by a chromosomal mutation. We therefore tested whether the suppressor segregated in a Mendelian manner as a single genetic locus. To do so, the suppressed strain was crossed to an unsuppressed, damage-sensitive rtt109Δ strain, and segregation of the damage-sensitivity phenotype was assessed. All the progeny of this cross were genotypically rtt109Δ and would therefore be damage-sensitive unless otherwise suppressed. We observed that two of every four spores were DNA damage-resistant, indicating that a single mutated locus was responsible for the suppressed phenotype (Figure 1A). We therefore referred to the mutated locus as sup−, assuming it represented an altered version of a gene termed SUP+.
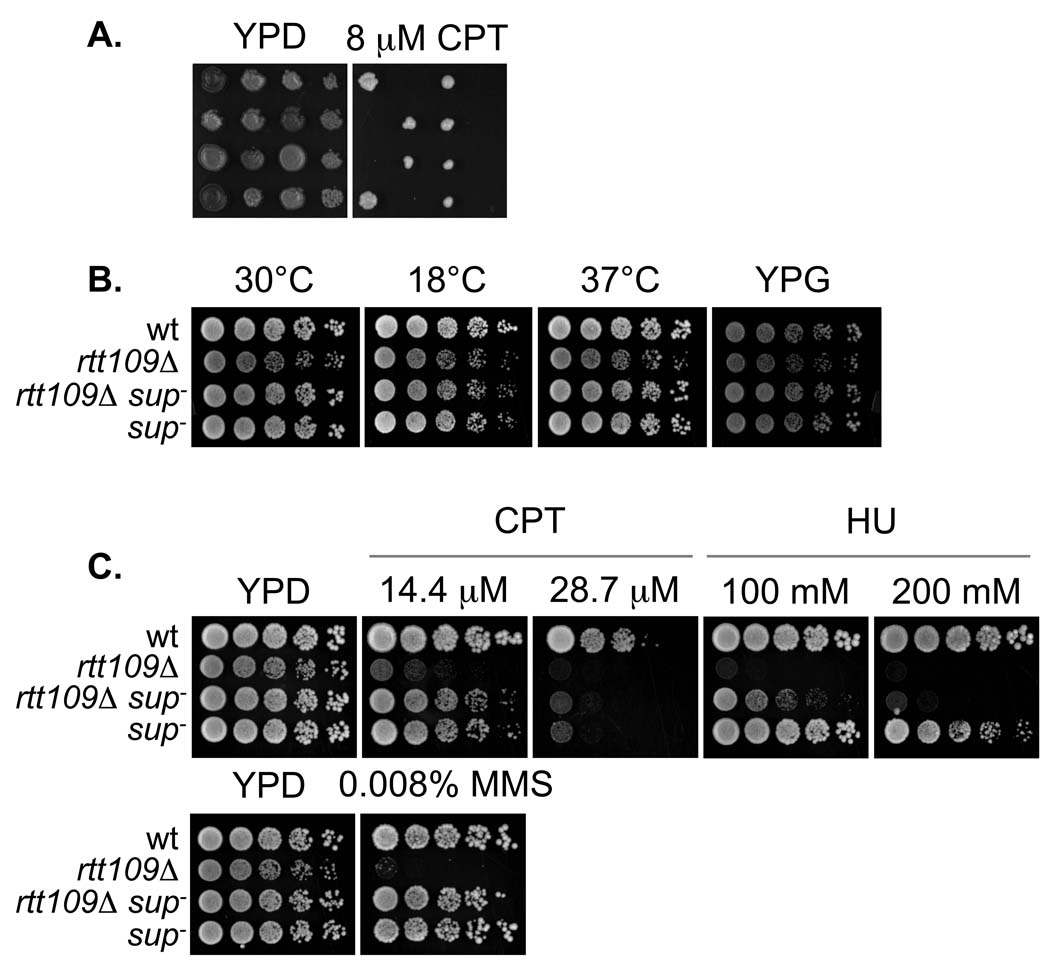
Figure 1.
Genetic characterization of the suppressor mutation. A. sup− segregates as a single genetic locus. The progeny of PKY4342 (rtt109Δ sup−) and PKY4176 (rtt109Δ) were spotted onto YPD or YPD with 8 µM CPT, and incubated at 30°C. Each horizontal row of four strains represents the progeny from a complete meiotic tetrad. B. Growth at low and high temperature and on YP glycerol (YPG). Five-fold serial dilutions of wt (PKY3310), rtt109Δ (PKY4170), rtt109Δ sup− (PKY4345) and sup− (PKY4347) strains were spotted onto either YPD (first three panels) or YPG. Unless otherwise indicated, the plates were incubated at 30°C. C. As in B, except the strains were spotted onto media containing CPT, HU or MMS.
While investigating the identity of the SUP gene, we wished to determine whether it displayed genetic interactions with other components of the Rtt109 damage tolerance pathway. To facilitate these efforts, we first tested whether sup− cells that had a wild-type RTT109 gene displayed any growth or damage sensitivity phenotypes. The sup− and rtt109Δ mutations were separated by crossing a rtt109Δ::kanMX sup− double mutant to a wild-type strain. We isolated the RTT109+ sup− (G418-sensitive) sisters from tetrads that yielded 2 out of 4 unsuppressed (that is, damage-sensitive and G418-resistant) rtt109Δ::kanMX SUP+ spores. We tested the resulting RTT109+ sup− strains for temperature and cold sensitivities, as well as growth on the alternate carbon source glycerol. However, the growth of the wt and sup− strains were indistinguishable (Figure 1B). We also compared growth of RTT109+ sup− and rtt109Δ sup− strains on medium containing a variety of genotoxic agents, including camptothecin (CPT), MMS and hydroxyurea (HU) (Figure 1C). We observed that rtt109Δ sup− strains were resistant to not only MMS, but also to CPT (as also demonstrated in Figure 1A) and HU, and that the RTT109+ sup− strains were largely insensitive to these compounds, except at very high CPT concentrations (Figure 1C). Together these results demonstrate that the sup− mutation alone does not significantly perturb cellular growth or resistance to DNA damage, but that in the absence of Rtt109, sup− restores resistance to a wide variety of genotoxic agents.
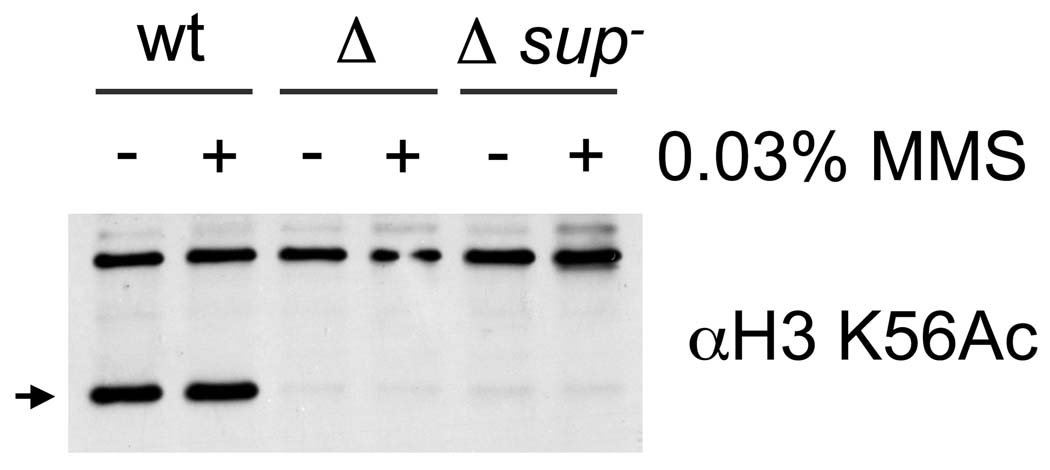
One possible, albeit unlikely, mechanism explaining suppression of rtt109Δ by sup− was the restoration of H3 K56 acetylation via an Rtt109-independent mechanism. To test this idea, extracts prepared from untreated and MMS-treated log phase cultures were probed for H3 K56ac by Western blotting. In neither case was H3 K56ac detected in the rtt109Δ sup− samples (Figure 2) indicating that suppression in this strain did not involve H3 K56 acetylation by a HAT other than Rtt109.
Figure 2.
H3 K56 acetylation is not restored in rtt109Δ sup− mutants. Alkaline lysis extracts [51] prepared from wt (PKY3310), rtt109Δ (PKY4170) and rtt109Δ sup− (PKY4342) strains were probed for H3 K56ac using an anti-H3 K56ac peptide antibody (21st Century). MMS indicates a prior 1 hr treatment of the cells with 0.03% MMS. The arrow marks the position of the H3 K56ac-specific band.
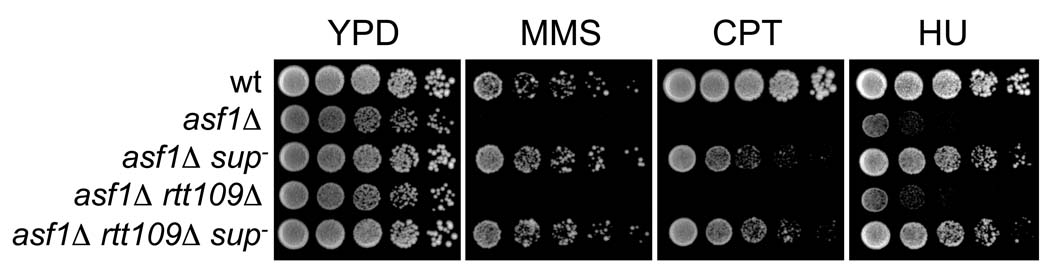
Given the established relationship between H3 K56ac and Rtt109 in damage tolerance [15,16], we predicted that the suppressor would restore damage resistance to other mutants that are defective in acetylating H3 K56. We tested this hypothesis by determining whether the damage sensitivity of asf1Δ mutants could be suppressed by sup−. Indeed, asf1Δ sup− double mutants were MMS-, CPT- and HU-resistant (Figure 3). To confirm that H3K56 acetylation was the target, we also tested the effects of an un-acetylatable H3 K56R mutant allele. To generate the strains needed for this experiment, an rtt109Δ sup− strain was crossed to a “histone shuffle” strain in which both chromosomal histone H3/H4 gene pairs (HHT1-HHF1 and HHT2-HHF2) were deleted and a point-mutated hht2(K56R)-HHF2 gene pair was carried on a plasmid. However, we found that none of the eleven rtt109Δ (hht1-hhf1)Δ (hht2-hhf2)Δ p[hht2(K56R)-HHF2] progeny isolated were MMS- or CPT-resistant. These results suggested that either sup− did not suppress the K56R mutation or that sup− itself resided at one of the two deleted histone gene loci and therefore segregated away from the marked histone gene deletions. Indeed, in twenty-one MMS-resistant rtt109Δ spore colonies isolated in this cross (all of which had retained the K56R plasmid (see below)), HHT1-HHF1 but not HHT2-HHF2 was deleted, indicating that the suppressor mutation was linked to the HHT2-HHF2 locus.
Figure 3.
sup− suppresses the DNA damage sensitivity of asf1Δ mutants. Five-fold serial dilutions of wt (PKY3310), asf1Δ (PKY3328), asf1Δ sup− (PKY4355), asf1Δ rtt109Δ (PKY4356) and asf1Δ rtt109Δ sup− (PKY4353) strains were spotted onto YPD plates containing 0.008% MMS, 8 µM CPT or 50 mM HU.
3.2 Distinct phenotypes of the suppressor, H3K56E
Sequencing the HHT2-HHF2 locus from the suppressed strain revealed a single nucleotide mutation in the HHT2 ORF, converting lysine 56 of H3 to glutamic acid. Therefore, an acidic residue replacement of lysine 56 substantially bypassed the cellular requirements for the Rtt109/Asf1 enzyme pair. Also, we noted that all of the rtt109Δ (hht1-hhf1)Δ hht2-K56E strains isolated during this cross had maintained the plasmid containing the hht2-K56R gene, although there had been no nutritional pressure to do so. Because a K56E allele causes slow growth when provided as the sole source of H3 [10] this suggested that the plasmid was maintained because the encoded H3-K56R molecules counteracted the deleterious effects of H3-K56E. To systematically test this idea, we constructed a series of related strains differing in histone gene and RTT109 status (see Methods).
The (hht1-hhf1)Δ hht2-K56E strains were indeed extremely slow growing as well as temperature- and DNA damage-sensitive, regardless of the presence of Rtt109 (Figure 4, strains 3 and 8). As expected, the slow-growth, ts, HU-, CPT-and MMS-sensitivity phenotypes were suppressed by introduction of a wild-type H3 gene, either in the chromosome or on a plasmid (Figure 4, strains 2 and 4). Similarly, providing H3-K56R to strains expressing H3-K56E resulted in strong resistance to high temperature and CPT, MMS and HU (Figure 4, strains 5 and 10). Therefore, co-expression of either wild-type H3 or H3-K56R suppresses the genotoxic hypersensitivity associated with H3-K56E; this in turn allows H3-K56E to bypass the phenotypes associated with loss of Rtt109. These observations are consistent with two sequential effects of H3-K56E, in which it bypasses the need for H3-K56 acetylation prior to deposition, but subsequently causes defects in chromosome function.
We then determined whether K56 glutamate and glutamine substitutions conferred similar phenotypes, because K56Q substitutions are known to partially suppress the damage sensitivities of asf1Δ and rtt109Δ mutant [24,45]. To compare strains with the same number of histone gene copies without having to replace chromosomal histone genes, we made episomal constructs carrying wild-type, K56E and K56Q alleles and analyzed the phenotypes caused by different pairwise allele combinations. In this manner, we made three observations regarding the CPT- and MMS- resistance phenotypes (Figure 5A): (1) Consistent with Figure 4, wild-type resistance to high CPT and MMS concentrations was observed only when Rtt109 was present and one or both of the two H3 alleles was a reversibly acetylatable lysine (Figure 5A, strains 1, 2, 4). (2) As above (Figure 4, strains 3 and 8), cells expressing only H3-K56E displayed little resistance to either CPT or MMS, regardless of the presence of Rtt109 (Figure 5, strains 5 and 11). (3) Intermediate but not full MMS or CPT resistance was observed for rtt109Δ mutants in which one histone gene was either K56E or K56Q (strains 8, 10 and 12) or both were K56Q (strain 9). We conclude that lysine 56 acetylation by Rtt109 is required for full CPT- and MMS-resistance, and that either an uncharged (Q) or negatively charged (E) substitution only partially compensates for this phenotype. Therefore, neither Q nor E appears to be a fully functional mimic of acetylated lysine. We note that K56Q/K56Q strains were moderately CPT- and MMS-sensitive (Figure 5A, strains 3 and 9), and displayed less temperature and damage sensitivity than the K56E/K56E strains. In light of these observations, we propose that K56Q and E bypass the need for H3 K56ac prior to histone deposition (see next section), but that chromatin made with these histones is not fully functional.
In these analyses, CPT and MMS generally caused similar phenotypes. In contrast, some different phenotypes were observed in the presence of HU. For example, the K56Q allele suppressed the HU-sensitivity of rtt109Δ cells much more efficiently than it suppressed CPT- or MMS-sensitivity (Figure 5, strains 9 and 12). The reason for these differences is not known; but, because it seems likely that they manifest after deposition of histones, we speculate that the mild chromatin perturbation in K56Q/K56Q cells may exacerbate replication fork malfunction in the presence of MMS and CPT, but not HU.
3.3 Biochemical analysis of H3K56E
Recently, it has been shown that H3 K56 acetylation stimulates the association of H3 with the histone chaperones CAF-1 and Rtt106, suggesting that K56 acetylation promotes genome stability by ensuring efficient histone chaperone loading during DNA replication/repair [36]. In support of this model, chromatin re-assembly at sites of repaired DNA damage is dependent on chromatin assembly proteins CAF-1 and Asf1 [33]. We therefore hypothesized that K56E suppresses rtt109Δ phenotypes by restoring the interaction between histones and CAF-1/Rtt106. To test this idea, we FLAG epitope-tagged the Cac1 subunit of CAF-1 in rtt109Δ and rtt109Δ hht2-K56E strains and examined the recovery of histones in anti-FLAG immunoprecipitates. As observed previously, CAF-1 complexes from wild-type cells contained easily-detected levels of H3, but those from rtt109Δ mutants did not ([36]; Figure 6A). Notably, H3 association with CAF-1 was restored by the hht2-K56E mutation. Equivalent results were observed when we examined H3 association with the Rtt106 histone deposition protein (Figure 6B). Combined, these results demonstrate that suppression of rtt109Δ by K56E is correlated with restored loading of histones onto histone chaperones. Because K56E restored levels of histone association resembling those observed in wild-type cells, these data support the idea that the damage sensitivity phenotypes of K56E cells reflect chromatin defects subsequent to histone deposition.
3.4 Role of the H3K56 deacetylases, Hst3/4
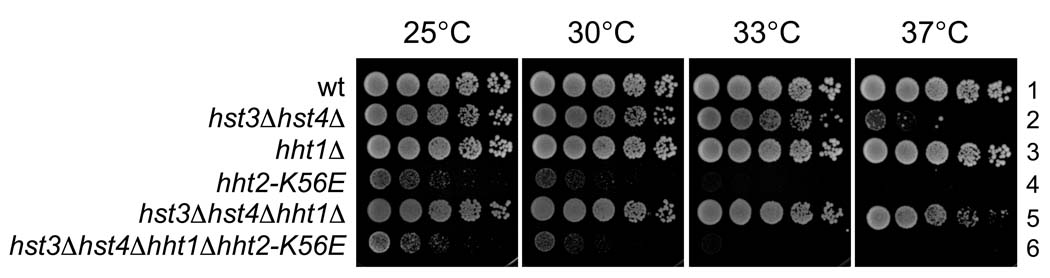
The sirtuin family deacetylases Hst3 and Hst4 downregulate H3 K56ac at the end of S phase, erasing this mark from histones deposited into chromatin during DNA replication [30–32]. Strains lacking both of these enzymes are extremely susceptible to DNA damaging agents and are dependent on DNA repair mechanisms for survival [31,46], suggesting that constitutive hyperacetylation of H3K56 in chromatin interferes with DNA damage repair or the subsequent recovery process. Because our data suggested that H3K56E also causes genome instability at a step subsequent to histone deposition, we therefore tested whether H3K56E displayed genetic interactions with hst3Δ hst4Δ gene deletions. We found that the K56E strains were more temperature- and damage-sensitive than hst3Δ hst4Δ double mutant cells (Figure 7, strains 4 and 2) and that combining these mutations did not exacerbate either of these phenotypes (strain 6). This suggests that these mutations affect the same process, and that chromosome instability observed in hst3Δ hst4Δ cells [31] is a less severe version of the phenotypes we observed in H3-K56E cells. These data are consistent with our previous conclusion that chromatin built solely with H3K56E functions more poorly than chromatin that is constitutively acetylated (hst3Δ hst4Δ) or contains only the H3K56Q acetyl mimic.
Figure 7.
Relationship between HST3 and HST4 gene deletions and H3 K56E. Five-fold serial dilutions of the indicated strains were spotted onto YPD plates and incubated at 25, 30, 33 or 37°C. wt (1: PKY3310), hst3Δ hst4Δ (2: PKY4220), hht1Δ (3: PKY388), hht2-K56E (4: PKY4375 without pRS414), hht1Δ hst3Δ hst4Δ (5: PKY4420) and hht1Δ hst3Δ hst4Δ hht2-K56E (6: PKY4415).
In the course of building the strains for these experiments, we also discovered that deleting one of the two H3/H4 gene copies strongly suppressed the temperature sensitivity of the hst3Δ hst4Δ double mutant strain (Figure 7, strain 5). Therefore, reducing histone copy number mitigates the effects of constitutive H3K56 acetylation, possibly by reducing the rate and/or improving the location of histone deposition.
To further explore the relationship between the K56E and hst3Δ hst4Δ phenotypes, we examined genes that function downstream of H3 K56 acetylation. Specifically, we determined whether a rtt101Δ deletion, a known suppressor of hst3Δ hst4Δ mutants [22], would also suppress the temperature sensitivity caused by H3-K56E. To do so, we deleted RTT101 from a “histone shuffle” strain, which lacks both chromosomal histone H3/H4-encoding genes and carries wild-type histone H3/H4 genes on a low copy plasmid. We then introduced H3-K56E/H4 on a plasmid, and screened for K56E transformants that had stochastically lost the wild-type histone plasmid. This event occurred with very low frequency, and the few resulting rtt101Δ H3-K56E double mutant strains were extremely slow growing, far more so than strains with only the K56E mutation (Figure 8). Thus, the rtt101Δ gene deletion can cause strikingly different phenotypes: it suppresses the temperature sensitivity of hst3Δ hst4Δ cells [22], but greatly exacerbates the growth defects of H3K56E cells. Therefore, we conclude that although the chromatin defects associated with K56E and constitutively acetylated H3 K56 are related (Figure 7), they are indeed distinct, because rtt101Δ suppresses the defects associated with constitutive acetylation but not K56E (Figure 8).
4. DISCUSSION
We summarize the major novel findings in this work here: 1. H3K56E suppresses the damage-sensitivity of rtt109 and asf1 cells. 2. This suppression requires a wild-type copy of H3 because H3K56E alone causes severe damage sensitivity. 3. H3K56E supports efficient chromatin assembly factor binding. 4. The temperature-sensitivity caused by H3K56E is not suppressed by deletion of RTT101. We discuss each of these in the following Discussion section.
4.1 Bypass of Rtt109 requires removing the positive charge from H3 residue 56
In our screen for suppressors of the DNA damage sensitivity of rtt109Δ cells, we isolated a strain containing a point mutation in the histone H3 gene HHT2, converting lysine 56 to glutamic acid. We found that H3-K56E strongly suppressed the damage sensitivity of cells lacking Rtt109 (Figure 1 and Figure 3) and also restored efficient histone binding to CAF-1 and Rtt106 (Figure 6). This correlation suggests that suppression by K56E involves facilitating chromatin assembly. Glutamic acid (negatively charged) and glutamine (neutral) substitution of H3 K56 both functionally mimic the acetylated form of this residue. Therefore, we propose that the purpose of lysine 56 acetylation, rather than to produce acetyl-lysine per se, is to eliminate the positive charge of lysine 56, which appears to impede binding to multiple chromatin assembly proteins. How this single charge neutralization affects binding to a variety of unrelated downstream proteins is an important question raised by these studies.
4.2 Dual effects of H3-K56E
Cells expressing H3-K56E as their only source of histone H3 are both temperature- and DNA damage-sensitive (Figure 4, Figure 5). Notably, suppression of rtt109Δ by H3-K56E is dependent on a second source of H3 to prevent these defects. Therefore, H3 K56E imparts seemingly antagonistic properties to H3: in cells expressing wild-type H3, H3-K56E promotes resistance to genotoxic agents by re-instating efficient chromatin assembly but causes sensitivity in the absence of other histones. Because chromatin assembly factor-H3 interactions appear normal in cells expressing H3-K56E (Figure 6), these data suggest that the deleterious effects of H3-K56E are manifest subsequent to assembly factor loading. We propose that nucleosomes containing H3-K56E malfunction in a manner that causes sensitivity to genotoxic agents, and that this defect is suppressed if wild-type histones are also present. Based on their proximity, we speculate that interactions between DNA and the H3 αN helix are perturbed by the negative charge of K56E, but that the chromatin fiber seems able to withstand this if half the H3 in cells is wild-type.
We made two other observations that further support the modulation of chromosome defects by altered histone supply. First, as was the case for H3-K56E, the damage sensitivity of H3 K56Q mutants was alleviated by introducing a second, wild-type H3 gene (Figure 4 and Figure 5). Second, the temperature sensitivity of hst3Δ hst4Δ cells was suppressed by deleting one of the two H3/H4 gene copies. We conclude that the genotoxic agent sensitivity associated with constitutively acetylated H3 K56 can be avoided by either reducing the amount of histones present (in hst3Δ hst4Δ cells) or by diluting them with wild-type H3 molecules that can be acetylated reversibly (in K56Q/E cells).
4.3 Comparison of H3K56E to H3K56Q
Multiple phenotypes caused by defects in H3 K56ac are suppressed by the uncharged acetyl mimic H3 K56Q, including damage sensitivity [24,45] and defects in transcription induction [47]. However, we found that unlike hst3Δ hst4Δ mutants, H3 K56Q mutants are not temperature sensitive (Figure 5 and Figure 7), suggesting that K56Q causes less severe chromosomal abnormalities than does constitutive K56 acetylation [30,31]. These and other data (Figure 4, Figure 5) reinforce our conclusion that K56E and K56Q are similar but not identical to H3 K56 acetyl, and that the differences between them are manifest subsequent to assembly factor loading. Likewise, the more severe growth defects of K56E cells compared to hst3Δ hst4Δ cells (Figure 7) suggests a constitutive negative charge at H3 residue 56 impedes chromosome function more than does constitutive acetyllysine. Notably, deletion of HST3 and HST4 cells does not exacerbate the growth defects of K56E cells (Figure 7), supporting the idea that K56 is the relevant substrate for these deacetylases with regard to the phenotypes we examined.
4.4 Signaling downstream of H3-K56ac
While it is evident that H3 K56 acetylation facilitates histone chaperone/histone interactions, several types of data suggest that the deposited H3-K56ac molecules subsequently signal to other proteins, and thereby contribute further to genome stability. For example, overexpression of RFC1, encoding the large subunit of the DNA replication clamp-loader complex RFC, suppresses the temperature- and genotoxic-sensitivity of hst3Δ hst4Δ mutant cells [46]. The suppression is not accompanied by reduced H3K56ac levels, suggesting instead that increasing the amount of Rfc1 allows cells to tolerate chromatin with constitutive H3 K56acetyl. Furthermore, deletion of the alternate clamp-loader subunits CTF18, RAD24 and ELG1 also suppresses hst3Δ hst4Δ phenotypes [46]. Because all these clamp loader subunits interact with the same Rfc2-5 subcomplex, these data suggest that the response to H3K56 acetylation affects clamp loader complex formation. Lastly, loss of Ctf4, a protein involved in sister chromatid cohesion that interacts with replication fork progression proteins also suppresses hst3Δ hst4Δ phenotypes [46]. Together, these data suggest that H3K56 acetylation can impact genome stability by alteration of DNA replication protein functions.
There are also well-established relationships between H3 K56 acetylation and components of the Rtt101Mms1/Mms22 ubiquitin ligase complex [22]. Deletion of any of these genes suppresses the temperature sensitivity caused by constitutive H3 K56 acetylation [22]. The Rtt101 cullin subunit is recruited to chromatin in response to DNA damage, in a way that is dependent on Rtt109 [43]. Therefore, Rtt101 carries out its genome protective function downstream of H3 K56ac, and is required for the pathological response that occurs in the presence of constitutive H3K56ac.
Our data had shown that the temperature sensitivity phenotypes caused by H3 K56E were more severe than those resulting from constitutive H3 K56 acetylation, and that K56E and hst3Δ hst4Δ mutations do not cause synergistic defects (Figure 7). We therefore tested whether the downstream response lead by Rtt101 would respond similarly to constitutive H3K56 acetylation and to H3K56E. However, we found that the temperature sensitivity of K56E cells was not suppressed by a rtt101Δ deletion, and in fact these double mutants were worse off than K56E cells (Figure 8). These data therefore show that Rtt101 is not required for the temperature sensitivity caused by H3 K56E, and are consistent with a H3 K56ac-independent role(s) for Rtt101 in cell metabolism. In fact, it has recently been reported that Rtt101 contributes to the destruction of non-functional ribosome complexes [48].
A major question raised by these studies is how alterations at residue 56 affect chromatin function after deposition. The negatively charged glutamate we recovered here may simply provide charge repulsion from the nearby DNA, enhancing the off rate of DNA at the nucleosome entry/exit points [49]. Alternatively, protein interactions with factors involved in histone turnover or remodeling could be affected. Indeed, a H3-K56R mutation promotes chromatin reassembly during repression of the PHO5 promoter [47] and reduces recruitment of the Snf5 remodeling protein to a histone gene promoter [11]. Therefore, the transient placement of H3K56ac into the genome [12,50] may provide a brief window of opportunity for the localized action of a large number of factors.
Acknowledgments
We thank the members of the Kaufman Lab for guidance with protocols and project direction, and Dave Allis for yeast strains. This work was funded by NIH grants 1 F32 GM075601-01 (JAE) and R01 GM55712 and NSF grant MCB-0641390 (PDK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest
References
- 1.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 4.Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation: a core domain modification associated with chromatin assembly. Mol. Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 7.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 9.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 10.Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 11.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 13.Game JC, Williamson MS, Baccari C. X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics. 2005;169:51–63. doi: 10.1534/genetics.104.028613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider J, Bajwa P, Johnson FC, Bhaumik SR. A. Shilatifard, Rtt109 is required for proper H3K56 acetylation: A chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 16.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- 19.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16:1503–1510. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, Marmorstein R. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat. Struct. Mol. Biol. 2008;15:738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 24.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 26.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 402;1999:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 27.Franco AA, Lam WM, Burgers PM, Kaufman PD. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramey CJ, Howar S, Adkins M, Linger J, Spicer J, Tyler JK. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 2004;24:10313–10327. doi: 10.1128/MCB.24.23.10313-10327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of Lysine 56 of Histone H3 Catalyzed by RTT109 and Regulated by ASF1 Is Required for Replisome Integrity. J. Biol. Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 30.Maas NL, Miller KM, Defazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 16;2006:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Thaminy S, Newcomb B, Kim J, Gatbonton T, Foss E, Simon J, Bedalov A. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J. Biol. Chem. 2007;282:37805–37814. doi: 10.1074/jbc.M706384200. [DOI] [PubMed] [Google Scholar]
- 33.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szyjka SJ, Aparicio JG, Viggiani CJ, Knott S, Xu W, Tavare S, Aparicio OM. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 2008;22:1906–1920. doi: 10.1101/gad.1660408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA Integrity Network in the Yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 39.Araki Y, Kawasaki Y, Sasanuma H, Tye BK, Sugino A. Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability. Genes Cells. 2003;8:465–480. doi: 10.1046/j.1365-2443.2003.00648.x. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin EL, Berger AC, Corbett AH, Osheroff N. Mms22p protects Saccharomyces cerevisiae from DNA damage induced by topoisomerase II. Nucleic Acids Res. 2005;33:1021–1030. doi: 10.1093/nar/gki246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr. Biol. 2006;16:786–792. doi: 10.1016/j.cub.2006.02.071. [DOI] [PubMed] [Google Scholar]
- 42.Duro E, Vaisica JA, Brown GW, Rouse J. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 2008;7:811–818. doi: 10.1016/j.dnarep.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Roberts TM, Zaidi IW, Vaisica JA, Peter M, Brown GW. Regulation of Rtt107 recruitment to stalled DNA replication forks by the cullin Rtt101 and the Rtt109 acetyltransferase. Mol. Biol. Cell. 2008;19:171–180. doi: 10.1091/mbc.E07-09-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas B, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;58:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 45.Miller A, Yang B, Foster T, Kirchmaier AL. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics. 2008;179:793–809. doi: 10.1534/genetics.107.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celic I, Verreault A, Boeke JD. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics. 2008;179:1769–1784. doi: 10.1534/genetics.108.088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009;23:963–974. doi: 10.1101/gad.1775609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 50.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]