Summary
Proteolysis is a key regulatory post-translational modification in diverse cellular processes including programmed cell death, immune function, and development. Tracking proteolytic events has become a focus of researchers assessing the downstream consequences of protease activation. In this review we summarize unbiased methods for identifying protease substrates and tracking the extent of cleavage, a field termed “degradomics”. These include one- and two-dimensional gel-based methods for identifying protease substrates, N-terminal peptide identification methods for simultaneously identifying substrates and cleavage sites, and approaches for quantitation of cleavage events during endogenous proteolysis. Individual methods have identified more than 300 caspase-cleaved targets during apoptosis suggesting broad future applications for these technologies.
Introduction: Proteolysis in Cellular Signaling
Proteolysis is both a catabolic process for metabolite recycling and energy generation[1] and an essential post-translational modification (PTM) that regulates intracellular and extracellular signal transduction.[2–4] Like other PTMs, proteolysis regulates protein function more rapidly than is possible via transcription and translation. Thus, its use is pivotal in responsive biological proccesses, for example in coagulation and the inflammatory response.[5–7] Site-specific proteolysis can also rapidly degrade numerous proteins in a controlled fashion, resulting in widespread morphological changes (e.g. in apoptosis or development).[8] In studies of other PTMs, such as phosphorylation or glycosylation, systems-level protein profiling has linked physiological stimuli to site-specific protein modifications.[9,10] However, while these modifications can be targeted by affinity chromatography, proteolyzed proteins have no unique chemical handle enabling their enrichment from complex mixtures. This review focuses on approaches to track proteolysis in complex mixtures by defining the sequence specificity of proteases, identifying their protein targets and sites of cleavage, and measuring the kinetics and extent of cleavage events. Apoptosis and the proteolytic activity of the caspase family of proteases are model systems that will be used as a semi-universal metric to compare the techniques.
Protease target identification: gel-based methods
Key efforts towards defining proteolytic substrates focus on the determination of primary sequence specificities. Diverse methods including substrate phage display [11,12] and mRNA display [13] identify small numbers of optimized cleavage sequences, while chemically- and enzymatically-derived peptide libraries interrogate the amino acid preferences for proteases at each subsite.[14–17] While highly successful, peptide-centric efforts suffer from an inability to define the roll of higher-order protein structure on substrate determination. To address these deficiencies, a number of gel-based techniques have been developed to identify proteases’ protein substrates.
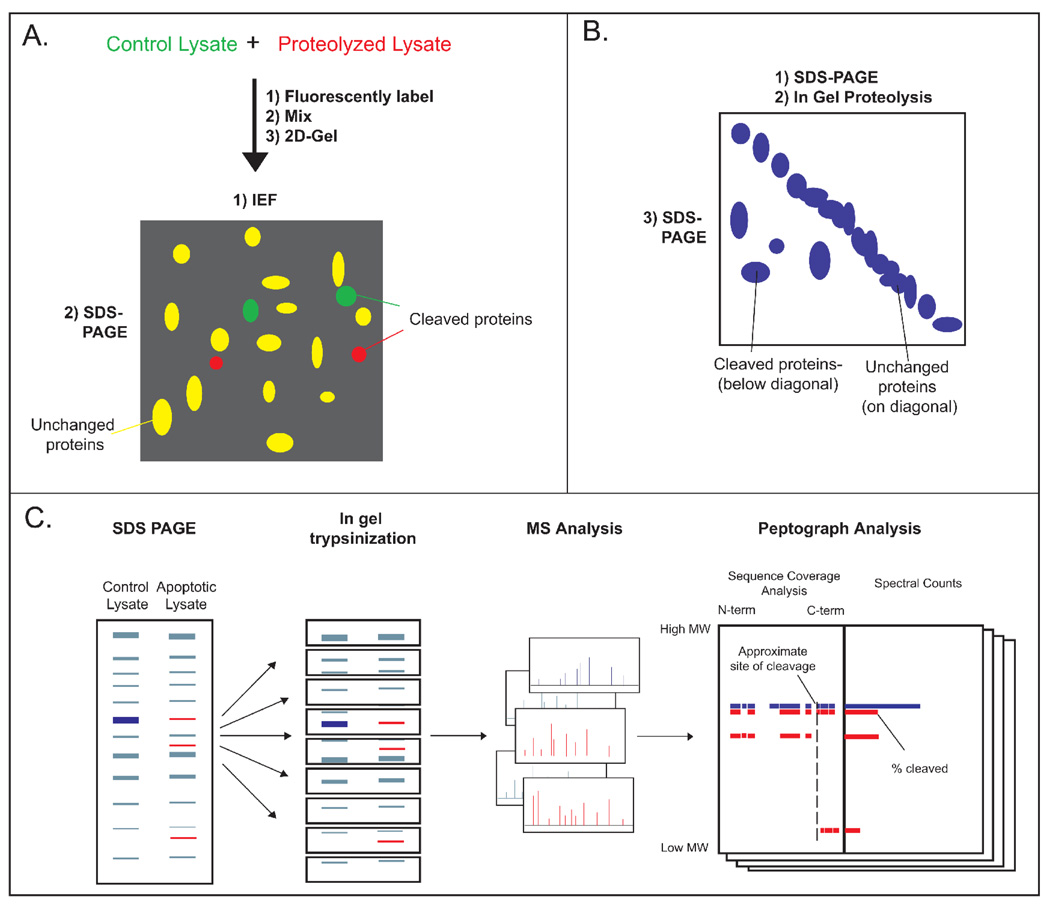
Two-dimensional differential gel electrophoresis (2D-DiGE) employs orthogonal electrophoresis methods to separate complex mixtures into resolvable spots.[18] Comparative staining reveals differences between proteolyzed and control samples that can be analyzed by MS (Figure 1a). A variation on this approach, diagonal electrophoresis, uses two dimensions of SDS-PAGE with an intermediate in-gel protease treatment to identify protease targets.[19] Proteins that migrate off the diagonal in the second, post-proteolysis dimension are likely substrates of the digesting protease (Figure 1b). 2D gel methods have been applied to caspase substrate identification, identifying 41, 13, and 15 putative substrates of caspases-1, -3, and -7 (Table 1).[19,20]
Figure 1. Gel-based methods for identifying protease substrates.
A. 2-D DIGE: A control and proteolyzed lysate are labeled with fluorescent dyes, mixed, and analyzed by 2-D gel electrophoresis. Spots with unequal fluorescence ratios are picked as proteolyzed proteins for MS analysis. B. 2-D SDS-PAGE: A lysate is resolved by SDS-PAGE, proteolyzed, and electrophoresed perpendicular to the initial migration. Spots occurring below the diagonal are identified as substrates. C. PROTOMAP: Control and proteolyzed cell lysates are analyzed side-by-side via SDS-PAGE. The gel is cut into bands, trypsinized, and peptides are identified by LC-MS/MS. For each protein, peptides are analyzed by peptographs (analysis tools that display the sequence coverage and intensity in each band), revealing the approximate site(s) and extent of cleavage.
Table 1.
Comparison of methods for identifying caspase substrates
| Method | Caspase(s) targeted |
Cleavage site? |
#of substrates |
Quantification | Reference |
|---|---|---|---|---|---|
| 2-D DIGE | Casp-7 | No | 15 | none | Jang et al (2008) |
| 2-D SDS-PAGE | Casp-1 | No | 41 | none | Shao et al (2007) |
| PROTOMAP | Apoptotic Cells | 26% | 261 | spectral counting | Dix et al. (2008) |
| COFRADIC | Apoptotic Cells | Yes | 92 | trypsin/H 2 18O | Van Damme et al. (2005) |
| COFRADIC | Casp-1/7 | Yes | 11/9 | SILAC | Lamkanfl et al. (2008) |
| Subtiligase | Apoptotic Cells | Yes | 333 | none | Mahrus et al. (2008) |
An alternative 1D gel-based method, termed PROTOMAP, has recently been described to identify cleavages that occur during Jurkat cell apoptosis.[21,22] Apoptotic and control cell lysates were separated by SDS-PAGE in adjacent lanes, and the gel lanes were sliced into bands (Figure 1c). Proteins in each band were identified by LC-MS/MS following in-gel trypsin digestion, and quantified by spectral counting. Proteins from apoptotic cells that decreased in intensity or shifted from higher to lower apparent molecular weight (261 of 1648 total proteins) were presumed to be caspase substrates. A caspase cleavage-derived peptide was detected directly in approximately one quarter of the proteolyzed proteins, and by overlaying the peptides of cleaved fragments onto a sequence map the authors could approximate the cleavage site for many others. However, without direct observation of a caspase-cleaved peptide, definitive cleavage sites cannot be assigned, and the possibility exists that non-caspase proteases are responsible for some of the observed proteolysis. A notable advantage of PROTOMAP is that an essentially infinite number of samples can be compared, limited only by the size of the gel. The authors exploited this to monitor a time course of apoptotic cleavage, and identified both early and late cleavage events during apoptosis and the relative stabilities of the resulting proteolytic fragments. Unfortunately, this technique does not enrich for proteolyzed proteins, and thus preferentially samples highly abundant proteins. Consequently, extensive MS instrument time is required for comprehensive peptide identification from low amounts (~100µg) of sample. It remains unclear whether the technique can be applied to study sparse proteolysis within a high background of unmodified proteins.
N-terminal identifications: Negative selections
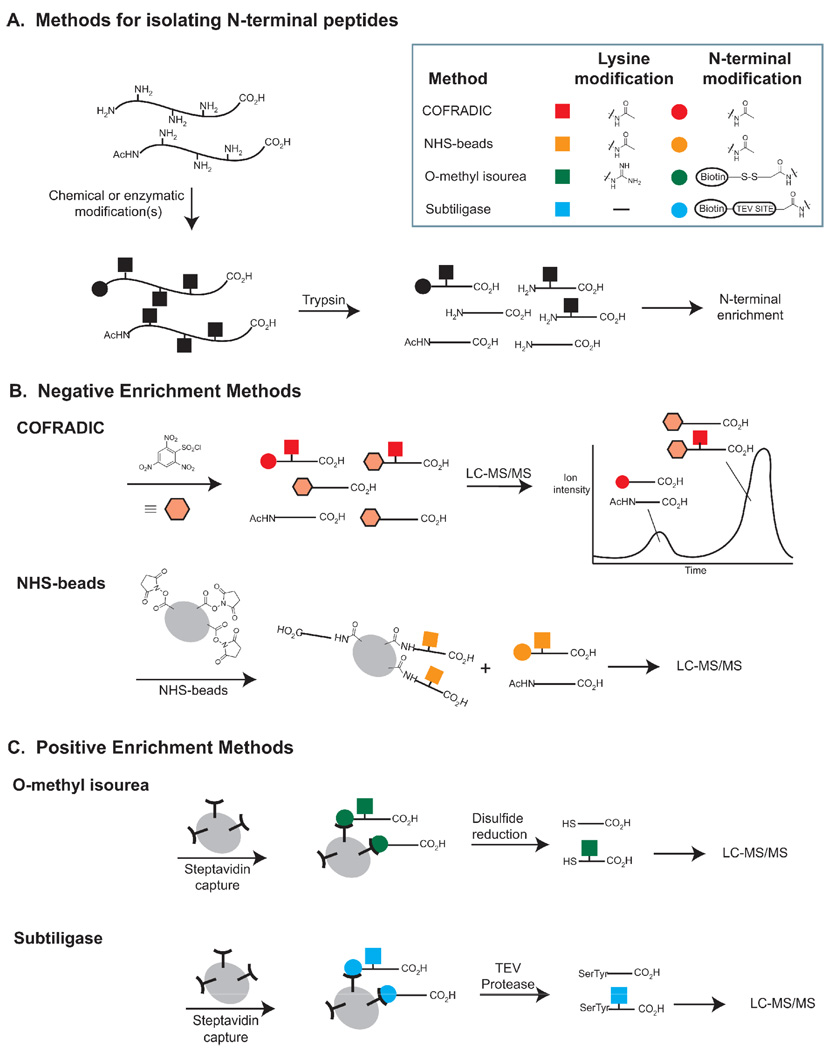
Several groups have taken advantage of the semi-unique nature of protein N-termini to identify both the proteolyzed protein and the site of cleavage. Full-length proteins have a limited number of reactive nitrogen atoms corresponding to lysine ε-amines and N-terminal α-amines. Upon digestion with a residue specific protease (e.g. trypsin), C-terminal and internal peptides gain an additional α-amine. Two teams of researchers have used this additional functionality to remove internal peptides and identify sites of proteolysis. One approach, termed COmbined FRActional DIagonal Chromatography (COFRADIC), treats full-length proteins with N-hydroxysuccinimide acetate to acetylate all lysines and N-termini (Figure 2a–b).[23–28] Subsequently, the proteins are trypsinized and the N-termini of internal peptides are capped with trinitrobenzenesulfonyl chloride, greatly increasing their hydrophobicity. The N-terminal peptides elute far earlier than the modified internal peptides on RP-HPLC, enabling their selective analysis by MS. By using an isotopically enriched amine-blocking group (e.g. trideuteroacetate) it is possible to differentiate blocked N-termini from endogenous N-terminal acetylation.[23] COFRADIC has identified 58 caspase-cleaved peptides from apoptotic Jurkat T lymphocytes,[23] 11 putative caspase-1 and 9 putative caspase-7 cleaved peptides in a caspase-1 treated lysate,[27] and 585 direct human granzyme B substrates from a granzyme B-treated lysate.[26] In a similar approach, Beynon and colleagues incubated an acetylated and trypsinized cell lysate with amine-reactive N-hydroxysuccinimide-activated beads, to capture internal peptides (Figure 2b).[29,30] Each of these techniques identifies proteins based on a single peptide. Inappropriate lengths (i.e. peptides that are too long or short) or poor ionizability make ~50% of tryptic peptides unsuitable for unambiguous identification via MS and thus limit the number of possible substrate identifications. Furthermore, incomplete capture of internal peptides can lead to a high background of false positives. On average, there are ~25 internal peptides for each N-terminal peptide in an acetylated lysate, so even small inefficiencies in their removal are highly problematic.
Figure 2. Methods for isolating N-terminal Peptides.
A. Lysates are treated with chemical reagents or enzymes to modify protein amines. Subsequent trypsinization reveals N-terminal peptides with derivatized N-termini and internal peptides with free amines. B. In negative enrichment methods, internal peptides are removed by reaction with an amine-reactive reagent followed by chromatography or solid phase extraction. The remaining peptides are analyzed by LC-MS/MS. C. In positive enrichment methods, N-terminally labeled peptides are isolated by bead capture, and chemically or enzymatically released for analysis by LC-MS/MS.
N-terminal identifications: Positive selections
An alternative to removing internal peptides for N-terminal peptide identification is to positively enrich for the peptides of interest. Salvesen and colleagues have developed a method for enrichment based on selective guanidinylation of lysine residues using O-methylisourea (Figure 2c). O-methylisourea inefficiently modifies protein α-amines, leaving them available for subsequent chemical biotinylation. After digestion with trypsin, N-terminal peptides can be isolated with immobilized streptavidin.[31] In contrast to the two previous methods for N-terminal modification, this approach discards endogenously N-acetylated N-termini (~80% of all N-termini)[32] and thus increases the sensitivity for detecting proteolytic cleavages. The authors used their technique to investigate mitochondrial transit peptides from yeast, mouse, and human cells. They found a total of 34 transit peptides from 27 proteins, only 10 of which had been previously annotated, demonstrating the utility of this method for N-terminal discovery. It should be noted that this method depends on highly efficient modification of lysine residues and highly selective biotinylation of the remaining α-amines. Biotinylation of serine, threonine, or histidine side chains could lead to false identifications. In principle, database matching in the MS-analysis should distinguish real N-terminal peptides from spurious identifications, but incomplete peptide coverage, particularly close to the N-terminus, can introduce false positives.
We have employed subtiligase, an engineered variant of the bacterial protease subtilisin BPN’, to selectively label N-terminal peptides in a single step without lysine derivatization. Created by mutating the catalytic serine to a cysteine and modifying the geometry of this active site residue with a second point mutation (P225A), subtiligase has negligible amidase activity but remains active as an esterase.[33–35] The thioester-enzyme intermediate formed with peptide ester substrates during the catalytic cycle is slowly hydrolyzed by water, but can be rapidly intercepted by N-terminal amines, allowing transfer of the N-terminal portion of the ester onto free amines. We have never observed transfer onto peptide or protein side chains, suggesting an enzymatic specificity thus far unachievable via small molecule approaches. Identification of N-terminal peptides was accomplished by treating cell lysates with subtiligase and a biotinylated-peptide ester (Figure 2c).[36,37] The resulting peptide mixture was trypsinized, N-terminal peptides were captured on streptavidin beads, and the desired peptides were released by cleavage of a TEV protease site in the original biotinylated peptide. TEV protease cleavage leaves a dipeptide tag that can be used to confirm true positives. In typical runs, >95% of identified peptides contain this tag. Using this method on apoptotic Jurkat cells, we identified 333 caspase-like cleavage sites on 292 protein substrates.[36] The broad scope of these substrates enabled the discovery of unexpected caspase enzymatic activities- most notably cleavage within helices and systematic cleavage of proteins within protein-protein complexes. Though highly specific, the efficiency of subtiligase N-terminal labeling is low, and thus this technique requires large amounts of material (typically 50–100 mg of a complex mixture per experiment) to identify numerous substrates. However, the highly enriched set of N-termini that result can be routinely analyzed in a single day via LC-MS/MS. Like all N-terminal identification procedures, the identification of one peptide per protein systematically excludes proteins with N-terminal peptides not readily identifiable via MS.
Quantitation of proteolysis
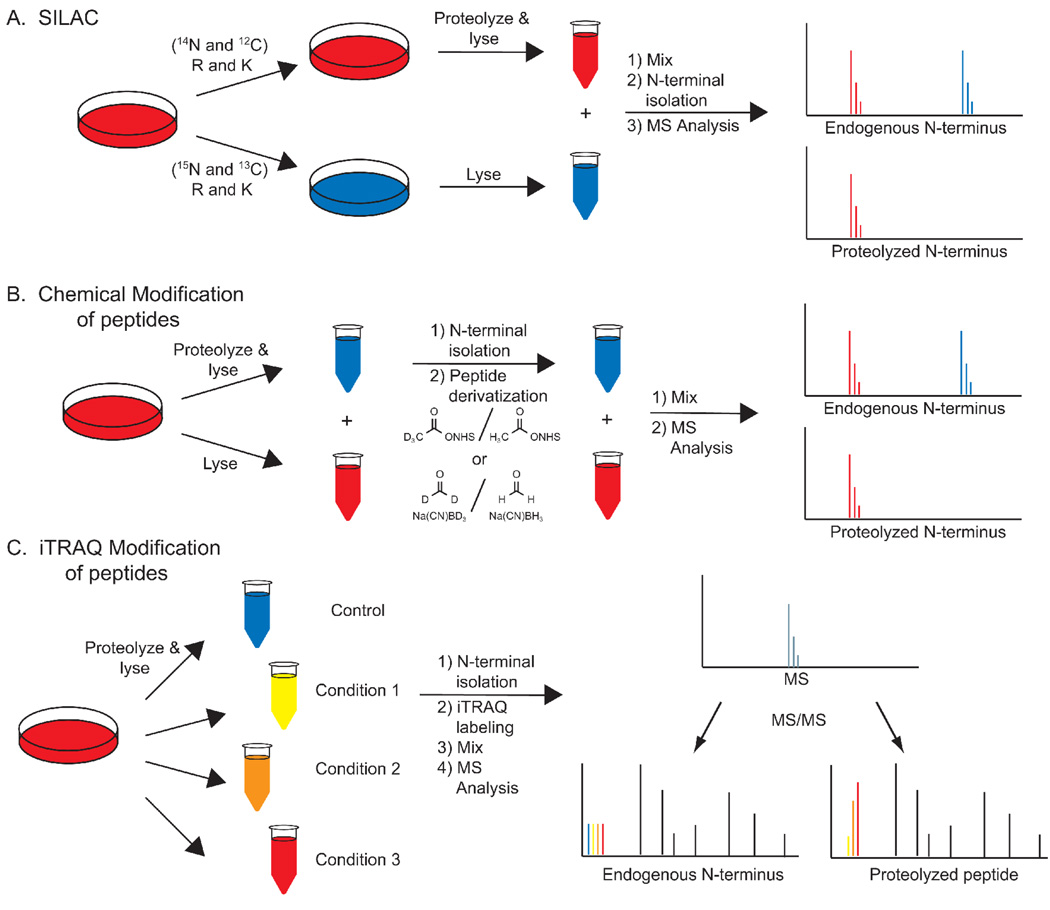
To date, the primary focus of protease-substrate classification techniques has been on identifying substrates and their cleavage sites. Efforts to understand the dynamics and extent of proteolysis could make significant contributions to our understanding of proteolysis in vivo. Simple, two-plex-quantitation can be metabolically, chemically, or enzymatically introduced into these techniques to evaluate relative levels of proteolysis in two samples (Figures 3a–b).[2,23,26,28,38] TMT-[38] and iTRAQ-based[39] quantitation allows for relative comparison of up to eight samples, enabling more detailed kinetic measurements or observations of proteolytic efficiency across multiple biological conditions (Figure 3c).[40] Application of these methods to N-terminal peptides, however, monitors only the appearance of new N-termini, leaving the extent of cleavage undetermined. PROTOMAP and other gel-based methods can theoretically analyze every peptide in the protein and thus can monitor both appearance of new peptides and the extent of cleavage.
Figure 3. Methods for relative peptide quantification.
A. SILAC. Duplicate cell cultures are grown in media containing light/heavy amino acids, proteolyzed separately, mixed, and then the N-termini are isolated. Relative peptide intensities are determined by comparing MS intensities. B. MS-based peptide quantification. Cells are lysed, protelyzed, and their N-termini are isolated and derivatized with isotopically labeled reagents. The modified peptides are mixed and quantified based on MS intensities. C. MSMS-based peptide quantification. Isolated N-termini are labeled with isobaric iTRAQ reagents, each with distinct isotopic compositions, and mixed. At the MS level of analysis each peptide is identical. At the MS/MS level the isobaric tag fragments to reveal the relative intensities of the four samples.
Conclusion
Recent technological developments have revolutionized the process of protease substrate identification. These advances have provided facile means to categorize a protease’s sequence specificity, in vitro substrates, and the in vivo proteolysis that results from biological stimuli. The two main approaches, gel-based methods and N-terminal identification, provide complementary information: gel-based methods do not enrich for the peptides of interest and thus may miss low abundance substrates, while protein identification based on enrichment of single peptides is more sensitive, but inherently limited by the nature of MS-database matching. The introduction of MS-based quantitation into these analyses will enable comprehensive and high-throughput profiling of substrate proteolysis. These advances should reveal the full scope of proteolysis in both normal and disease states.
Acknowledgements
We would like to thank Juan Diaz, J. T. Koerber, Sami Mahrus, and David Wildes for their helpful comments on the manuscript. This work is funded by the National Institute of Health F32-AI077177-01 (NJA), and R01 GM081051 (JAW), and from the Hartwell Foundation (JAW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicholas J. Agard, Department of Pharmaceutical Chemistry, University of California, San Francisco, UCSF MC 2552, 1700 4th St, San Francisco, CA 94158, Nicholas.Agard@ucsf.edu
James A. Wells, Department of Pharmaceutical Chemistry, University of California, San Francisco, UCSF MC 2330, 1700 4th St, San Francisco, CA 94158, Jim.Wells@ucsf.edu
References
- 1.Muntz K. Protein dynamics and proteolysis in plant vacuoles. J Exp Bot. 2007;58:2391–2407. doi: 10.1093/jxb/erm089. [DOI] [PubMed] [Google Scholar]
- 2.Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 5.Roberts HR, Monroe DM, Hoffman M. Molecular biology and biochemistry of the coagulation factors and pathways of hemostasis. In: Lichtman MA, Kipps JA, Kaushansky K, Beutler E, Seligsohn U, editors. Williams Hematology. edn 7th. Prchal JT: McGraw-Hill; 2006. pp. 1665–1695. [Google Scholar]
- 6.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 7.Scheidereit C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 8.Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Matthews DJ, Wells JA. Substrate phage: selection of protease substrates by monovalent phage display. Science. 1993;260:1113–1117. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- 12.Hansen M, Wind T, Blouse GE, Christensen A, Petersen HH, Kjelgaard S, Mathiasen L, Holtet TL, Andreasen PA. A urokinase-type plasminogen activator-inhibiting cyclic peptide with an unusual P2 residue and an extended protease binding surface demonstrates new modalities for enzyme inhibition. J Biol Chem. 2005;280:38424–38437. doi: 10.1074/jbc.M505933200. [DOI] [PubMed] [Google Scholar]
- 13.Baggio R, Burgstaller P, Hale SP, Putney AR, Lane M, Lipovsek D, Wright MC, Roberts RW, Liu R, Szostak JW, et al. Identification of epitope-like consensus motifs using mRNA display. J Mol Recognit. 2002;15:126–134. doi: 10.1002/jmr.567. [DOI] [PubMed] [Google Scholar]
- 14.Backes BJ, Harris JL, Leonetti F, Craik CS, Ellman JA. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat Biotechnol. 2000;18:187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- 15.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 16.Diamond SL. Methods for mapping protease specificity. Curr Opin Chem Biol. 2007;11:46–51. doi: 10.1016/j.cbpa.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 17. Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. The authors create a peptide library by digesting lysates with residue-specific proteases. Subsequent treatment with proteases of interest followed by MS analysis enables the rapid identification of protease sequence specificities.
- 18.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem. 2007;282:36321–36329. doi: 10.1074/jbc.M708182200. [DOI] [PubMed] [Google Scholar]
- 20.Jang M, Park BC, Kang S, Lee do H, Cho S, Lee SC, Bae KH, Park SG. Mining of caspase-7 substrates using a degradomic approach. Mol Cells. 2008;26:152–157. [PubMed] [Google Scholar]
- 21. Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. The authors use 1D gels coupled with quantitative MS to track apoptosis in human Jurkat cells. The use of label-free quantitation enabled a detailed kinetic evaluation of proteolysis during apoptosis.
- 22.Simon G, Dix MM, Cravatt BF. Comparative assessment of large-scale proteomic studies of apoptotic proteolysis. ACS Chem Biol. 2009 doi: 10.1021/cb900082q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. The authors present the first example of N-terminal peptide identification for tracking proteolysis during apoptosis. This paper demonstrated the utility of globally tracking N-terminal identifications during apoptosis and set the stage for evaluating numerous additional proteases.
- 24.Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 25.Van Damme P, Impens F, Vandekerckhove J, Gevaert K. Protein processing characterized by a gel-free proteomics approach. Methods Mol Biol. 2008;484:245–262. doi: 10.1007/978-1-59745-398-1_16. [DOI] [PubMed] [Google Scholar]
- 26.Van Damme P, Maurer-Stroh S, Plasman K, Van Durme J, Colaert N, Timmerman E, De Bock PJ, Goethals M, Rousseau F, Schymkowitz J, et al. Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol Cell Proteomics. 2009;8:258–272. doi: 10.1074/mcp.M800060-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Lamkanfi M, Kanneganti TD, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J, Vandenabeele P, Gevaert K, Nunez G. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staes A, Van Damme P, Helsens K, Demol H, Vandekerckhove J, Gevaert K. Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC) Proteomics. 2008;8:1362–1370. doi: 10.1002/pmic.200700950. [DOI] [PubMed] [Google Scholar]
- 29.McDonald L, Beynon RJ. Positional proteomics: preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat Protoc. 2006;1:1790–1798. doi: 10.1038/nprot.2006.317. [DOI] [PubMed] [Google Scholar]
- 30.McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- 31.Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown JL, Roberts WK. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem. 1976;251:1009–1014. [PubMed] [Google Scholar]
- 33.Abrahmsen L, Tom J, Burnier J, Butcher KA, Kossiakoff A, Wells JA. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry. 1991;30:4151–4159. doi: 10.1021/bi00231a007. [DOI] [PubMed] [Google Scholar]
- 34.Chang TK, Jackson DY, Burnier JP, Wells JA. Subtiligase: a tool for semisynthesis of proteins. Proc Natl Acad Sci USA. 1994;91:12544–12548. doi: 10.1073/pnas.91.26.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson DY, Burnier J, Quan C, Stanley M, Tom J, Wells JA. A designed peptide ligase for total synthesis of ribonuclease A with unnatural catalytic residues. Science. 1994;266:243–247. doi: 10.1126/science.7939659. [DOI] [PubMed] [Google Scholar]
- 36. Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. Using enzymatic rather than chemical methods for N-terminal peptide identification, the authors discover the largest current collection apoptotic cleavages. These substrates reveal unexpected biology of the caspases including a preference for cleavage within protrein-protein complexes.
- 37.Yoshihara HA, Mahrus S, Wells JA. Tags for labeling protein N-termini with subtiligase for proteomics. Bioorg Med Chem Lett. 2008;18:6000–6003. doi: 10.1016/j.bmcl.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 39.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Hardt M, Witkowska HE, Webb S, Thomas LR, Dixon SE, Hall SC, Fisher SJ. Assessing the effects of diurnal variation on the composition of human parotid saliva: quantitative analysis of native peptides using iTRAQ reagents. Anal Chem. 2005;77:4947–4954. doi: 10.1021/ac050161r. [DOI] [PubMed] [Google Scholar]