Abstract
Labeling of neurons and monitoring their activity with genetically encoded fluorescent reporters have been a staple of neuroscience research for several years. More recently, photoswitchable ion channels and pumps, such as channelrhodopsin (ChR2), halorhodopsin (NpHR), and light-gated glutamate receptor (LiGluR), have been introduced that allow for the remote optical manipulation of neuronal activity. The expanding optogenetic toolbox is finding important applications in the translucent brains of zebrafish. Enhancer and gene trapping approaches have generated hundreds of Gal4 driver lines in which the expression of UAS-linked effectors can be targeted to subpopulations of neurons. Local photoactivation of genetically targeted LiGluR, ChR2, or NpHR has uncovered novel functions for specific areas and cell types in zebrafish behavior. Because the manipulation is restricted to times and places where genetics (cell types) and optics (beams of light) intersect, this method affords superior resolving power for the functional analysis of neural circuitry.
Introduction
Optogenetic technologies, which combine the use of light-controlled reporters and manipulators of neuronal activity with genetic targeting, are revolutionizing behavioral neuroscience. Zebrafish are particularly well suited for this approach, due to their small size, translucency, and the emergence of complex behaviors at early life stages. Additionally, transgenic techniques that are staples of research in Drosophila are tractable in zebrafish, including the use of the Gal4/UAS and targeted recombination systems, as well as enhancer (ET) and gene trapping (GT) strategies [1–3]. It is now straightforward to direct expression of foreign proteins to many different cell types in the zebrafish brain.
Targeting subpopulations of neurons is, of course, also possible in other systems, including mammals [4]. But we wish to argue here that direct optical access to intact nervous tissue, as is possible in zebrafish larvae, enables unmatched control over the locale and duration of manipulations. The marriage of its inherent optical properties with a suite of genetically encoded probes has already allowed for experiments into the anatomy, activity, and behavioral roles of specific neurons in the zebrafish brain and spinal cord. Here, we will review the state of the optogenetic toolbox in zebrafish and some of the biological insights that have been gained from this approach over the past few years.
Trapping neurons in their natural habitat
Transgenesis has become efficient in zebrafish, largely due to the introduction of the Tol2 transposon system by K. Kawakami and collaborators [5]. Transgenic fish are obtained by injection of plasmid DNA, together with Tol2 transposase mRNA, at the embryonic stage, followed by raising of the founder fish and establishment of stable lines. Marker genes can be targeted to particular cell types through the use of endogenous regulatory sequences. This approach has the advantage that a desired pattern can be targeted relatively quickly and easily, but the selected regulatory region may yield broader (if it lacks repressor elements) or narrower (if it lacks enhancers) expression than the endogenous gene. An alternate approach is to undertake ET or GT screens, where random insertions of a marker gene lead to expression if they “trap” (i. e., come under control of) endogenous regulatory elements. ET and GT screens are not efficient approaches for targeting a preselected cell type of interest, because the integration events are stochastic and a great number of expression patterns must be screened before a suitably sized assortment of interesting lines is established.
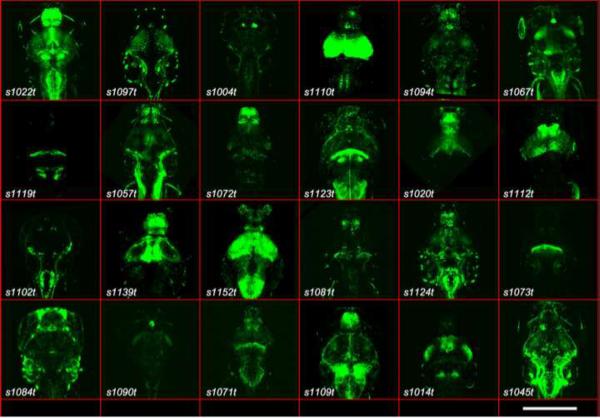
Characterizing a cell's anatomy, monitoring its function, and manipulating its activity require different probes. This provides a strong incentive for a combinatorial system that allows for more flexible targeting of transgenes. The Gal4/UAS system, which is drawn from yeast and has been used extensively in Drosophila [6], provides such flexibility. It comprises the Gal4 transcription factor and its DNA binding site, called Upstream Activating Sequence (UAS). In cells where the Gal4 gene is expressed, the Gal4 protein targets UAS, thus driving expression of any downstream open reading frame. In principle, this allows for any genetically encodable probe (linked to UAS) to be expressed in any pattern (expressing Gal4). The same concept holds for other bipartite systems currently in use in zebrafish and elsewhere, such as LexPR/LexA [7] and Cre/LoxP [8–11]. A collection of hundreds of Gal4 lines from GT or ET screens is currently being developed as a community resource in zebrafish [••12][••13, 14][••15]. Fig. 1 shows a subset of Gal4 patterns generated by our group in a recent ET screen [•16].
Figure 1. Examples of Gal4-VP16 driver lines labeling subsets of neurons in the zebrafish brain.
Panels show twenty-four examples of UAS:Kaede expression patterns picked up in a recent ET screen, with the Gal4-VP16 line number indicated. All are dorsal images of 5 or 6 dpf live larvae, mounted in agarose. Scale bar is 200 m. Reprinted from [•16]
Painting neurons with light: Kaede, Dendra, Dronpa
While methods for complete reconstruction of neural circuits [17–19] may become feasible in the future, work on the cellular anatomy of the CNS will continue to rely on the ability to restrict the labeling to single or few neurons, akin to a “genetic Golgi stain”. Several methods are available for mosaic expression of reporters in vivo, including transient transgenesis, single-cell electroporation, and blastomere transplantation of labeled donor cells into unlabeled host embryos. A new generation of photoconvertible fluorescent reporters promises further refinements to the analysis of single neurons.
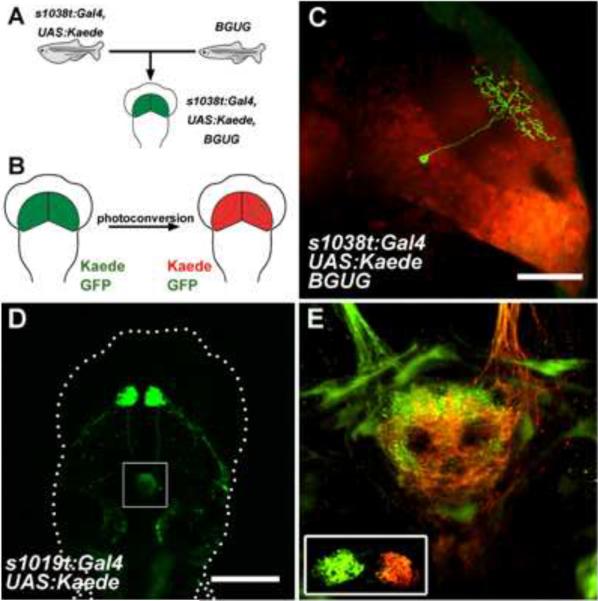
Kaede protein [20] initially fluoresces in a green wavelength. With exposure to violet or UV light, it irreversibly converts into a red-emitting fluorophore. Thus, individual neurons can be visualized either by spatially restricted illumination in a dense pattern [21] or by broad photoconversion if a subset of the neurons carry a non-convertible fluorophore in addition to Kaede [••15]. We have used this method by exploiting the variegated expression of membrane-targeted GFP in the Brn3c:Gal4, UAS:GAP43-GFP (BGUG) transgenic line, revealing the cellular architecture of the zebrafish optic tectum (Fig. 2a–c) [•16]. Other approaches for observing single neuron morphologies combine Gal4/UAS targeting with stochastic Cre/LoxP recombination [••22] or multicolor labeling with the Brainbow method [23], which shows great promise in zebrafish (E. Robles, pers. communication).
Figure 2. Cellular architecture of the zebrafish brain revealed by Kaede photoconversion.
a–c, Combining Kaede and BGUG. Observing single cells in the context of a broader expression pattern requires three transgenes, 1) a Gal4 line, in this case the tectum-specific ET line s1038t, 2) UAS:Kaede (or a red-fluorescent protein), and 3) the BGUG transgene, which leads to highly variegated expression of GFP in Gal4-positive neurons (a). GFP-positive cells are made visible by red-conversion of Kaede (b). A tectal periventricular interneuron with laminated dendritic arbor is revealed using this approach (c). d, e, Labeling long-range projections. ET line s1019t drives expression in habenular neurons (d), with projections to the interpeduncular nucleus (IPN, box in d). Targeted photoconversion of the right habenula (inset, e) labels right axons red and leaves left axons green, showing that each habenula targets different parts of the IPN (e). Scale bars are 50μm (c) and 200μm (d).
Photoconversion is also useful to trace long-range projections. After focusing violet light on the right habenula of an ET line with a composite expression pattern, red-converted Kaede diffused down the axonal shafts, revealing a strikingly asymmetric innervation of the interpeduncular nucleus, the major target of habenular axons (Fig. 2d). Dendra is another green-to-red convertible fluorophore [24], but, unlike the tetrameric Kaede, it is functional as a monomer. This means it should have an even greater potential for tracing neuronal connections, because it can be used to tag synaptic proteins and label membranes [••25].
Fluorescent emission of Dronpa can be switched off by blue and on by violet [26]. Using successive cycles of blue and violet illumination, Aramaki et al. visualized the morphologies and relative positions of individual Dronpa-expressing neurons in small networks of the hindbrain and spinal cord [27], a procedure that could potentially be scaled up and automated to reconstruct an entire circuit. The ability to extrinsically modulate emission of a fluorescent label on fast time scales might be employed to amplify a tiny signal and enhance its contrast against a noisy or bright background [•28].
Watching neurons in action
Since this field was last reviewed [29], genetically encoded Ca++ indicators, such as Cameleon [30], GCaMP [31, 32], and Inverse Pericam [33], have found a number of novel applications to monitor cellular physiology in both normal and mutant zebrafish. These could be complemented in the future by Clomeleon, a Cl− indicator, useful for the measurement of synaptic inhibition [34]. However, the use of synthetic calcium dyes still predominates because of their superior signal strength compared to protein-based indicators. Two recent imaging studies, for instance, revealed the function of identifiable spinal interneurons in defined motor behaviors [•35][•36]. This work established a specialized contribution of the glycinergic CoLo neurons to the escape response.
In a particularly elegant series of experiments, M. Orger and colleagues [••37] identified a set of bilaterally symmetric neurons in the hindbrain whose Ca++ activity was strongly predictive of turning movements, as part of the fish's innate optomotor response to drifting visual cues. Following laser ablation of these neurons, the animals were unable to turn. This work is one of only a handful of studies that show a necessary function for identified neurons in vertebrate behavior.
Selective chemical and optical poisoning of neurons
Various genetically encoded probes are available that perturb neuronal activity in the CNS [••38]. Tetanus toxin light chain (TeTxLC) has been used in Drosophila, zebrafish [••13], and mouse to block synaptic exocytosis. In a recent example, a specific zebrafish Gal4 GT line was combined with UAS:TeTxLC to demonstrate that feeding behavior in response to amino acids was dependent on a subset of olfactory sensory neurons, specifically those that carry microvilli and project to glomeruli in the lateral olfactory bulb [••39]. While TeTxLc blocks only one specific neuronal function and leaves the circuit otherwise intact, zebrafish cells can also be ablated entirely by targeted expression of Nitroreductase, a bacterial enzyme, which converts a cell-permeable prodrug into a cell-autonomous toxin [40][••13]. This provides inducibility, since Nitroreductase-expressing cells are healthy until the prodrug is introduced.
The introduction of optical methods offers rapid inducibility with added spatial resolution. KillerRed is a photosensitive relative of GFP that kills cells by generating reactive oxygen species upon illumination with red light [41]. When combined with genetic targeting by Gal4/UAS, this ablation method has extraordinary potential for circuit analysis in zebrafish (F. Del Bene, C. Wyart, E. Y. Isacoff & H. B., unpublished results).
Switching neurons on and off with microbial opsins
The introduction of Channelrhodopsin (ChR2) from the green alga Chlamydomonas reinhardtii has sparked excitement not only in the neurons that express it, but also in many neuroscientists. ChR2 is a light-gated cation channel that activates neurons in response to pulses of blue light [42, 43]. Photoactivation experiments with ChR2 have linked discrete sets of neurons to specific animal behaviors (reviewed in [44]). In zebrafish, ChR2 has been deployed to activate individual mechanosensory neurons. Strikingly, single spikes in single Rohon-Beard (RB) cells could trigger a full-blown escape response [••45]. Halorhodopsin (NpHR), a light-activated chloride pump from the bacterium Natronomas pharaonis, is used to hyperpolarize neurons with yellow light [46, 47]. In zebrafish, NpHR has been used, alone and in combination with ChR2, to assign behavioral functions to small areas of the brain. This work identified two non-overlapping groups of hindbrain neurons that were necessary and sufficient to elicit forward swimming or rapid eye movements, respectively [••25].
Remote optical control of behavior with LiGluR
iGluR(M439C) is an ionotropic glutamate receptor of the AMPA type, which has been genetically engineered to carry a surface-exposed cysteine near the ligand binding pocket. This cysteine will react with a synthetic tethered agonist, maleimide azobenzene glutamate (MAG), yielding a light-gated ion channel, LiGluR [48]. The tether is shifted into a cis alignment by ultraviolet light, bringing the agonist into contact with the channel's binding pocket. This opens the channel, allowing for Na+ influx, depolarization, and spiking of the affected neurons. Blue light reverses this alignment and leads to closure of the channel. The inherent advantage of this configuration is that LiGluR acts as a bistable switch. This feature allows for the photostimulation to be turned off, while spike activity persists, for example, during light-sensitive behavioral measurements. Szobota et al. [••49] used a specific Gal4 ET line to drive LiGluR in RB cells of the dorsal spinal cord. MAG was simply added to the bath shortly before the experiment. Activation of the channel with 380nm light prevented the normal escape response from a touch stimulus, and this response was returned to normal by irradiation with 488nm light [••49].
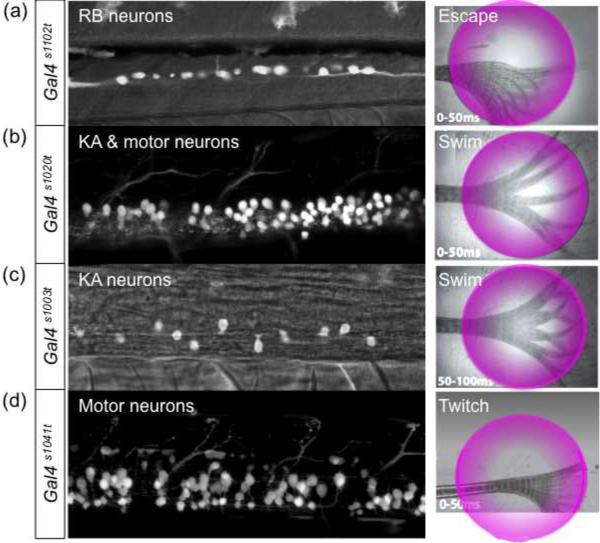
LiGluR-based dissection of neuronal circuits has now been taken beyond proof of principle by selecting ET lines with spinal cord expression from a recent Gal4 ET screen [•16]. Photostimulation of RB cells on only one side of the animal evoked a strong, hindbrain-dependent escape response (Fig. 3a), whereas activation of a hitherto enigmatic class of cerebrospinal fluid contacting neurons, the Kolmer-Agduhr (KA) cells triggered prolonged forward swimming (Fig. 3b, c) [••50]. Stimulating just a handful of KA cells at any position along the spinal cord or on only one side was sufficient to drive a complete swimming response. This behavior was reliably generated even in the absence of descending projections from the brain, suggesting that KA cells provide local sensory input to the central pattern generator circuitry underlying swimming. Activation of motor neurons resulted in a short-lived twitch if any visible behavior at all (Fig. 3d). The availability of several Gal4 ET lines with distinct but partially overlapping expression patterns was instrumental in assigning specific behavioral functions to neuronal types [••50].
Figure 3. Photostimulation of LiGluR-expressing subsets of spinal neurons elicits distinct motor responses.
The Gal4 ET lines indicated on the left were crossed to UAS:iGluR(M439C). Each line drives the UAS-linked transgene to a different, occasionally overlapping subset of spinal neurons (side views of spinal cord, four to five segments shown). MAG was added to the bath. The fish larvae were then mounted in agarose, with their tails free to move. At least three distinct types of tail movements were observed (superimposed high-speed video frames on the right) in response to application of UV light (circular illumination area). a, Stimulation of RB cells in Gal4s1102t evoked a “C bend”, indicative of an attempted escape. b, Combined stimulation of motor neurons and KA cells in Gal4s1020t evoked tail beats similar to swimming. c, Stimulation of the KA cell population alone in Gal4s1003t also evoked swimming. d, Stimulation of motor neurons alone in Gal4s1041t evoked a twitch.
Options for delivering light to a transparent brain
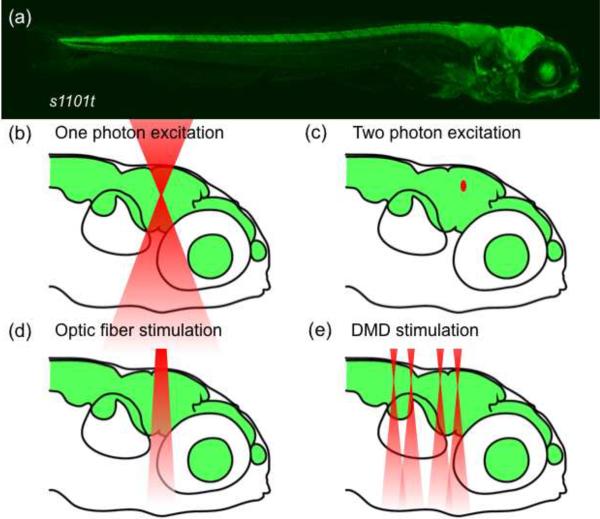
Depending on the experimental design, light can be applied to the intact nervous system by various means. Global illumination of the dish containing freely moving animals (with light-emitting diodes, for example) is useful only under specific circumstances, e. g., when the light-gated probe is expressed in a specific pattern that does not necessitate the use of targeted light beams and when sensory stimulation of the visual system is of no concern for the experiment at hand. However, often it is desirable to restrain the animal and restrict the illumination volume. Options for spatially confined light stimulation are illustrated in Fig. 4 for a fish larva that expresses Gal4 in most neurons (Fig. 4a). The laser beam of a confocal microscope produces two narrow cones of excitation whose tips fuse in a small spot, where the light intensity is highest (Fig. 4b).
Figure 4. Options for the spatially confined delivery of light into the brain of a zebrafish larva.
a, Side view of a 6 dpf zebrafish larva, carrying the pan-neuronal Gal4s1101t driver and UAS:Kaede. b–e, Four methods for the local excitation of fluorophores and photoswitches. The images show the zebrafish head with a schematic depiction of illumination volumes (red) for each method.
With multi-photon excitation, the photostimulated volume can be substantially reduced (Fig. 4c). Moreover, the wavelengths of the individual beams are in the infrared range of the electromagnetic spectrum, thus reducing scatter, improving depth penetration, and avoiding inadvertent stimulation of the animal's light-sensitive organs. These advantages have been exploited in the imaging of visual response properties (by Ca++ recordings) in the zebrafish and Xenopus optic tectum and reticular formation [51, 52] [••32, 37]. Two-photon excitation of ChR2-expressing cells, while possible in the culture dish [53], however, has not been achieved in vivo. Fiber optics provides a versatile and inexpensive route of applying light locally to the zebrafish brain (Fig. 4d). Thin optic fibers (10–100 μm tip diameter), coupled to laser light sources, were used to scan the brains of NpHR- or ChR2-expressing zebrafish for areas whose photoactivation could alter elementary motor responses [••25]. In the near future, digital micromirror device (DMD) arrays, based on the spatial modulation technology used in digital light projection (DLP) video systems, will afford superior control over spatiotemporal patterns of illumination (Fig. 4e) [54]. Because with DMDs groups of neurons can be manipulated either simultaneously or in any desired order, entire network dynamics could, in principle, be reconstituted by photostimulation.
Conclusion
Genetic targeting of fluorescent reporters and indicators to specific subpopulations of neurons has led to major advances in neuroscience. A new generation of photoswitchable ion channels and pumps, such as microbial opsins and LiGluR, now allow loss-of-function and gain-of-function approaches to animal behavior at the level of cell types or small groups of neurons. The zebrafish brain, being both genetically and optically accessible, is slated to become an important system in this new era of “neural enlightenment”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50(6):391–9. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 2.Halpern ME, et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5(2):97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott EK. The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits. J Neurochem. 2009;110(2):441–56. doi: 10.1111/j.1471-4159.2009.06161.x. [DOI] [PubMed] [Google Scholar]
- 4.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–44. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Emelyanov A, Parinov S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev Biol. 2008;320(1):113–21. doi: 10.1016/j.ydbio.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 8.Boniface EJ, et al. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47(7):484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hans S, et al. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4(2):e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thummel R, et al. Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn. 2005;233(4):1366–77. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- 11.Langenau DM, et al. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2005;102(17):6068–73. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••12.Asakawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105(4):1255–60. doi: 10.1073/pnas.0704963105. This study, together with refs. ••13, ••15, and •16 describes the generation and characterization of Gal4 enhancer trap lines in zebrafish using the Tol2 transposon system. The authors also create a UAS:TeTxLc transgenic line, which they cross into select Gal4 lines to show that block of synaptic transmission in the hindbrain-spinal cord circuitry results in defects in the fish's touch response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••13.Davison JM, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304(2):811–24. doi: 10.1016/j.ydbio.2007.01.033. Together with refs. ••12, ••15, and •16, this paper demonstrates the utility of Gal4 enhancer trap lines in zebrafish. An added innovation is the introduction of UAS:Nitroreductase, which enables the authors to chemically ablate Gal4-expressing cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogura E, et al. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn. 2009;238(3):641–55. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- ••15.Scott EK, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4(4):323–6. doi: 10.1038/nmeth1033. The authors employ here for the first time the Tol2 transposition method to create Gal4 enhancer-trap lines with expression patterns restricted to subsets of zebrafish neurons. They also demonstrate the use of Kaede as a tool to analyze neuronal morphologies and projection patterns. [DOI] [PubMed] [Google Scholar]
- •16.Scott EK, Baier H. The cellular architecture of the larval zebrafish tectum, as revealed by Gal4 enhancer trap lines. Frontiers in Neural Circuits. 2009 doi: 10.3389/neuro.04.013.2009. (in press). This work reports 156 Gal4 lines with CNS expression, which are made publicly available through the Zebrafish International Resource Center in Eugene (Oregon, USA). The paper also presents a catalog of neuronal cell types in the optic tectum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2(11):e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu J, Fiala JC, Lichtman JW. Semi-automated reconstruction of neural processes from large numbers of fluorescence images. PLoS One. 2009;4(5):e5655. doi: 10.1371/journal.pone.0005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55(1):25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ando R, et al. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99(20):12651–6. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44(3):136–42. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- ••22.Sato T, et al. Genetic single-cell mosaic analysis implicates ephrinB2 reverse signaling in projections from the posterior tectum to the hindbrain in zebrafish. J Neurosci. 2007;27(20):5271–9. doi: 10.1523/JNEUROSCI.0883-07.2007. In a technical tour de force, taking advantage of the Gal4/UAS and Cre/loxP systems, the authors label multiple neurons in the optic tectum with different colors. They demonstrate an orderly map of axonal connections from the optic tectum to the premotor reticular system, which likely forms the structural basis for visuomotor transformations. The axon guidance molecule EphrinB2a, acting within the tectal axons, is important for the development this projection. This work illustrates the power of elegant genetics and multi-color in vivo imaging for zebrafish neuro-anatomy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 24.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24(4):461–5. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- ••25.Arrenberg AB, Baier H. Optical control of zebrafish behavior with Halorhodopsin. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0906252106. (in press). This study adapts Channelrhodopsin-2 and enhanced Halorhodopsin to zebrafish. Both microbial opsins are driven by the Gal4/UAS system and activated by thin optic fibers positioned above the head of a semi-restrained fish larva. The authors use this configuration to scan the brain of pan-neuronally expressing fish for areas whose photostimulation evokes or inhibits behavior. A small group of neurons in the caudal hindbrain is shown to be necessary and sufficient for forward swimming. Electrophysiology and subcellular localization studies of NpHR in vivo support the conclusions. This is the first demonstration of a behavioral change resulting from photoactivation of NpHR in the brain of any animal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habuchi S, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci U S A. 2005;102(27):9511–6. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aramaki S, Hatta K. Visualizing neurons one-by-one in vivo: optical dissection and reconstruction of neural networks with reversible fluorescent proteins. Dev Dyn. 2006;235(8):2192–9. doi: 10.1002/dvdy.20826. [DOI] [PubMed] [Google Scholar]
- •28.Marriott G, et al. Optical lock-in detection imaging microscopy for contrast-enhanced imaging in living cells. Proc Natl Acad Sci U S A. 2008;105(46):17789–94. doi: 10.1073/pnas.0808882105. Dronpa is a fluorescent protein whose emission can be turned on and off with different wavelengths. Fast modulation of the on and off states and detection of the resulting signal oscillation was used here to enhance the contrast against a noisy background. Proof of principle that this new method, termed optical lock-in detection (OLID), works was provided by imaging of Dronpa-labeled neurons in Gal4-transgenic zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fetcho JR, Bhatt DH. Genes and photons: new avenues into the neuronal basis of behavior. Curr Opin Neurobiol. 2004;14(6):707–14. doi: 10.1016/j.conb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Fan X, et al. New statistical methods enhance imaging of cameleon fluorescence resonance energy transfer in cultured zebrafish spinal neurons. J Biomed Opt. 2007;12(3):034017. doi: 10.1117/1.2745263. [DOI] [PubMed] [Google Scholar]
- 31.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumbre G, et al. Entrained rhythmic activities of neuronal ensembles as perceptual memory of time interval. Nature. 2008;456(7218):102–6. doi: 10.1038/nature07351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. Early development of functional spatial maps in the zebrafish olfactory bulb. J Neurosci. 2005;25(24):5784–95. doi: 10.1523/JNEUROSCI.0922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berglund K, et al. Imaging synaptic inhibition throughout the brain via genetically targeted Clomeleon. Brain Cell Biol. 2008;36(14):101–18. doi: 10.1007/s11068-008-9031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •35.Liao JC, Fetcho JR. Shared versus specialized glycinergic spinal interneurons in axial motor circuits of larval zebrafish. J Neurosci. 2008;28(48):12982–92. doi: 10.1523/JNEUROSCI.3330-08.2008. See ref. •36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •36.Satou C, et al. Functional role of a specialized class of spinal commissural inhibitory neurons during fast escapes in zebrafish. J Neurosci. 2009;29(21):6780–93. doi: 10.1523/JNEUROSCI.0801-09.2009. This paper, as well as ref. •35 and other studies, continue the authors' elegant work on the dissection of spinal cord circuits underlying elementary motor behaviors. A powerful combination of electrophysiology, optophysiology, labeling of cell types with tissue-specific promoters, and laser ablation, is used to assign specific functions to spinal interneurons in swimming, struggling, and escape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••37.Orger MB, et al. Control of visually guided behavior by distinct populations of spinal projection neurons. Nat Neurosci. 2008;11(3):327–33. doi: 10.1038/nn2048. This work identifies reticulospinal cell assemblies involved in the optomotor response to drifting gratings. A small number of neurons (labeled retrogradely from the spinal cord with a synthetic Ca++ indicator coupled to dextran tracer) “lights up” when the fish attempts to turn. Turning movements are inhibited following laser ablation of these neurons. This is the first report of the function of identified hindbrain neurons in a specific swimming maneuver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••38.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57(5):634–60. doi: 10.1016/j.neuron.2008.01.002. This is an authoritative recent review of the field, with an emphasis on animal model systems other than zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••39.Koide T, et al. Olfactory neural circuitry for attraction to amino acids revealed by transposon-mediated gene trap approach in zebrafish. Proc Natl Acad Sci U S A. 2009;106(24):9884–9. doi: 10.1073/pnas.0900470106. This study continues an exciting line of research from the authors' laboratory that establishes zebrafish olfaction as a genetically tractable system. It is also one of the first demonstrations of the use of gene-trap Gal4 lines with Tetanus toxin silencing for the functional annotation of sensory pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 41.Bulina ME, et al. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24(1):95–9. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 42.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100(24):13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyden ES, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, et al. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8(8):577–81. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- ••45.Douglass AD, et al. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18(15):1133–7. doi: 10.1016/j.cub.2008.06.077. In this elegant study, the authors show that single spikes in Rohon-Beard touch-sensitive neurons, evoked by photostimulation of ChR2, may elicit escape responses, i. e., coordinated muscle contractions several synapses away of the sensory surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36(14):129–39. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao S, et al. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36(14):141–54. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volgraf M, et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2(1):47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••49.Szobota S, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54(4):535–45. doi: 10.1016/j.neuron.2007.05.010. This Neurotechnique paper introduces LiGluR, a photoswitchable ion channel, based on the ionotropic glutamate receptor, for use in neuroscience. The authors use zebrafish to provide proof of principle that the photoswitch can be targeted to subpopulations of neurons. Photoactivation of sensory neurons resulted in reversible behavioral changes. [DOI] [PubMed] [Google Scholar]
- ••50.Wyart C, et al. Optogenetic dissection of a behavioral module in the vertebrate spinal cord. Nature. 2009 doi: 10.1038/nature08323. (in press). The authors use a small collection of Gal4 enhancer-trap lines to drive LiGluR (see ref. ••49) in subsets of spinal cord neurons. They find that photoactivation of an identified GABAergic cell type, the Kolmer-Agduhr (KA) cells, evokes robust swimming behavior. The KA cells are unusual in that they contact the cerebrospinal fluid. This approach is not only technically innovative, but also reveals an underappreciated intraspinal, presumably sensory input to the central pattern generator underlying swimming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bollmann JH, Engert F. Subcellular topography of visually driven dendritic activity in the vertebrate visual system. Neuron. 2009;61(6):895–905. doi: 10.1016/j.neuron.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niell CM, Smith SJ. Functional imaging reveals rapid development of visual response properties in the zebrafish tectum. Neuron. 2005;45(6):941–51. doi: 10.1016/j.neuron.2005.01.047. [DOI] [PubMed] [Google Scholar]
- 53.Mohanty SK, et al. In-depth activation of channelrhodopsin 2-sensitized excitable cells with high spatial resolution using two-photon excitation with a near-infrared laser microbeam. Biophys J. 2008;95(8):3916–26. doi: 10.1529/biophysj.108.130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, et al. All optical interface for parallel, remote, and spatiotemporal control of neuronal activity. Nano Lett. 2007;7(12):3859–63. doi: 10.1021/nl072783t. [DOI] [PubMed] [Google Scholar]