Abstract
Background and Purpose
The cerebral volume of T2-hyperintense white matter (HWM) is an important neuroimaging marker of cerebral integrity. Pathophysiology studies identified that subcortical and ependymal HWM are produced by two different mechanisms but shared a common risk factor: high arterial pulse pressure. Recent studies have demonstrated high heritability of the whole-brain (WB) HMW volume and reported significant and suggestive evidence of genetic linkage. We performed heritability and whole-genome linkage analysis to replicate previous reported findings and to study shared genetic variance, and possible overlap for specific loci, between subcortical and ependymal HWM volumes in a population of healthy Mexican Americans.
Methods
The volumes of subcortical and ependymal HWM regions were measured from high-resolution (1mm3), 3D-FLAIR images acquired for 459 (283/176 females/males) active participants in the SAFH study. Subjects ranged in age from 19 to 85 years of age (47.9±13.5years) and were part of 49 families (9.4±8.5 individuals/family).
Results
The volumes of WB, subcortical and ependymal HWM were highly heritable (h2=.72;.66;.73 respectively). The subcortical and ependymal HWM volumes shared 21% of genetic variability indicating significant pleiotropy. Genome-wide linkage analysis showed only a suggestive bivariate linkage for subcortical and ependymal HWM volumes (LOD = 2.12) on chromosome 1 at 288cM.
Conclusion
We replicated previous findings of high heritability for the WB-HWM volume. We also showed that subcortical and ependymal volume shared a significant portion of genetic variability and the bivarate linkage analysis produced a suggestive linkage near the locus previously identified in a study of WB-HWM volume and arterial pulse pressure.
Keywords: Brain Imaging, Genetics, MRI, White Matter Disease, Aging, Hyperintense White Matter, Structural imaging
Background and Purpose
The volume of T2-hyperintense white matter (HWM) regions is an important neuroimaging marker of cerebral integrity. In normal aging, HWM regions begin to form during mid-adulthood (4–5thth decade of life) and their onset and progression is associated with declines in other indices of cerebral integrity1. Increasing volume of HWM regions is highly correlated with cerebral blood flow decline2 and reduced glucose metabolism3. In addition, increasing numbers and volume of HWM regions is linked to age related cognitive declines, particularly in executive functioning4, processing speed and general cognitive status5. Although the pathogenic mechanisms of HWM are unclear, histopathological and imaging findings indicate that whole-brain HWM (WB-HWM) volume includes at least two distinct mechanisms with distinct etiologies6. Subcortical HWM regions are more closely associated with ischemic factors6. In contrast, subependymal, periventricular HWM are thought to be of non-ischemic origin and potentially produced by pulse-wave encelophaty7–9.
WB-HWM volumes are significantly influenced by genetic factors, with heritability estimates ranging between .55–.7310–13. Two recent genome-wide linkage studies reported significant and suggestive evidence of linkage for WB-HWM13, 14. DeStefano and colleagues reported significant linkage (LOD=3.69) for the WB-HWM volume at 4cM on the chromosome 4 in 747 healthy individuals from 237 families14. It was proposed that genes responsible for mitochondrial functioning, located near this region, may modulate volume of HWM during normal aging process. The second study performed whole-genome linkage analysis for WB-HWM in a population of 488 hypertensive adults13. Univariate linkage analysis only reached suggestive significance and did not overlap with genetic loci reported DeStefano and colleagues14, possibly due to its focus on individuals with hypertension. Multivariate analysis reported many highly significant loci for WB-HWM and quantitative measurements of hypertension, suggesting a high degree of pleiotropy.
Neither of the previous studies separated the WB-HWM volume into the subcortical and ependymal components. In the current manuscript we pursued three goals: 1) to replicate findings of high heritability for WB-HWM volume; 2) to perform a novel analysis of shared genetic variance, and possible overlap for specific loci, between subcortical and ependymal HWM volumes; and 3) to search for chromosomal regions influencing HWM volume in a Mexican American population.
Methods
Subjects and measurements
459 (283/176 females/males; average age=47.9±13.5years, range=19–85years) Mexican American participants in the San Antonio Family Heart Study (SAFHS)15 were recruited for this study. Recruited subjects were from large extended pedigree composed of 49 families with the average family size of 9.4±8.5 individuals (range=2–38). At the time of the imaging, 144 subjects were treated for hypertension, 80 subjects were treated for type II diabetes and 52 subjects had both hypertension and diabetes. Subjects were excluded for MRI contraindications, history of neurological illnesses or major neurological event (stroke). Subject’s diagnosis status for hypertension and diabetes were coded as binary covariates. All subjects provided written informed consent on forms approved by the Internal Review Board of the University of Texas Health Science Center at San Antonio.
Imaging was performed at the Research Imaging Center, UTHSCSA, using a Siemens 3T Trio scanner and an eight-channel head coil. 3D T2-weighted imaging data were acquired using a high-resolution (isotropic 1mm), turbo-spin-echo Fluid Attenuated Inversion Recovery (FLAIR) sequence with the following parameters: TR/TE/TI/Flip angle/ETL=5sec/353 ms/1.8s/180°/221. This 3D FLAIR protocol was designed to overcome limitation of 2D, thick-slice (2–5mm), imaging methods reported in prior studies of genetics of HWM10–13. Chosen protocol allowed for sensitive detection of smaller lesions and accurate tracing of lesion boundaries. 3D FLAIR sequence applied in the current study uses a non-selective inversion RF pulse to suppress long T1-relaxation time tissue signal, producing images with improved lesion contrast16 that is highly advantageous over to a dual-echo sequence used in previous studies. Finally, the non-selective inversion RF pulse in 3D FLAIR sequence prevents ventricular CSF pulsation artifacts commonly seen as false-negative hyperintense regions in the 2D FLAIR sequences17.
Measurement of HWM volume from FLAIR images is discussed elsewhere18. In short, FLAIR images were preprocessed by removal of non-brain tissue, registration to the T1-weighted images/Talairach frame and RF inhomogeneity correction. HWM regions were manually delineated in 3D-space using an in-house software (http://ric.uthscsa.edu/mango) by an experienced neuroanatomist with high (r2>0.9) test-retest reproducibility. HWM regions were coded as ependymal regions, contiguous with CSF structures, and subcortical in accordance with9. The WB-HWM volume and the volumes of subcortical and ependymal HWM were measured for each subject.
Genotyping
The details of the genotyping procedure can be found in19. After DNA was extracted from lymphocytes, fluorescently labeled primers from the MapPairs Human Screening set (versions 6 and 8 (Research Genetics, Huntsville, AL, USA)) and PCR were used to amplified 417 microsatellite markers spaced at approximately 10-cM intervals across 22 autosomes. An automated DNA sequencer (ABI Model 377 with Genescan and Genotyper software; Applied Biosystems, Foster City, CA, USA) was used. The average heterozygosity index for these markers was approximately .76. The sex-averaged marker map was confirmed by deCODE genetics20 and markers not on this map were placed by interpolation based on physical location21.
Genotypes were subjected to extensive data cleaning with SimWalk2 software package22. The computation was based on maximum likelihood marker allele frequencies23. This statistical procedure is designed to detect inconsistencies and unlikely genotypes. An iterative process was followed to eliminate genotypes that are likely to be erroneous until no more inconsistencies or possible errors remained. Following this, the multipoint identity-by-descent matrices were estimated using the Markov chain Monte Carlo methods implemented in Loki24. The probabilities of multipoint identity-by-descent allele sharing among all possible pairs of related individuals were computed using the genotypes at all linked markers jointly in the computations.
Heritability and quantitative trait linkage analysis
Quantitative genetic analyses were performed using a variance components methods implemented in the SOLAR25. SOLAR employs maximum likelihood variance decomposition methods to determine the relative importance of genetic and environmental influences on a trait by modeling the covariance among family members as a function of genetic proximity (kinship). Heritability (h2), the portion of phenotypic variance accounted for by additive genetic variance (h=σ2g/σ2p) was assessed by contrasting observed covariance matrices with the covariance matrix predicted by kinship. Bivariate genetic correlation analyses were performed to decompose phenotypic correlations (ρp) between regional HWM measurements into the genetic (ρg) and environmental (ρe) correlations, accounting for kinship: ρp=ρg(h21)1/2(h22)1/2+ρe(1−h21)1/2(1−h22)1/2.
Quantitative trait linkage analysis was performed to localize potential genes influencing phenotypic variation to specific chromosomal locations 25. Model parameters were estimated using maximum likelihood. The hypothesis of significant linkage was assessed by comparing the likelihood of a classical additive polygenic model to that of a model allowing for both a polygenic component and a variance component due to linkage at a specific chromosomal location. The LOD score given by the log10 of the ratio of the likelihood of the linkage and the polygenic model served as the test statistic for genetic linkage. For this exact pedigree structure and density of markers, a LOD of 1.67 is required for suggestive significance (likely to happen by chance less than once in a genome-wide scan) and a LOD of 2.88 is required for genome-wide significance at the 0.05 level.
Similar to previous studies 10–13, HWM volumes were positively skewed. An inverse Gaussian transformation was utilized to assure normal range for kurtosis and skewness25. All genetic analyses were conducted with age, sex, age*sex, age2, age2*sex and diagnostic status for hypertension, diabetes and heart disorder (encoded as 0 or 1) included as covariates.
Results
Heritability Analysis
Whole brain (WB) HWM volume increased exponentially with age (Figure 1), consistent with previous findings by this center and others10, 18, 26. All measures of HWM volume were significantly influence by genetic factors. Heritability estimate for WB-HWM volume was .72±0.11, p=1.0×10−14. Consistent with previous reports, the age and age2 covariates were nominally significant (p<0.10) for the WB-HWM volume10, 11. Lobar, ependymal and sublobar HWM volumes were also determined to be highly heritable (Table 1). Age was a significant covariate for both regional HWM volume measurements and age2 approached significance for the ependymal HWM volume. Binary covariates that coded diagnosis for hypertension and diabetes did not reach statistical significance for any of the traits.
Figure 1.
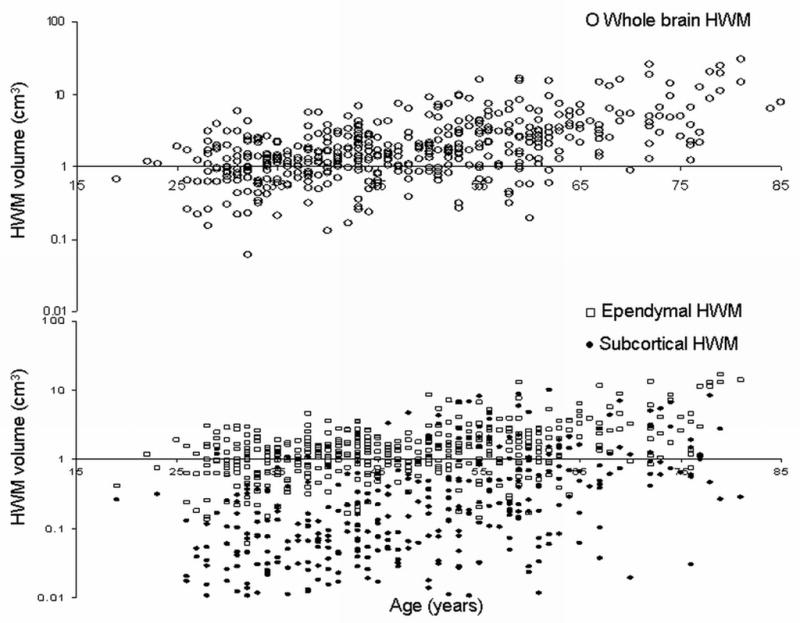
Whole brain HWM raw data (top) and Ependymal ( □ ) and Subcortical (●) HWM volumes (bottom) versus age.
Table 1.
Heritability (h2) Estimates for HWM Volume
| Trait | % of Total Volume | h2 | p | Significant Covariates | Variance Explained by Covariates |
|---|---|---|---|---|---|
| Whole Brain | 100.00 | .72 | 1E-14 | Age (5E-14), Age2 (0.08) | 28% |
| Subcortical | 23.57 | .66 | 4E-11 | Age (3E-16) | 27% |
| Ependymal | 76.43 | .73 | 1E-9 | Age (2E-9), Age2 (0.06) | 20% |
Bivariate Correlation Analysis
Genetic correlation analysis indicated that the subcortical and ependymal HWM volumes shared 21% of genetic variance (ρG =.46±0.12; p=.001), suggesting that some degree of pleiotropy. In contrast, the environmental correlation between HWM measurements was not significant (ρE=−.07±.24; p=.90). This result suggests that the observed phenotypic correlation between these two traits is driven overwhelmingly by shared genes.
Linkage Analysis
Genome-wide linkage analysis did not reveal a genome-wide significant localization for any of the HWM traits. However, suggestive linkage for subcortical and ependymal HWM volumes combined (bivariate) was found (LOD = 2.12) on chromosome 1 at 288cM near the p-terminus. Ependymal volume alone also showed suggestive linkage (LOD=1.72) at this chromosomal location. WB and subcortical HWM volumes were nominally significant (LOD=1.51 and 1.63 respectively) at this location.
Discussion
WB-HWM volume is a complex trait with a large genetic component. Histopathological findings suggest two distinct forms of HWM lesions, subcortical and ependymal 6–8. In normal aging, subcortical HWM are though to result from ischemic and/or neuroinflamatory etiologies. Therefore, formation of subcortical HWM is thought to be the product of age-related loss of permeability of small vessels, age-related free-radical damage to oligodendrocytes and immune-system mediated gliosis6, 27–30. In contrast, ependymal HWM appears to be of nonischemic origin. Histopathological findings indicate that the subependymal HWM is formed by the gliosis of periventricular WM due to the disruptions of the ependymal lining of cerebral ventricles, a condition also commonly observed in traumatic brain injuries7–9. The ependymal gliosis in normal aging is thought to be produced by a condition called the pulse-wave encephalopathy 7–9, which refers to the mechanical damages caused to the ependymal lining by the pulsatile movements of CSF due to the intra-cranial pulse pressure waves that produce “traumatic” micro-tears in the ependymal lining7–9. The magnitude of the CSF intra-cranial pulse pressure waves is linked to the gradient between systolic and diastolic arterial pressure. High arterial pulse pressure was shown to be a major risk factor for vascular damage and small vessel disease and was associated with higher ependymal HWM volumes31, 32. Pulse pressure is thereby could be a mechanism partially responsible for production of both subcortical and ependymal HWM. In fact, a recent whole-genome study of HWM in hypertensive individuals found evidence of significant multivariate linkage between two traits13.
Our results in 459 generally healthy Mexican Americans individuals confirmed previous reports of significant genetic control over variation in the WB-HWM volumes 10, 11. In addition, the heritability estimates for subcortical and ependymal HWM volume measurements were also highly significant. A significant genetic correlation supported the hypothesis that these two distinct forms of HWM lesions are partially influenced by common genetic factors. The results of the univariate whole-genome linkage analysis for the WB and regional HWM volumes did not reach statistical significance for linkage. Traits with high heritability estimates do not always produce significant linkages because heritability estimates do not provide information concerning the complexity of the underling genetic architecture33. The lack of significant linkage in the presents of significant heritability implies that the whole-brain and regional HWM volumes are polygenic traits with many QTLs each exerting only moderate effects. For example, normal variation in adult height is highly heritable (h2=0.89–0.93), but current estimates suggest that up to 44 independent loci are associated with normal stature34. In contrast, the heritability of the neuregulin 1 transcript is somewhat lower (h2~.50) but linkage analysis indicated a single locus (LOD=15.8) on chromosome eight21. However, results from bivariate linkage analysis for subcortical and ependymal volumes achieved suggestive linkage at marker 288 on chromosome 1. This finding suggests that some of the shared genetic variance between two traits reported may be related to this chromosomal region. Turner and colleagues13 reported a suggestive linkage association for bivariate linkage analysis for the WB-HWM volume and pulse pressure (difference between systolic and diastolic BP) market 274 on chromosome 1. This hints on the nature of common genetic variance between two pools of HWM. It also supports the previously suggested hypothesis regarding the formation of ependymal gliosis and the link between ependymal and lobar HWM volumes. In contrast, the current study did not replicate findings of significant linkage on the chromosome 4 as reported in14. Indeed, at this chromosomal location, our peak LOD score was only ~0.3.
It is important to note that lack of overlap in genetic loci among this and other studies may be due to a number of potential issues. Genetic factors vary across different ethnicities. Prior studies of HWM were focused on populations of European ancestry while our study is the first to examine Mexican Americans, a population with significant Native American admixture. If relatively rare variants are involved in the determination of quantitative variability, we may expect considerable differences in the localization of the most important genetic loci across populations35. Additionally, this study and two other studies of HWM are relatively underpowered and are likely to miss important chromosomal localizations. Also, in general, linkage studies of such complex phenotypes cannot be used to exclude genetic regions for important QTLs. Thus lack of concordance cannot be interpreted as evidence against the hypothesis that a QTL exists in a particular genomic region. Finally, the cross-sectional nature of these data is suboptimal. There are well-known limitations to the conclusions that can be made about longitudinal processes, such as aging, from cross-sectional measurements. The longitudinal studies often fail to confirm the age-related trends observed in the cross-sectional samples36, 37 due to heterogeneity of individual aging trends. Longitudinal data from the Australian Stroke Prevention Study (ASPS) reported large intersubject differences in the rates of accumulation of HWM lesions38. Older subjects, subjects with higher baseline HWM volume and subjects diagnosed with neurological disorders were found to have accelerated rates of accumulation of HWM volume39. It is unclear to what degree longitudinal study design can confound the genetic analysis of HWM volume. Age and age2 accounted for up to 27% of the variability in this study and others10–13. The individual rates of accumulation of the HWM volume rates greatly vary from a subject to subject, possibly due to individual genetic responses to aging (e.g. a genotype by age interaction). Hence, it may be useful to explicitly allow for the potential influences of genotype by age interactions. While advanced statistical genetic methods for family-based data allow for the formal detection of such interactions within cross-sectional data35, longitudinal family studies will have much greater power to localize and ultimately identify the specific genes involved in intersubject differences in accumulation rates of HWM volume.
Figure 2.
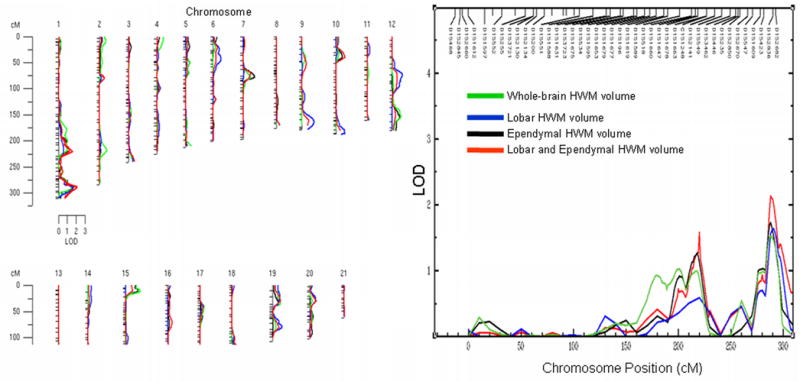
Genome-wide LOD scores for the whole-brain and regional HWM volumes (left). Distribution of LOD scores for chromosome 1 is plotted along with the locations of the genetic markers (right)
Acknowledgments
This research was supported by National Institute of Biomedical Imaging and Bioengineering (K01 EB006395) grant to P.K. and the National Institute of Mental Health (RO1s MH078111, MH0708143 and MH083824) to J.B. and D.G.
Footnotes
Conflicts of Interest
Authors have no conflicts of interest to disclose.
References
- 1.Kochunov P, Thompson PM, Coyle TR, Lancaster JL, Kochunov V, Royall D, Mangin JF, Riviere D, Fox PT. Relationship among neuroimaging indices of cerebral health during normal aging. Hum Brain Mapp. 2008;29:36–45. doi: 10.1002/hbm.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraut MA, Beason-Held LL, Elkins WD, Resnick SM. The impact of magnetic resonance imaging-detected white matter hyperintensities on longitudinal changes in regional cerebral blood flow. J Cereb Blood Flow Metab. 2008;28:190–197. doi: 10.1038/sj.jcbfm.9600512. [DOI] [PubMed] [Google Scholar]
- 3.Kochunov P, Ramage AE, Lancaster JL, Robin DA, Narayana S, Coyle T, Royall DR, Fox P. Loss of cerebral white matter structural integrity tracks the gray matter metabolic decline in normal aging. Neuroimage. 2009;45:17–28. doi: 10.1016/j.neuroimage.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochunov P, Robin D, Royall D, Lancaster J, Kochunov V, Coyle T, Schlosser A, Fox P. Can structural mri cerebral health markers track cognitive trends in executive control function during normal maturation and adulthood? Hum Brain Mapp. 2008 doi: 10.1002/hbm.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi S, Lanni C, Pantoni L, Filippi M, Frisoni GB. White matter lesions in the elderly: Pathophysiological hypothesis on the effect on brain plasticity and reserve. J Neurol Sci. 2008;273:3–9. doi: 10.1016/j.jns.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Fazekas F, Kleinert R, Offenbacher H, Schmidt R, Kleinert G, Payer F, Radner H, Lechner H. Pathologic correlates of incidental mri white matter signal hyperintensities. Neurology. 1993;43:1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- 7.Bateman GA. Pulse-wave encephalopathy: A comparative study of the hydrodynamics of leukoaraiosis and normal-pressure hydrocephalus. Neuroradiology. 2002;44:740–748. doi: 10.1007/s00234-002-0812-0. [DOI] [PubMed] [Google Scholar]
- 8.Bateman GA. Pulse wave encephalopathy: A spectrum hypothesis incorporating alzheimer’s disease, vascular dementia and normal pressure hydrocephalus. Med Hypotheses. 2004;62:182–187. doi: 10.1016/S0306-9877(03)00330-X. [DOI] [PubMed] [Google Scholar]
- 9.Henry Feugeas MC, De Marco G, Peretti II, Godon-Hardy S, Fredy D, Claeys ES. Age-related cerebral white matter changes and pulse-wave encephalopathy: Observations with three-dimensional mri. Magn Reson Imaging. 2005;23:929–937. doi: 10.1016/j.mri.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Atwood LD, Wolf PA, Heard-Costa NL, Massaro JM, Beiser A, D’Agostino RB, DeCarli C. Genetic variation in white matter hyperintensity volume in the framingham study. Stroke. 2004;35:1609–1613. doi: 10.1161/01.STR.0000129643.77045.10. [DOI] [PubMed] [Google Scholar]
- 11.Carmelli D, DeCarli C, Swan GE, Jack LM, Reed T, Wolf PA, Miller BL. Evidence for genetic variance in white matter hyperintensity volume in normal elderly male twins. Stroke. 1998;29:1177–1181. doi: 10.1161/01.str.29.6.1177. [DOI] [PubMed] [Google Scholar]
- 12.Reed T, Kirkwood SC, DeCarli C, Swan GE, Miller BL, Wolf PA, Jack LM, Carmelli D. Relationship of family history scores for stroke and hypertension to quantitative measures of white-matter hyperintensities and stroke volume in elderly males. Neuroepidemiology. 2000;19:76–86. doi: 10.1159/000026242. [DOI] [PubMed] [Google Scholar]
- 13.Turner ST, Fornage M, Jack CR, Jr, Mosley TH, Kardia SL, Boerwinkle E, de Andrade M. Genomic susceptibility loci for brain atrophy in hypertensive sibships from the genoa study. Hypertension. 2005;45:793–798. doi: 10.1161/01.HYP.0000154685.54766.2d. [DOI] [PubMed] [Google Scholar]
- 14.DeStefano AL, Atwood LD, Massaro JM, Heard-Costa N, Beiser A, Au R, Wolf PA, DeCarli C. Genome-wide scan for white matter hyperintensity: The framingham heart study. Stroke. 2006;37:77–81. doi: 10.1161/01.STR.0000196987.68770.b3. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in mexican americans. The san antonio family heart study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 16.De Coene B, Hajnal JV, Gatehouse P, Longmore DB, White SJ, Oatridge A, Pennock JM, Young IR, Bydder GM. Mr of the brain using fluid-attenuated inversion recovery (flair) pulse sequences. AJNR Am J Neuroradiol. 1992;13:1555–1564. [PMC free article] [PubMed] [Google Scholar]
- 17.Bakshi R, Caruthers SD, Janardhan V, Wasay M. Intraventricular csf pulsation artifact on fast fluid-attenuated inversion-recovery mr images: Analysis of 100 consecutive normal studies. AJNR Am J Neuroradiol. 2000;21:503–508. [PMC free article] [PubMed] [Google Scholar]
- 18.Kochunov P, Thompson PM, Lancaster JL, Bartzokis G, Smith S, Coyle T, Royall DR, Laird A, Fox PT. Relationship between white matter fractional anisotropy and other indices of cerebral health in normal aging: Tract-based spatial statistics study of aging. Neuroimage. 2007;35:478–487. doi: 10.1016/j.neuroimage.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 19.Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O’Connell JR, Perez R, Dyer TD, Almasy L, Blangero J, Bauer RL, Mitchell BD. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in mexican americans. J Bone Miner Res. 2003;18:2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 20.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 21.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression qtls using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 22.Sobel E, Lange K. Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 23.Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- 24.Heath SC. Markov chain monte carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw FE, Barkhof F, Scheltens P. Progression of cerebral white matter lesions in alzheimer’s disease: A new window for therapy? J Neurol Neurosurg Psychiatry. 2005;76:1286–1288. doi: 10.1136/jnnp.2004.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 28.Bartzokis G, Cummings JL, Markham CH, Marmarelis PZ, Treciokas LJ, Tishler TA, Marder SR, Mintz J. Mri evaluation of brain iron in earlier- and later-onset parkinson’s disease and normal subjects. Magn Reson Imaging. 1999;17:213–222. doi: 10.1016/s0730-725x(98)00155-6. [DOI] [PubMed] [Google Scholar]
- 29.Bartzokis G, Sultzer D, Lu PH, Nuechterlein KH, Mintz J, Cummings JL. Heterogeneous age-related breakdown of white matter structural integrity: Implications for cortical “Disconnection” In aging and alzheimer’s disease. Neurobiol Aging. 2004;25:843–851. doi: 10.1016/j.neurobiolaging.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Bartzokis G, Tishler TA, Shin IS, Lu PH, Cummings JL. Brain ferritin iron as a risk factor for age at onset in neurodegenerative diseases. Ann N Y Acad Sci. 2004;1012:224–236. doi: 10.1196/annals.1306.019. [DOI] [PubMed] [Google Scholar]
- 31.Nair GV, Chaput LA, Vittinghoff E, Herrington DM. Pulse pressure and cardiovascular events in postmenopausal women with coronary heart disease. Chest. 2005;127:1498–1506. doi: 10.1378/chest.127.5.1498. [DOI] [PubMed] [Google Scholar]
- 32.Miura K, Soyama Y, Morikawa Y, Nishijo M, Nakanishi Y, Naruse Y, Yoshita K, Kagamimori S, Nakagawa H. Comparison of four blood pressure indexes for the prediction of 10-year stroke risk in middle-aged and older asians. Hypertension. 2004;44:715–720. doi: 10.1161/01.HYP.0000145108.23948.7b. [DOI] [PubMed] [Google Scholar]
- 33.Devlin B, Daniels M, Roeder K. The heritability of iq. Nature. 1997;388:468–471. doi: 10.1038/41319. [DOI] [PubMed] [Google Scholar]
- 34.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blangero J. Statistical genetic approaches to human adaptability. Hum Biol. 1993;65:941–966. [PubMed] [Google Scholar]
- 36.Sliwinski M, Buschke H. Cross-sectional and longitudinal relationships among age, cognition, and processing speed. Psychol Aging. 1999;14:18–33. doi: 10.1037//0882-7974.14.1.18. [DOI] [PubMed] [Google Scholar]
- 37.Royall DR, Palmer R, Chiodo LK, Polk MJ. Normal rates of cognitive change in successful aging: The freedom house study. J Int Neuropsychol Soc. 2005;11:899–909. doi: 10.1017/s135561770505109x. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt R, Schmidt H, Kapeller P, Fazekas F. Slow progression of white-matter changes. Int Psychogeriatr. 2003;15 (Suppl 1):173–176. doi: 10.1017/S1041610203009153. [DOI] [PubMed] [Google Scholar]
- 39.Teodorczuk A, O’Brien JT, Firbank MJ, Pantoni L, Poggesi A, Erkinjuntti T, Wallin A, Wahlund LO, Gouw A, Waldemar G, Schmidt R, Ferro JM, Chabriat H, Bazner H, Inzitari D. White matter changes and late-life depressive symptoms: Longitudinal study. Br J Psychiatry. 2007;191:212–217. doi: 10.1192/bjp.bp.107.036756. [DOI] [PubMed] [Google Scholar]


