Abstract
Lipidomics is a logical outcome of the history and traditions of lipid biochemistry and advances in mass spectrometry are at the heart of a renaissance in understanding the roles of lipids in cellular functions. Our desire to understand the complexity of lipids in biology has led to new techniques that allow us to identify over 1000 phospholipids in mammalian cell types and tissues. Improvements in chromatographic separation and mass spectrometry have positioned us to determine not only the lipid composition (i.e., parts list) of cells and tissues, but also address questions regarding lipid substrates and products that previously overwhelmed traditional analytical technologies. In the decade since lipidomics was conceived much of the efforts have been on new methodologies, development of computer programs to decipher the gigabytes of raw data, and struggling with the highly variable nature of biological systems where absolute quantities of a given metabolite may be less important than its relative change in concentration. It is clear that the technology is now sufficiently developed to address fundamental questions about the roles of lipids in cellular signaling and metabolic pathways.
Introduction
Genomic and proteomic innovations revealed the need to explore metabolic processes at the system level and lead inevitably to the development of lipidomics. Our laboratory initiated efforts to develop a lipidomics platform in the late 1990s. A driving force was the recognition that cells generate phosphatidic acid, a lipid second messenger, via multiple pathways. Thin layer chromatography and high performance liquid chromatography (HPLC) were proving insufficient to adequately address questions of sources of lipid molecular species. The focus of the work was to integrate changes in cellular lipids into the larger network of cell surface receptor signaling pathways. Much of the early efforts were designed to define how pattern changes in cellular lipids influenced the cellular response to G protein coupled receptor activation. Thus, lipidomics began with a focus on identifying lipid species that act as cellular messengers and how these molecules integrate signaling and metabolic processes of cells.
As originally conceived computational lipidomics was a mass spectrometry based profiling approach that includes the resolution, detection, and identification of lipid species [1, 2]. However, it was intended to be more than comprehensive lipid analysis and to include a systems-biology approach to the study of lipids, their interaction with other molecules, their cellular functions, and determination of pattern changes in membrane lipid composition following signal transduction events or other important biological processes [3–5]. Characterization of lipid species by MS has evolved with advancements in instrumentation and technology. The variety of ionization methods used in the current instrumentation has the ability to generate gas phase ions from nonvolatile samples and has expanded the capabilities for detection and analysis of a wide range of lipids of all sizes and structures, described in recent reviews [6, 7]. The many facets of lipidomics reflect both the diversity of lipid species in biology and the plethora of functions mediated by lipids in physiology and disease. Owing to lipidomics technology a precise phospholipid composition of E. coli was recently reported [8], the critical role of lipids in HIV replication was unveiled [9], and the spatial and temporal differences in phospholipid composition during embryo implantation were revealed [10]. Using lipidomics technology to examine phospholipid composition of liver extract in a hypercholesterolemia study potential biomarkers were recently identified [11]. Other uses that further illustrate the diversity of applications include differentiating roles of two diacylglycerol kinase isoenzymes in lipid metabolism [12], defining lipid changes in brain regions of a mouse model of Parkinson’s disease [13] and use of lipid MS as a screen for development of inhibitors of phospholipases [14].
Mass spectrometric techniques for glycerophospholipid identification and quantitation
The two predominant methods for phospholipid identification and quantitation are shotgun lipidomics and LC/MS. These approaches have distinct strengths and weaknesses, but can be used most effectively in combination.
Identification of lipids by collision-induced dissociation
Tandem mass spectrometry (MS/MS or MS2) is an essential tool in the identification of glycerophospholipids. In excess of 1000 phospholipids are present in mammalian cell types. This complexity leads to isobaric inter-class species (i.e. 34:0 PC and 34:1 PS in positive-ion mode), which are inseparable by direct infusion MS analysis. An even more complicated situation arises when samples have intra-class isobaric compounds (i.e. 38:4 PI, which can be composed of 18:0/20:4, 18:1/20:3, or 16:0/22:4 fatty acid combinations, to name a few examples). Mixtures of isobaric species such as these are extremely difficult to separate by any MS1 chromatographic technique.
Unambiguous structural characterization of the glycerophospholipids requires an understanding of the fragmentation processes involved in each lipid type. Hsu and Turk have recently completed an in-depth study of the major phospholipid classes [15–22]. Product ions arising from both positive and negative-ion mode fragmentatio n processes have been investigated yielding a wealth of information on fatty acid, lysolipid and headgroup-related fragments expected for each lipid type. In addition to the predominant diacyl phospholipids, fragmentation processes for ether- and vinyl ether-containing phospholipids have also been reported [21]. When chemically pure samples are analyzed, the sn-1 and sn-2 fatty acid composition can be determined following analysis of the lysolipid fragment ratios.
Shotgun lipidomics
Important innovations in ESI intrasource separation of lipids by direct infusion MS without prior chromatographic separation was described by Han and Gross over the last several years [23–27]. This approach, now termed “shotgun lipidomics”, has gained popularity [28–31]. Cell extracts are analyzed by direct infusion MS using precursor ion scans (PIS) and neutral loss scans (NL) to identify key lipid fragments. Using this method, lipid class (headgroup identification) is accomplished using PIS and/or NL scans in positive- and/or negative-ion modes (see Table 1). The fatty acid content of individual lipids is then identified by PIS analysis in negative-ion mode. For example, 38:4 PI (18:0/20:4 PI) would be identified by a precursor ion scan of 241 m/z in negative ion mode (PI headgroup) as well and PIS scans of 283 m/z (18:0 FA) and 303 m/z (20:4 FA). In its current form, shotgun lipidomics now widely utilizes automated nanospray techniques. This facilitates extended analysis of low volume samples that would not be practical using other analytical methods. Overall, this technique is excellent for identifying the major pools of phospholipids (ca. 90% of the total phospholipid pool by mass). However, this technique in its current form is not ideal for identifying trace level phospholipids and there are significant limitations in the ability to achieve absolute quantitation except for the most abundant species.
Table 1.
Summary of MS/MS methods for phospholipid headgroup analysis
| Lipid Class | Precursor Ion | MS/MS Mode | Fragment |
|---|---|---|---|
| PA | [M-H]− | PIS, 153 m/z | glycerol phosphate -H2O |
| PC | [M+H]+ | PIS, 184 m/z | phosphocholine |
| [M+Li]+ | NL, 189 m/z | Li cholinephosphate | |
| [M+Na]+ | NL, 205 m/z | Na cholinephosphate | |
| [M+Li/Na]+ | NL, 59 m/z | trimethylamine | |
| [M+Li/Na]+ | NL, 183 m/z | phosphocholine | |
| [M+Cl]− | NL, 50 m/z | methylchloride | |
| PE | [M-H]− | PIS, 196 m/z | glycerol phosphoethanolamine -H2O |
| PG | [M-H]− | PIS, 153 m/z | glycerol phosphate -H2O |
| PIS, 227 m/z | glycerol phosphoglycerol -H2O | ||
| PI | [M-H]− | PIS, 153 m/z | glycerol phosphate -H2O |
| PIS, 241 m/z | cyclic inositol phosphate | ||
| PS | [M-H]− | PIS, 153 m/z | glycerol phosphate -H2O |
| NL, 87 amu | serine |
HPLC/MS lipid identification and quantitation
The application of ESI-MS as a soft ionization technique, originally developed for macromolecules [32], was an important breakthrough in the analysis of glycerophospholipids. Although shotgun or direct infusion mass spectrometry offers some advantages for analysis of phospholipids from complex mixtures there are limitations in its use. The presence of isobaric species, ion suppression, and exact lipid identification requires a different analytical approach. Some of these problems can be solved by interfacing HPLC with on-line ESI-MS. Initial separation of phospholipids by class can be achieved by normal phase LC/MS [33–36] resulting in less ion suppression, high ionization yield and increased sensitivity for minor components. A gradient as well as isocratic elution can be applied [35, 36]. An important factor for lipid quantitation is the use of internal standards which have a similar instrumental response to the one of the analytes since it depends on the head group chemistry, acyl chain length and degree of unsaturation. [37]. The use of several standards per class ensures greater number of minor species identified in a complex lipid extracts as this relaxes the requirement for low lipid concentrations needed for linearity [35,37].
A combination of reverse phase HPLC and MS allows detailed analysis of individual molecular species with a high precision in a focused approach applicable for some limited categories of molecules, including polyphosphoinositides [38–41]. Another useful technique based on LC/MS is the focused analyses of specific groups of phospholipids by way of employing headgroup specific scans [39, 40].
The practical results from comprehensive lipidomics profiling of different cells or tissues is the discovery of novel lipid species previously not identified. The strategy for MS based novel lipid identification and characterization is presented in Fig. 1. Based on this strategy we have recently identified a unique ether phosphatidylinositol species during lipidomics profiling of human cirrhotic liver (unpublished data). The application of lipidomics profiling have led to discovery of N-acyl phosphatidylserine in mouse brain [42, 43] and n-acyl phosphatidylethanolamine and phosphatidylserylglutamate in E. coli [44, 45].
Figure 1.
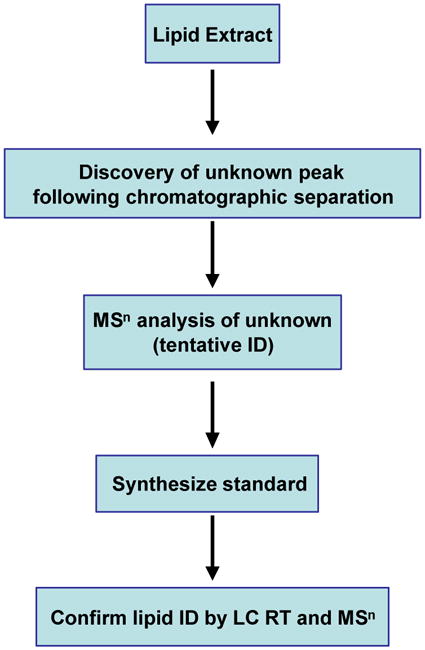
Strategy for MS-based novel lipid identification. Glycerophospholipids from biological extracts are separated by HPLC chromatography. Unknown peaks are subjected to MS/MS or MSn analysis to tentatively identify lipids. Synthetic standards are then used to confirm the lipid identity using HPLC retention time (RT) and MSn fragmentation patterns.
Issues in mass spectrometry-based data analysis and quantification
Independent of the analytical method used, the rate limiting step in lipidomics is still the data analysis that can impede the screen of large sets of samples. Nonetheless, the sine qua non of scientific research is quantitation, and state-of-the-art lipid data analysis has been greatly transformed by the rise of algorithms, tools, and standards for use in quantification. Generally, this is accomplished through the judicious use of ESI-LC/MS for high quality separation of lipid extractions along gradients that allow for area under curve (AUC) peak integrations, or by use of headgroup-specific mass scanning techniques. The workflow in a typical quantitative analysis system is summarized in Figure 2.
Figure 2.
Workflow in a quantitative lipid analysis determination. Fragmentation of member species are conducted to determine the molecular species. Varying degrees of quantitation (e.g., absolute or relative) can be performed on data coming from either full or headgroup-specific scans in systems with or without an LC column. A series of data processing steps including but not limited to baseline subtractions, deisotoping, and peak matching to lipids positively identified by MS/MS are standard elements in a typical lipidomics system. The sophisticated use of higher order statistical analysis, often multivariate by nature, is growing in applications and importance.
There are several complications that must be dealt with in order to accomplish a comprehensive lipidomics analysis. Readers are referred to a detailed overview of the data analysis methodologies currently in use in the field [46], though several challenges for future improvements exist. Here, we focus on a few common features of state-of-the-art quantification systems currently in use. At the beginning of the data analysis pipeline, more than one strategy exists for automated ESI-MS/MS identification [30, 47], but none is ideal. Once full- or headgroup-specific-scans by ESI-MS have been acquired, visualization of the raw data is a critical step to ascertain quality control, to error check AUC integrations, which may often be semi-automated, and to ensure that physical separation on LC columns has been accomplished as intended. Instrument manufacturer software is currently inadequate, and eve n open source solutions [48] may require considerable customization for use with complex lipid mixtures. This is followed by the use of background baseline corrections and signal-to- noise criteria in most analysis systems [e.g., 48]. As the ionization efficiency varies across carbon number, degrees of unsaturation and headgroup composition under normal ESI MS conditions, robust quantitation methods take this factor into account. When appropriate (e.g., for lipid classes with large heterogeneity of ionization efficiency), multiple internal standards per class are typically used [35, 37]. Particularly when analyzing classes with large numbers of species, deisotoping is essential to accurate quantitation, and algorithms to accomplish this exist [49]. The application of such methods in portions of the spectrum where lipid species exist at nearly every m/z or where there are severe isobaric overlaps are still a major challenge. In some samples (e.g., headgroup scanning conducted with a large number of heterogeneous acyl chain lengths and number of double bonds, or direct inject MS), relative quantitation within similarly ionizable class groupings is reported. Such profiling analyses have the greatest potential to be informative when the lipid subclass of interest can be measured over the broadest number of identified spesies possible and with sufficient numbers of replicates to draw robust conclusions.
Association of measured lipid changes with biological pathways of interest is an emerging area in systems biology. Various methodologies are in active states of development and use [47, 50]. The number of lipids simultaneously measured continues to grow and the appreciation of increased complexity of lipid species is likely to increase dramatically as emerging techniques better define positional specificity of double bonds on fatty acids [51]. The need for application of modern methods to limit the false discovery rate [52] in high-dimensional statistical comparisons is also essential. With thousands of lipid analytes per experiment, such issues are as important in lipidomics as in other omics endeavors.
Conclusions
The combination of highly sensitive ESI-based mass spectrometric techniques and the ability to identify and quantitate thousands of lipid species has made mass spectrometry an essential tool for lipid biochemistry. Results from lipidomics profiling provides insights into the roles of lipids in cellular networks and is being used to identify prognostic or diagnostic markers of disease progression. An extensive database on the lipid composition of macrophages, mass spectra, CID-fragmentation spectra, and useful resources for lipidomics research can be found at the LIPID MAPS website, http://www.lipidmaps.org/.
Acknowledgments
This work was supported in part by a large scale consortium grant from the National Institutes of Health (U54 GM069338), a program project grant (ES013125) and the Vanderbilt Institute for Chemical Biology. We thank Avanti Polar Lipids for the synthesis of high quality lipid molecules used as internal and external standards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review are highlighted as:
* of special interest
** of outstanding interest
- 1.Ivanova PT, Cerda BA, Horn DM, Cohen JS, McLafferty FW, Brown HA. Electrospray ionization mass spectrometry analysis of changes in phospholipids in RBL-2H3 mastocytoma cells during degranulation. Proc Natl Acad Sci USA. 2001;98(13):7152–7157. doi: 10.1073/pnas.131195098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrester JS, Milne SB, Ivanova PT, Brown HA. Computational Lipidomics: A multiplexed analysis of dynamic changes in membrane lipid composition during signal transduction. Mol Pharmacol. 2004;65:813–821. doi: 10.1124/mol.65.4.813. [DOI] [PubMed] [Google Scholar]
- 3 *.Wenk M. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. An overview of the phospholipids analytical methods, systems biology approach in lipid research and applications of lipidomics. [DOI] [PubMed] [Google Scholar]
- 4.Van Meer G. Cellular lipidomics. The EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson AD. Lipidomics—a global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Murphy RC, Fiedler J, Hevko J. Analysis of nonvolatile lipids by mass spectrometry. Chem Rev. 2001;101:479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 7 **.Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. This excellent review offers a detailed perspective on utilizing mass spectrometry for phospholipid analysis. [DOI] [PubMed] [Google Scholar]
- 8.Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapi Commun Mass Spectrom. 2007;21:1721–728. doi: 10.1002/rcm.3013. [DOI] [PubMed] [Google Scholar]
- 9.Brügger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kräusslich H-G. The HIV lipidome: a raft with and unusual composition. Proc Natl Acad Sci USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res. 2009 doi: 10.1194/jlr.M900100-JLR200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Bennett R, Strum J, Ellsworth BB, Hamilton D, Tomlinson M, Wolf RW, Housley M, Roberts BA, Welsh J, Jackson BJ, Wood SG, Banka CL, Thulin CD, Linford MR. Screening phosphatidylcholine biomarkers in mouse liver extracts from a hypercholesterolemia study using ESI-MS and chemometrics. Anal Bioanal Chem. 2009;393:643–654. doi: 10.1007/s00216-008-2504-z. [DOI] [PubMed] [Google Scholar]
- 12.Milne SB, Ivanova PT, Armstrong MD, Myers DS, Lubarda J, Shulga YV, Topham MK, Brown HA, Epand RM. Dramatic differences in the roles in lipid metabolism of two isoforms of diacylglycerol kinase. Biochemistry. 2008;47:9372–9379. doi: 10.1021/bi800492c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rappley I, Myers DS, Milne SB, Ivanova PT, LaVoie MJ, Brown HA, Selkoe DJ. Lipidomic Profiling in Mouse Brain Reveals Differences Between Ages and Genders, with Smaller Changes Associated with α-Synuclein Genotype. J Neurochem. 2009;111(1):15–25. doi: 10.1111/j.1471-4159.2009.06290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5(2):108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu FF, Turk J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation: A mechanistic proposal. J Am Soc Mass Spectrom. 2000;11:797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 16.Hsu FF, Turk J. Charge-re mote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: A mechanistic proposal. J Am Soc Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 17.Hsu FF, Turk J. Characterization of Phosphatidylinositol, Phosphatidylinositol-4-phosphate, and Phosphatidylinositol-4,5-bisphosphate by Electrospray Ionization Tandem Mass Spectrometry: A Mechanistic Study. J Am Soc Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 18.Hsu FF, Turk J. Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: Fragmentation processes and structural characterization. J Am Soc Mass Spectrom. 2001;12:1036–1043. [Google Scholar]
- 19.Hsu FF, Turk J. Electrospray Ionization/Tandem Quadrupole Mass Spectrometric Studies on Phosphatidylcholines: The Fragmentation Processes. J Am Soc Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 20.Hsu FF, Turk J. Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J Am Soc Mass Spectrom. 2005;16:1510–1522. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Hsu FF, Turk J. Differentiation of 1-O-alk-1′-enyl-2-acyl and 1-O-alkyl-2-acyl glycerophospholipids by multiple-stage linear ion-trap mass spectrometry with electrospray. J Am Soc Mass Spectrom. 2007;18:2065–2073. doi: 10.1016/j.jasms.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu FF, Turk J. Electrospray ionization with low-energy collisionally activated dissociation tandem mass spectrometry of glycerophospholipids: Mechanisms of fragmentation and structural characterization. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2673–2695. doi: 10.1016/j.jchromb.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two -dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Gross RW. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 26.Han X. Neurolipidomics: challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X, Yang K, Gross RW. Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekroos K, Chernushevich IV, Simons K, Schevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 29.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzhchalia T, Schevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 30.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 31.Ståhlman M, Ejsing CS, Tarasov K, Perman J, Borén J, Ekroos K. High-throughput shotgun lipidomics by quadrupole time -of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(26):2664–2672. doi: 10.1016/j.jchromb.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 32.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi R, Hayakawa J, Takeuchi Y, Ishida M. Two-dimensional analysis of phospholipids by capillary liquid chromatography/electrospray ionization mass spectrometry. J Mass Spectrom. 2000;35:953–966. doi: 10.1002/1096-9888(200008)35:8<953::AID-JMS23>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:654–666. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- 35 **.Hermansson M, Uphoff A, Käkelä R, Somerharju P. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. This paper describes and justifies the use of more than one standard for a true quantification of the identified phospholipids. [DOI] [PubMed] [Google Scholar]
- 36.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 37 **.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS. effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J Lipid Res. 2001;42:663–672. This report discusses the instrument response dependency on many lipid structural features, lipid concentration, solvent composition and instrument settings. [PubMed] [Google Scholar]
- 38.Ogiso H, Suzuki T, Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal Biochem. 2008;375:124–131. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 39.Retra K, Bleijerveld OB, van Gestel RA, Tielens AGM, van Hellemond JJ, Brouwers JF. A simple and universal method for the separation and identification of phospholipid molecular species. Rapid Commun Mass Spectrom. 2008;22:1853–1862. doi: 10.1002/rcm.3562. [DOI] [PubMed] [Google Scholar]
- 40.Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T. Focused lipidomics by tandem mass spectrometry. J Chrom B. 2005;823:26–36. doi: 10.1016/j.jchromb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Ogiso H, Taguchi R. Reverse-phase LC/MS method for polyphosphoinositides analysis: changes in molecular species level during epidermal growth factor activation in A431 cells. Anal Chem. 2008;80:9226–9232. doi: 10.1021/ac801451p. [DOI] [PubMed] [Google Scholar]
- 42 *.Guan Z, Shengrong L, Smith DC, Shaw WA, Raetz CHR. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 2007;46:14500–14513. doi: 10.1021/bi701907g. A presentation of the strategy for using lipidomics in the discovery and identification of novel phospholipids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan Z. Discovering novel brain lipids by liquid chromatography/tandem mass spectrometry. J Chrom B. 2009;877(26):2814–2821. doi: 10.1016/j.jchromb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. Phosphatidic acid and N-acylphosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrett TA, Raetz CR, Richardson T, Kordestani R, Son JD, Rose RL. Identification of phosphatidylserylglutamate: a novel minor lipid in Escherichia coli. J Lipid Res. 2009;50(8):1589–1599. doi: 10.1194/jlr.M800549-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46 *.Niemelä PS, Castillo S, Sysi- Aho M, Oresic M. Bioinformatics and computational methods for lipidomics. J Chromatography B. 2009;877(26):2855–2862. doi: 10.1016/j.jchromb.2009.01.025. Contains overview of several infrastructure components in a comprehensive quantitative data analysis system. [DOI] [PubMed] [Google Scholar]
- 47.Lu Y, Hong S, Tjonahen E, Serhan CN. Mediator-lipidomics: databases and search algorithms for PUFA-derived mediators. J Lipid Res. 2005;46:790–802. doi: 10.1194/jlr.D400020-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Katajamaa M, Oresic M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics. 2005;6:179–190. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haimi P, Uphoff A, Hermansson M, Somerharju P. Software tools for analysis of mass spectrometric lipidome data. Anal Chem. 2006;78:8324–8331. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- 50.Yetukuri L, Mikko Katajamaa M, Medina-Gomez G, Seppänen-Laakso T, Vidal-Puig A, Oresic M. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Systems Biology. 2007:1. doi: 10.1186/1752-0509-1-12. article 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown HA, Murphy RC. Working towards an exegesis for lipids in biology. Nat Chem Biol. 2009;5(9):602–606. doi: 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]



