Abstract
The screening of a >9000 compound library of synthetic DNA binding molecules for selective binding to the consensus sequence of the transcription factor LEF-1 followed by assessment of the candidate compounds in a series of assays that characterized functional activity (disruption of DNA–LEF-1 binding) at the intended target and site (inhibition of intracellular LEF-1 mediated gene transcription) resulting in a desired phenotypic cellular change (inhibit LEF-1 driven cell transformation) provided two lead compounds: lefmycin-1 and lefmycin-2. The sequence of screens defining the approach assures that activity in the final functional assay may be directly related to the inhibition of gene transcription and DNA binding properties of the identified molecules. Central to the implementation of this generalized approach to the discovery of DNA binding small molecule inhibitors of gene transcription was: (1) the use of a technically non-demanding fluorescent intercalator displacement (FID) assay for initial assessment of the DNA binding affinity and selectivity of a library of compounds for any sequence of interest, and (2) the technology used to prepare a sufficiently large library of DNA binding compounds.
Introduction
Fundamental to opportunities for modulating aberrant gene transcription is a detailed understanding of integrated gene expression and the development of molecules that can selectively modulate it. Typically, genes with complementary functions are synchronized by highly specific and tightly controlled upstream transcription regulators under normal physiological states, although aberrant signaling or activation of downstream transcription factors can lead to deregulated gene expression associated with tumor transformation or progression. Historically, insights into how small molecule therapeutic intervention can be utilized in such cases emerged first from functional screens of natural products whose biological effects often could be traced to their DNA binding properties and subsequent impact on gene transcription.1,2 Based on these observations, subsequent and extensive efforts have been directed at the discovery of small molecules that selectively bind DNA and predictably inhibit gene expression.3 This effort to design compounds that interact with targeted DNA sequences or structural motifs requires not only the identification of therapeutically exploitable DNA sequences, but also that the underlying principles by which small molecules recognize and interact with DNA be understood. However, the discovery of such agents has been slow due to the complexity associated with understanding small molecule–DNA interactions, the effort required to design individual compounds that target specific sequences, and the technically demanding techniques involved in the determination of their DNA binding affinity and selectivity, while simultaneously addressing the requirement for functional activity in subsequent cell-based and organism-based assays. Moreover, the design of sequence-specific DNA binding agents that are selective for not just a single sequence, but a collection of sequences or a desired subset of sequences constituting a targeted transcription factor consensus binding site constitutes a challenging problem especially when their individual functional impact on integrated gene expression is not yet known or available.
Herein, we report an additional approach to the discovery of such lead compounds and their functional activity and provide the tools for such studies. This entails the synthesis and rapid throughput screen of a library of DNA binding molecules for binding to a sequence or ensemble of sequences of interest, the identification of those sufficiently selective for the sequence(s) of interest using tools introduced to establish their intrinsic selectivity, followed by implementation of a series of selection assays that characterize functional activity (disruption of a protein–DNA binding interaction) at the intended target and site (intracellular gene transcription) resulting in a desired phenotypic cellular change (cell transformation). Central to these studies was introduction of (1) a technically non-demanding fluorescent intercalator displacement (FID) assay as the screen for rapidly assessing the DNA binding affinity of libraries of compounds and comprehensively defining their DNA binding selectivity,4,5 as well as (2) technology for the preparation of a useful and sufficiently large library of DNA binding compounds.6
The system chosen to exemplify the approach was inhibition of LEF-1-mediated gene transcription. The majority of colorectal tumors arise from mutations in the tumor suppressor protein adenomatous polyposis coli (APC)7 or its binding partner β-catenin that result in the release and nuclear accumulation of β-catenin.8–13 The unregulated -catenin binds to and activates transcription factors including LEF-1 (lymphoid enhancer binding factor 1).14–16 This results in upregulated and aberrant gene expression which is the key transformation step in the development of colon cancer (Figure 1).17–19 The LEF-1 (TCF) transcription factors18–24 share an identical DNA-binding domain referred to as the high mobility group (HMG) domain recognizing the sequences 5’-CTTTGWW-3’ (W = A or T).25,26 Importantly, LEF-1 binds the minor groove through its HMG domain making this DNA–protein interaction an ideal target for libraries of minor groove binding ligands.27,28
Figure 1.
Screening protocol used for identification of DNA binding compounds that selectively bind the LEF-1 consensus sequence, inhibit LEF-1 responsive gene transcription and LEF-1 DNA binding, and inhibit LEF-1 driven cell transformation.
Results and Discussion
Library of DNA Binding Molecules
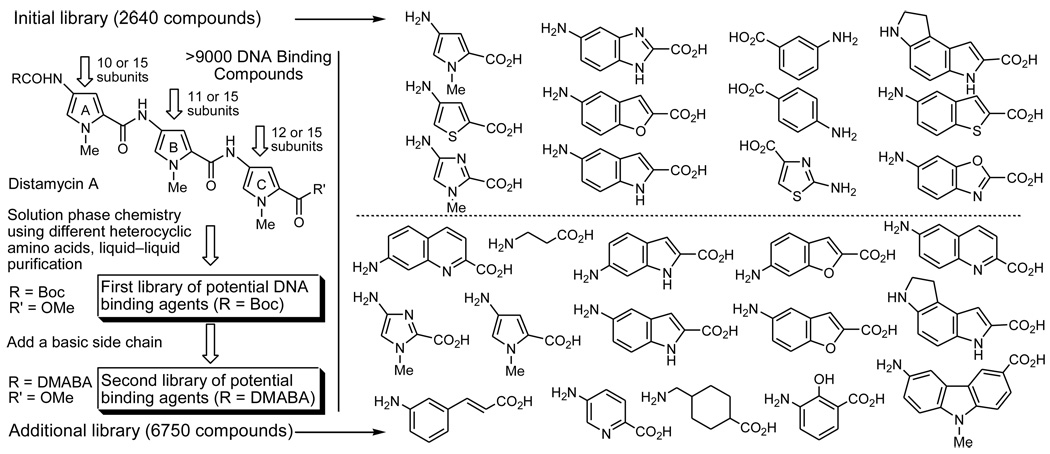
An additional library of 6750 compounds complementary to an initial library of 2640 compounds6 of candidate DNA minor groove binders was prepared enlisting the solution-phase library synthesis techniques disclosed previously29–31 (Figure 2). These combined libraries provide >9000 compounds containing systematic replacements for each of the three repeating pyrrole subunits of the minor groove binding natural product distamycin. The inclusion of many of the replacement subunits was inspired by the design elements that have emerged from the Dervan polyamides,32–35 represent repeating subunits found in additional minor groove binding compounds (e.g., CC-1065, duocarmycin, yatakemycin),36,37 or represent conservative departures from such structural skeletons. In total, they represent a unique collection of compounds that can be expected to exhibit a systematically altered composite of DNA minor groove binding properties (affinity and selectivity)38 that is now available39 for these and related studies.
Figure 2.
Libraries of candidate DNA binding compounds (>9000 compounds) prepared and screened.
For ease of synthesis and convenience in screening, the libraries were prepared and are initially screened as small mixtures where the B and C subunits are fixed (132 combinations for library 1 and 225 combinations for library 2), but contain all subunits at position A (132 and 225 mixtures of 10 or 15 compounds, respectively).38 Additionally, each library was capped as both its Boc derivative (neutral) and dimethylaminobutanoyl amide (protonated amine, DMABA) enhancing cell permeability or DNA binding affinity and solubility, respectively. Thus, the initial library screening occurs with 714 wells (2 × 132 and 2 × 225) of compounds (nine 96-well plates), a number easily manageable even without automation. Initial screening hits in the DNA binding assay (FID assay) are rescreened enlisting the individual compounds making up the active mixtures to identify those with the desired binding characteristics and the resulting individual compounds are progressed through the subsequent screening assays.38
Screening and Selection Paradigm
This combined library of minor groove binding compounds was screened sequentially for: (1) selective binding to the LEF-1/β-catenin sequence (5’-CCTTTGATC-3’, Oligo 1) versus a closely related nonbinding sequence (5’-CCTTTGGCC-3’, Oligo 2) using the fluorescent intercalator displacement (FID) assay, (2) for inhibition of LEF-1/β-catenin mediated gene transcription enlisting a cell-based luciferase reporter assay disclosed by Clevers19 (TOPFLASH), (3) for inhibition of the interaction between LEF-1 and an oligonucleotide containing a consensus binding site for LEF-1 (gel shift assay),19,23 and (4) for their effects on oncogenic transformation induced by a retrovirus expressing LEF-1 in chicken embryo fibroblasts (CEF), Figure 1. Only the successful ligands identified in the sequential screens were progressed into the next assays. Notably, the FID assay was also utilized to define the DNA binding selectivity of the leads and the sequence of screens assures that activity in the final functional assay may be directly related to the inhibition of gene transcription and DNA binding properties of the identified molecules.
Initial FID Screen and Identification of Ligands Selective for the LEF-1 Consensus Sequence
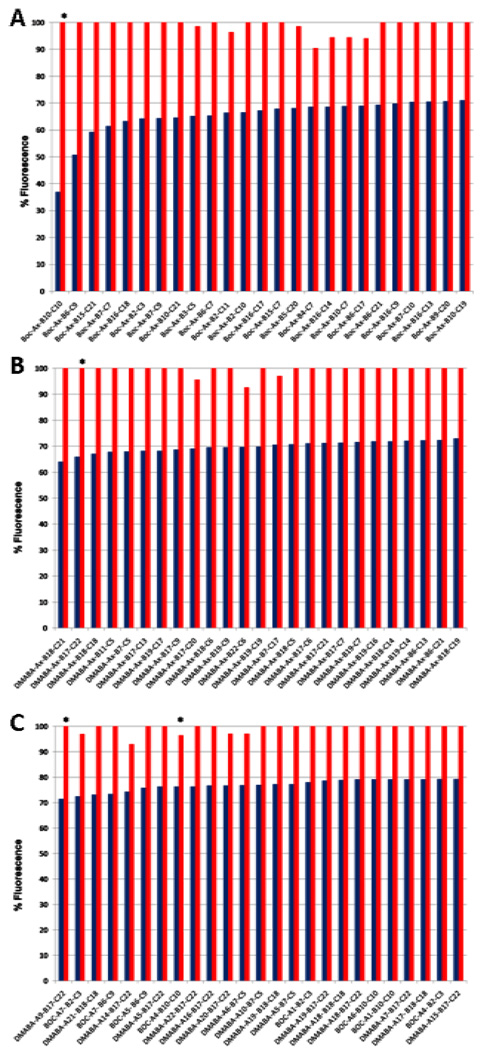
The initial screen and an essential component of the study is the assessment of the library for selective binding to the DNA sequence of interest and this was conducted with the FID assay.4,5 This assay simply entails measuring the fluorescent decrease derived from ligand displacement of bound ethidium bromide from hairpin deoxyoligonucleotides bearing the sequence(s) of interest. The assay is technically non-demanding, nondestructive, does not require modification, labeling, or immobilization of the candidate ligands or the DNA, and can be conducted simply in a 96-well plate format. In addition, the initial screen was set up to measure binding to the LEF-1/β-catenin sequence of 5’-CCTTTGATC-3’ (consensus = 5’-CTTTGWW-3’, W = A or T) versus a closely related nonbinding sequence of 5’-CTTTGGCC-3’ where the affinity of ligands for the desired sequence as well as their potential selectivity could be initially assessed. Thus, the library composed of 714 mixtures was screened and the top five Boc and top five DMABA mixtures were deconvoluted into their individual compounds (Figure 3a and 3b). This entailed their individual synthesis from the archived two subunit precursors conducting the last step of the library synthesis with the individual subunits, and the individual compounds (125 compounds) were screened in the FID assay for binding to the two DNA sequences (Figure 3c). From this, the top 30 binders were selected for advancement through the subsequent functional assays. Like the mixtures from which they were derived, most exhibited surprisingly selective binding for the desired (5’-CCTTTGATC-3’, Oligo 1) versus altered (5’-CCTTTGGCC-3’, Oligo 2) sequence. The two active compounds (denoted with * in Figure 3) that were selected by the functional screens that follow were titrated against the screening hairpin deoxyoligonucleotides containing the LEF-1 consensus sequence and its altered nonbinding sequence using the FID assay and found to exhibit binding constants reflecting their relative affinity and selectivity (Figure 4).
Figure 3.
A and B: Results of the FID assay (1.5 µM DNA, 1.0 µM compound mixture, 6.0 µM ethidium bromide) of the entire compound library (>9000 compounds). The top 25 mixtures in each series are shown (A: Boc, and B: DMABA). Blue: binding to LEF-1 consensus sequence (Oligo 1, affinity). Red: binding to the altered LEF-1 sequence (Oligo 2, selectivity). * Denotes library mixtures from which the two final compounds were identified. C: Results of the FID assay (1.5 µM DNA, 1.0 µM compound, 6.0 µM ethidium bromide) of the individual compounds composing the top 10 compound mixtures (top 25 compounds shown). * Denotes the final active compounds identified in the subsequent functional screens.
Figure 4.
aAssociation constants (Ka) established by FID titration (1.5 µM DNA, 6.0 µM ethidium bromide) for the final two compounds identified in the subsequent functional assays. Oligo 1 contains the LEF-1 consensus sequence and Oligo 2 contains the altered LEF-1 nonbinding sequence. bInhibition of LEF-1 mediated gene transcription in luciferase reporter (TOPFLASH)16,19 assay, IC50.
Inhibition of Intracellular LEF-1-mediated Gene Transcription
These top 30 compounds were examined for inhibition of LEF-1/β-catenin mediated gene transcription enlisting a cell-based luciferase reporter assay introduced by Clevers19 (TOPFLASH) in which luciferase gene expression is placed under the control of a minimal c-fos promoter and four copies of the LEF-1 consensus binding sequence in human embryonic kidney (HEK-293) cells (Figure 5).16 Additionally and as a control, the active compounds were examined and shown to not alter luciferase activity in the analogous reporter assay containing an altered LEF-1 sequence (CCTTTGGCC, FOPFLASH19). These reporter assays not only assess the functional activity of the selected compounds that the preceding FID assay would suggest is derived from inhibition of LEF-1 DNA binding, but it also selects for compounds that can access the target in a cell-based assay. Nine of the 30 compounds exhibited superb inhibition (>65% inhibition) of luciferase activity in the TOPFLASH, but not FOPFLASH, assay at a screening concentration of 30 µM and were selected for further study (Figure 5). The two active compounds (denoted with * in Figure 5) that were selected by the following functional screen were examined over a range of concentrations providing well-defined dose-response curves and IC50’s of 6.2 µM and 0.15 µM (Figure 4). Interestingly, all but one of the compounds in this top tier emerged from the Boc versus DMABA library suggesting that the preferential selection of this series may represent, in part, the diminished cell permeability of the protonated and charged DMABA derivatives. Concurrent with these studies, the 30 compounds were assayed for cytotoxic activity against chicken embryonic fibroblast (CEF) cells in order to eliminate those candidates whose subsequent activity might arise from cell growth inhibition or possess otherwise unwanted toxicity (data not shown). Of the nine compounds selected from the reporter assay, four were found to be cytotoxic to CEF cells and were eliminated from further consideration.
Figure 5.
Repression of luciferase activity in the reporter assay, % inhibition at 30 µM compound concentration. * Denotes the final two active compounds identified in the subsequent functional assays.
Inhibition of LEF-1 Driven Oncogenic Transformation
Overexpression of LEF-1 induces oncogenic transformation in cultures of CEF infected with the RCAS virus expressing chicken cellular LEF-1.40 Because this transforming activity depends on LEF-1/β-catenin-mediated gene transcription, we examined the five candidate inhibitors for their effects on oncogenicity in cell culture (Figure 6). Unlike a negative control compound (DMABA-A18-B18-C18), two of the five candidate compounds, Boc-A4-B10-C10 (named lefmycin-1) and DMABA-A9-B17-C22 (named lefmycin-2), completely inhibited colony formation induced by overexpressing LEF-1 at concentrations of 10 and 20 µM, respectively.
Figure 6.
The effect of lefmycin-1 (Boc-A4-B10-C10) and lefmycin-2 (DMABA-A9-B17-C22) on soft agar colony formation induced by RCAS-LEF-1 in CEF. LEF-1-induced transformation is completely inhibited at 10 and 20 µM, respectively.
Inhibition of LEF-1 DNA Binding
To confirm that the compounds inhibit LEF-1/β-catenin DNA binding, EMSA assays were performed with immunoprecipitated β-catenin and a 32P-labeled double-stranded deoxyoligonucleotide (5’-CCCTTTGATCTTACC-3’) containing the LEF-1 consensus binding site in the presence of varied concentrations of the compounds. Under the conditions of the assay, LEF-1 binds DNA resulting in a single retarded band in the gel shift electrophoresis assay.19,23 Consistent with expectations, both compounds inhibited LEF-1 DNA binding with lefmycin-1 (Boc-A4-B10-C10) being roughly four-fold more effective than lefmycin-2 (DMABA-A9-B17-C22). Dose-response curves using EMSA were determined and yielded IC50 values of 31 µM and 115 µM for lefmycin-1 and lefmycin-2, respectively. Since LEF-1 exhibits significant nonspecific DNA binding affinity25,26 with single base mismatches exhibiting Ka’s within 2-fold of the consensus sequence, the disruption of its sequence specific DNA binding is complicated by its natural residual DNA binding. Thus, while the absolute potencies of the inhibitors in the artificial gel shift assay may not reflect the potency of their impact on gene transcription, their relative potencies may. In this regard, it is notable that the four-fold differences in the behavior of lefmycin-1 (Boc-A4-B10-C10) and lefmycin-2 (DMABA-A9-B17-C22) parallels the two-fold differences observed in the focus assay.
DNA Binding Selectivity of Lefmycin-1 and Lefmycin-2
The complete DNA binding profiles of lefmycin-1 (Boc-A4-B10-C10) and lefmycin-2 (DMABA-A9-B17-C22) were established by examining their binding to a library of hairpin deoxyoligonucleotides containing all possible five base-pair sequences5 enlisting the FID assay (Figure 7). This study, which establishes a rank order binding of the compounds to all possible five base-pair sites, provides a high resolution definition of their selectivity and qualitatively displays their relative selectivity. Even without digestion of the binding data, the overall DNA binding profile of lefmycin-1 is clearly more selective than that of lefmycin-2. Additionally and for lefmycin-1, two of the top eleven sequences in the 512 hairpin library (no. 7 and 11, and four of the top 25 with no. 21 and 24) constitute those found in the LEF-1 consensus sequence examined and these exhibit strong binding relative to the remainder of the hairpin deoxyoligonucleotides. In contrast, lefmycin-2 bound only two LEF-1 sites among its top 25 sequences (sequence no. 3 and 20) indicating that it is less selective for the LEF-1 sites. More quantitatively, the summed average location of a LEF-1 site in the profile was 111 (median = 24) for lefmycin-1 versus 139 (median = 99) for lefmycin-2 and the summed average relative binding affinity (average score)41 was 0.55 for lefmycin-1 versus 0.41 for lefmycin-2 indicating that lefmycin-1 is more selective than lefmycin-2 for the composite sequences of interest. These observations along with those indicating that four LEF-1 sites are high affinity binding sites for lefmycin-1 parallel the relative activities of lefmycin-1 versus lefmycin-2 observed in the soft agar focus assay.
Figure 7.
A: DNA binding profiles (merged bar graph) of the two active compounds established by measuring their binding to all possible 5 base-pair duplex sequences5 using the FID assay (1.5 µM DNA, 1.5 µM and 2.0 µM compound, 6.0 µM ethidium bromide) and B: an expanded view showing their top 25 sequences.
Although on the surface this may appear to be a rather nonselective behavior for even lefmycin-1, its comparison with the performance of the highly selective Dervan polyamides (avg. location = 16–60, avg. score = 0.85−0.43)41 suggests lefmycin-1 is surprisingly good for an initial screening lead without any effort at optimization. In addition to confirming that lefmycin-1 is more selective that lefmycin-2 for the target sequences of interest, the studies also define the competitive and nonproductive sequences against which future lefmycin-1 analogues should be counterscreened in continued studies.
A digestion of the lefmycin-1 binding data following protocols described earlier41 revealed that it preferentially binds the three base-pair sequence of ATC with an average rank of 34 and average score of 0.50 with all 34 five base-pair sequences containing ATC located (ranked) in the top 65 sequences, Figure 8. Notably, the best that could be observed for a three base-pair sequence would be an average rank of 17, so the behavior of lefmycin-1 appears to be quite selective. Consistent with this, thirteen of the top 25 oligos contain this sequence. Extending this analysis to four base-pair sites revealed an additional preference for GATC with an average rank of 20 and an average score of 0.54 with three of the four such sequences in the library being located in the top 25. Beautifully, this represents the palindromic sequence found in the hairpin deoxyoligonucleotide screened (Oligo 1) that contains the LEF-1 consensus sequence.
Figure 8.
Location of all lefmycin-1 binding sequences in the 512 oligo library that contain (A) ATC or (B) GATC.
Conclusions
Adopting a simple screening and selection approach conducted first with the screening of focused libraries of candidate DNA binding molecules for selective binding to targeted DNA sequences, we have shown that it is possible to discover lead compounds that inhibit a targeted transcription factor binding to DNA, inhibit intracellular gene transcription under the control of the targeted transcription factor, and produce (reverse) phenotypic cellular changes driven by the targeted transcription factor (inhibit oncogenic transformation). In addition to providing attractive leads (lefmycin-1 and -2) for further optimization against the target with which this was exemplified (unregulated LEF-1-mediated gene transcription, colon cancer), the studies define a general approach to the discovery of molecules capable of modulating aberrant gene transcription utilizing a modestly sized library of DNA binding molecules and provide the experimental tools with which to implement the approach. The order of screens is designed to assure that the final functional activity of the identified molecules may be related to preselected DNA binding properties, and the implementation of the approach relies on two key features: (1) introduction of a technically non-demanding FID assay used for the initial assessment of the DNA binding affinity and selectivity of a library of compounds for any DNA sequence of interest, and (2) the assemblage of a useful and sufficiently large library of candidate DNA binding molecules. Impressively, The FID assay permits the initial and rapid screening of a large library of compounds against any defined DNA sequence for selection of candidate ligands as well as the subsequent screening of the identified candidate ligands against a comprehensive library of DNA sequences used to define their intrinsic selectivity and sequence selectivity. The expansion of the current library of >9000 DNA binding compounds continues, and we are unaware of any comparable compound library. As such, it represents a unique resource that we encourage the community to request39 for similar screening endeavors.
Experimental
Library of DNA Binding Molecules
Full details of the preparation and characterization of the 6750 compound library is provided in Supporting Information and that of the 2640 compound library has been reported.6 Full characterization of individual compounds including lefmycin-1 and lefmycin-2 is provided.
FID Assay
The FID assay was conducted as described5 using purified hairpin deoxyoligonucleotides (Oligo 1 = 5’- CGATCAAAGGCAAAAAGCCTTTGATCG-3’, Oligo 2 = 5’-CGGCCAAAGGCAAAAAGCCTTTGGCCG-3’) purchased from Invitrogen. The full DNA binding profiles of lefmycin-1 and lefmycin-2 were established using a library of 512 hairpin deoxyoligonucleotides as described.5
Luciferase Reporter Assay
Reporter assays were performed using the human kidney cell line 293 (HEK-293) maintained in Dulbecco's modified Eagle's medium (DMEM) / 10% fetal bovine serum (FBS). Cells were seeded into MP-24 tissue culture plates at 4.0 × 104 cells per well. On the next day, the cultures were transfected with 100 ng of TOPFLASH firefly-luciferase reporter,16 300 ng of pcDNA3 expression vector carrying LEF constructs or empty pcDNA3 vector. Compounds were added to the cells after 24 h transfection. After 48 h incubation, the cultures were lysed in 120 µL of Passive Lysis Buffer and firefly luciferase activity was measured (Promega) according to the manufacturer's protocol. Firefly luciferase activities were normalized by protein concentration.
Inhibition of LEF-1 Driven Oncogenic Transformation
CEF were seeded at 5 × 105 cells per 6-well tissue culture plate in HAM's F10 containing 10% FBS. One day after seeding, the cells were infected with 10-fold serial dilutions of the RCAS viral vector expressing chicken cellular LEF-1. The cultures were overlaid every 2–3 days with nutrient agarose consisting of 57.5% (vol/vol) of media (75% F-10 2×, 5% FBS, 2% chicken serum, 15% tryptose-phosphate broth, 1.5% of L-glutamine/penicillin/streptomycin solution, screening compounds in DMSO, final concentration of DMSO: 0.1%) and 42.5% (vol/vol) of 1.5% Sea Plaque Agarose. When foci developed (14 days), they were stained with 2% (wt/vol) crystal violet (in 20% methanol). Soft agar colony assays were performed as described previously.40 In brief, transfected cells were grown for 10–14 days; they then were trypsinized and reseeded in nutrient agar consisting of 0.3% Sea Plaque agar in F-10 supplemented with 10% donor calf serum, 4% chicken serum, 3 mg/mL tryptose phosphate broth, 1.4× minimal essential medium vitamin solution, 12 µg/mL folic acid, 1.6 mM L-glutamine, 80 units/mL penicillin, 80 µg/mL streptomycin, and 0.2% DMSO. Cells were fed with the same nutrient agar every 2–3 days. Agar colonies were counted after 3 weeks.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extract was prepared using CEF infected with RCAS virus expressing LEF-1 protein using Dounce homogenization.16 Gel retardation was performed as previously described.23 Briefly, 3 µg of nuclear protein and 60 pg/µL of 32P labeled probe (duplex CCCTTTGATCTTACC) in 20 µL binding buffer (20 mM Hepes pH 7.5, 60 mM MgCl2, 1 mM EDTA, 1 mM DTT, 50 ng Salmon testis DNA (Sigma), and 10% Glycerol) were incubated in the presence of various compounds (in DMSO at a final concentration 2.5%) for 30 min at 4 °C. The candidate compounds and the negative control were tested at the following concentrations in 25 µM increments: Boc-A4-B10-C10 (lefmycin-1), 0 200 µM; and DMABA-A9-B17-C22 (lefmycin-2), 0–200 µM. The DNA protein complexes were resolved on 5% non-denaturing acrylamide gels, and autoradiography was performed on the dried gels. The relative quantity of the shifted complexes was determined using a phosphoimager (Molecular Dynamics, Model SF). Control contained only DMSO (no compounds), and specific supershift bands were detected by using anti-LEF antibody (Transduction Laboratories). Concentrations of compounds at which 50% inhibition of LEF-1/DNA binding were obtained (IC50) using Sigma Plot by extrapolating the inhibition values recorded in the experiments.
Supplementary Material
Full details of the compound library synthesis and full characterization of the individual compounds examined are provided. This material is available with the online version, at http://pubs.acs.org.
Acknowledgement
This work was supported by the National Institute of Health (CA 78045, CA 41986).
References
- 1.Tse WC, Boger DL. Chem. Biol. 2004;11:1607–1617. doi: 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Chem. Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dervan PB, Edelson BS. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 4.Tse WC, Boger DL. Acc. Chem. Res. 2004;37:61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 5.(a) Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]; (b) Boger DL, Tse W. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 6.Boger DL, Fink BE, Hedrick MP. J. Am. Chem. Soc. 2000;122:6382–6394. [Google Scholar]
- 7.Approximately 50% of the Western population develop colorectal adenomas by the age of 70: Ransohoff D, Lang C. N. Engl. J. Med. 1991;325:37–41. doi: 10.1056/NEJM199107043250107.. At least 85% of these tumors contain APC mutations, see: Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. Nature. 1992;359:235–237. doi: 10.1038/359235a0. Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Hum. Mol. Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. Jen J, Powell SM, Papadopoulos N, Smith KJ, Hamilton SR, Vogelstein B, Kinzler KW. Cancer Res. 1994;54:5523–5526.
- 8.Novak A, Dedhar S. Cell. Mol. Life Sci. 1999;56:523–537. doi: 10.1007/s000180050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrens J. Cancer Metastasis Rev. 1999;18:15–30. doi: 10.1023/a:1006200102166. [DOI] [PubMed] [Google Scholar]
- 10.Eastman Q, Grosschedl R. Curr. Opin. Cell. Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 11.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 12.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 13.Munemitsu S, Albert I, Rubinfeld B, Polakis P. Mol. Cell. Biol. 1996;16:4088–4094. doi: 10.1128/mcb.16.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlsson P, Waterman ML, Jones KA. Genes Dev. 1994;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 15.Giese K, Grosschedl R. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Proc. Natl. Acad. Sci. U.S.A. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogt PK, Aoki M, Bottoli I, Chang HW, Fu SL, Hecht A, Iacovoni JS, Jiang BH, Kruse U. Cell Growth Diff. 1999;10:777–784. [PubMed] [Google Scholar]
- 18.Aoki M, Sobek V, Maslyar DJ, Hecht A, Vogt PK. Oncogene. 2002;21:6983–6991. doi: 10.1038/sj.onc.1205796. [DOI] [PubMed] [Google Scholar]
- 19.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler K, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 20.Travis A, Amsterdam A, Belanger C, Grosschedl R. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 21.Waterman ML, Jones KA. New Biol. 1990;2:621–636. [PubMed] [Google Scholar]
- 22.Waterman ML, Fischer WH, Jones KA. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 23.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Embo J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 25.Giese K, Amsterdam A, Grosschedl R. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 26.Ham YW, Tse WC, Boger DL. Bioorg. Med. Chem. Lett. 1993;13:3805–3807. doi: 10.1016/j.bmcl.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Love JJ, Li X, Case D, Giese K, Grosschedl R, Wright PE. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson LA, Gulizia RJ, Trauger JW, Baird EE, Mosier DE, Gottesfeld JM, Dervan PB. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12890–12895. doi: 10.1073/pnas.95.22.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng S, Comer DD, Williams JP, Boger DL. J. Am. Chem. Soc. 1996;118:2567–2573. [Google Scholar]
- 30.Cheng S, Tarby CM, Comer DD, Williams JP, Caporale LH, Boger DL. Bioorg. Med. Chem. 1996;4:727–737. doi: 10.1016/0968-0896(96)00069-7. [DOI] [PubMed] [Google Scholar]
- 31.Boger DL, Desharnais J, Capps K. Angew. Chem. Int. Ed. 2003;42:4138–4176. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- 32.Dervan PB, Doss RM, Marques MA. Curr. Med. Chem.: Anti-Cancer Agents. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- 33.Dervan PB, Edelson BS. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 34.Dervan PB. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 35.Dervan PB, Bürli RW. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 36.Boger DL, Johnson DS. Angew. Chem. Int. Ed. Engl. 1996;35:1438–1474. [Google Scholar]
- 37.Boger DL, Garbaccio RM. Acc. Chem. Res. 1999;32:1043–1052. [Google Scholar]; Tichenor MS, Boger DL. Natural Prod. Rep. 2008;25:220–226. doi: 10.1039/b705665f. [DOI] [PubMed] [Google Scholar]
- 38.It is possible that a single uniquely active compound in a mixture may be missed by screening even such small mixtures (10 or 15 compounds). However, since two of the subunits of each compound in a given mixture are the same (same B and C subunits), it is more likely that each compound in a mixture exhibits a progressive range of related binding affinities and selectivities rather than exhibiting a unique behavior. As such, the screening of mixtures containing such common elements not only minimize the chances of missing a uniquely active compound, but it can often provide a first level SAR that distinguishes it from randomly combined 10–15 compound mixtures that are commonly used to expedite HTS. For our purposes and as shown herein, the approach provided useful lead structures utilizing equipment, reagent amounts, and time accessible in any academic laboratory. For more detailed documentation of testing such mixtures see: Ambroise Y, Yuspan B, Ginsberg MH, Boger DL. Chem. Biol. 2002;9:1219–1226. doi: 10.1016/s1074-5521(02)00246-6. Boger DL, Dechantsreiter MA, Ishii T, Fink BE, Hedrick MP. Bioorg. Med. Chem. 2000;8:2049–2057. doi: 10.1016/s0968-0896(00)00137-1. Boger DL, Lee JK, Goldberg J, Jin Q. J. Org. Chem. 2000;65:1467–1474. doi: 10.1021/jo9916481.
- 39.This library is available upon request for screening.
- 40.Bister K, Hayman MJ, Vogt PK. Virology. 1977;82:431–448. doi: 10.1016/0042-6822(77)90017-4. [DOI] [PubMed] [Google Scholar]
- 41.Tse W, Ishii T, Boger DL. Bioorg. Med. Chem. 2003;11:4479–4486. doi: 10.1016/s0968-0896(03)00455-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full details of the compound library synthesis and full characterization of the individual compounds examined are provided. This material is available with the online version, at http://pubs.acs.org.