Abstract
The salinization of irrigated lands is increasingly detrimental to plant biomass production and agricultural productivity as most plant species are sensitive to high concentrations of sodium (Na+), which causes combined Na+ toxicity and osmotic stress. Plants have multiple Na+ transport systems to circumvent Na+ toxicity. Essential physiological functions of major Na+ transporters and their mechanisms mediating salinity resistance have been identified in Arabidopsis, including the SOS1, AtNHX and AtHKT1;1 transporters. As we discuss here, recent studies have demonstrated that a class of xylem–parenchyma-expressed Na+-permeable plant HKT transporters represent a primary mechanism mediating salt tolerance and Na+ exclusion from leaves in Arabidopsis, and that major salt tolerance QTL in monocot crop plants are also based on this HKT-mediated mechanism. Sodium toxicity and salt tolerance in plants
Physiological studies have shown that salinity stress in plants is multifactorial, including osmotic stress [1] and cellular sodium (Na+) toxicity, such as inhibition of vital enzymes and metabolic processes [2–14]. Photosynthetic processes are among the most sensitive to salinity and, therefore, salinity stress directly reduces carbon fixation and biomass production in plants [5,15–18]. Sodium transport processes have major roles in salinity tolerance, including organellar Na+ sequestration [4,8,9,15,19,20]; Na+ extrusion by plasma membrane Na+–H+ exchange transporters, such as AtSOS1 [21,22] and exclusion of Na+ from leaves and shoots [11,23–29]. In addition, reducing Na+ uptake or increasing cytoplasmic potassium (K+) levels relative to Na+ increases Na+ tolerance in plants [30–33]. However, given that multiple independent cationic nutrient uptake transporters mediate Na+ uptake from the soil into roots (reviewed in Refs [6,34]), engineering of reduced Na+ influx into plant roots is likely a more challenging endeavor. The identification and characterization of Na+-permeable transporters is therefore pivotal to understanding plant Na+ toxicity and tolerance [13,35–38].
Recent research has demonstrated that members of the high-affinity K+ transporter (HKT) transporter/channel family mediate important Na+ tolerance mechanisms in plants. The TaHKT2;1 gene from wheat (Triticum aestivum) (previously named HKT1), was the first HKT transporter gene found in plants [39]. It was shown to mediate high-affinity Na+–K+ co-transport and also preferred Na+-selective low-affinity Na+ transport in the presence of a millimolar [Na+] in Xenopus laevis oocytes and yeast [40,41]. Members of the HKT transmembrane transporter family are among the most-studied Na+-permeable plant transporter proteins to date and have been identified and characterized in many other plant species [11,24–29,42–52]. The Arabidopsis thaliana genome includes only one HKT gene, AtHKT1;1 (previously named AtHKT1) [42]. Multiple HKT genes are found in monocot genomes [28,29,49,53]. Studies of Arabidopsis plants carrying mutations in AtHKT1;1 revealed that Na+-selective transport via AtHKT1;1 has an essential role in Na+ exclusion from leaves and K+ homeostasis in leaves during salinity stress [11,24,26,27,54–56]. Here, we review findings of the functions of AtHKT1;1 and its orthologs in rice (Oryza sativa) and durum wheat (Triticum turgidum durum), which mediate a major salinity-resistant mechanism both in dicots and monocots via Na+ exclusion from leaves. We also review findings of OsHKT2;1 in rice, which functions in mediating Na+ influx into roots of K+-starved rice plants [51].
Sodium exclusion from leaves mediated by AtHKT1;1 and OsHKT1;5
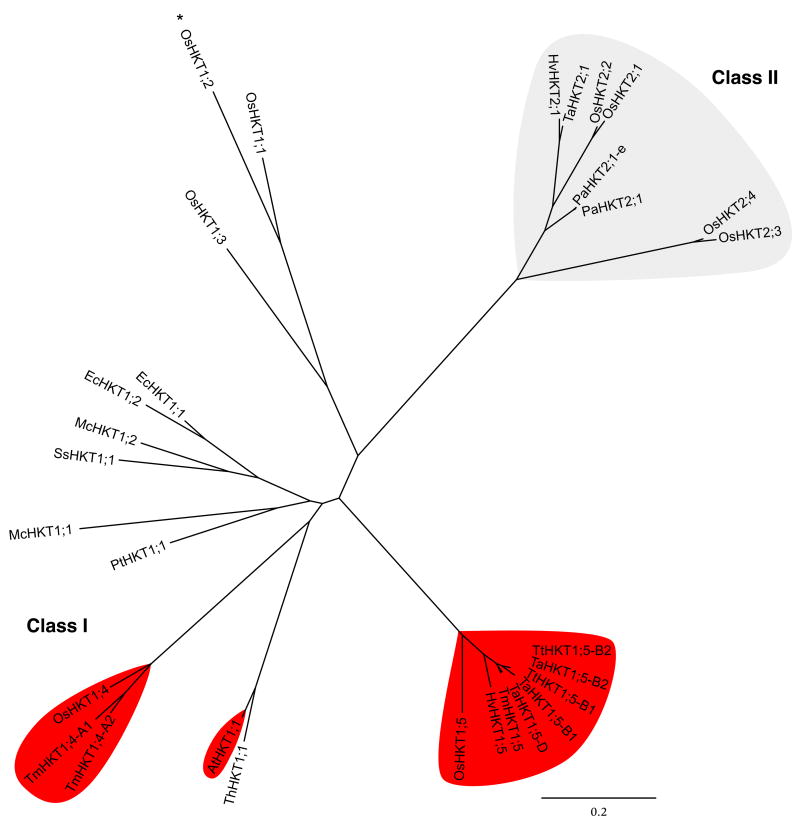
Biophysical transport and phylogenetic analyses showed that HKT transporters can be classified into two subgroups, class 1 and class 2 [57,58] (Figure 1). Class 1 HKT transporters show a preference for Na+ transport over other cations, whereas class 2 HKT transporters show a larger K+ permeability as well as Na+ permeability in heterologous expression systems, with some exceptions [43,46,48,59].
Figure 1.
Phylogenetic tree of angiosperm HKT transporter proteins illustrating the two major HKT transporter classes. The class 1 HKT transporters that have been shown or suggested to mediate Na+ removal from xylem vessels are shaded in red. The proteins shaded in grey represent class 2 HKT transporters and the remaining HKT transporters correspond to class 1 HKT transporters. The tree was constructed using phyml on a gblocks curated muscle alignment [116–118]. The asterisk marks the OsHKT1;2 pseudogene of rice cv. Nipponbare [49]. The three internal stop codons were overridden to create a putative full-length OsHKT1;2 protein solely for comparison here. A third class of HKT transporters found in Physcomitrella patens and related organisms is not shown here. Abbreviations: At, Arabidopsis thaliana; Ec, Eucalyptus camaldulensis; Hv, Hordeum vulgare; Mc, Mesembryanthemum crystallinum; Os, Oryza sativa; Pa, Phragmites australis; Pt, Populus trichocarpa; Ss, Suaeda salsa; Ta, Triticum aestivum; Th, Thellungiella halophila; Tm, Triticum monoccocum; Tt, Triticum turgidum.
AtHKT1;1 was identified as encoding a relatively Na+-selective class 1 transporter in Arabidopsis, with additional small K+ uptake activity when expressed in an Escherichia coli K+-uptake mutant [42]. Mutations by T-DNA insertion, ethylmethane sulphonate (EMS) treatment or deletions in AtHKT1;1 caused severe Na+ overaccumulation in leaves, leading to leaf chlorosis under salinity stress [11,24,54]. Furthermore, athkt1;1 T-DNA disruption mutations caused greatly reduced Na+ concentrations in roots [11]. However, the total Na+ content in whole Arabidopsis plants was similar in athkt1;1 mutant and wild-type control plants, and, therefore, AtHKT1;1 was concluded to regulate the Na+ distribution between roots and shoots [11]. AtHKT1;1 is expressed in the vasculature in roots and shoots of Arabidopsis [11,24]. athkt1;1 EMS mutant plants accumulate significantly less Na+ in the phloem sap compared with wild-type plants [24]. Together with findings of phloem tissue-specific expression of AtHKT1;1 mRNA, a phloem Na+ re-circulation model for AtHKT1;1 was proposed in which Na+ is loaded into phloem cells by AtHKT1;1 and transferred from shoots to roots via a downward stream of phloem, preventing Na+ overaccumulation in shoots [24].
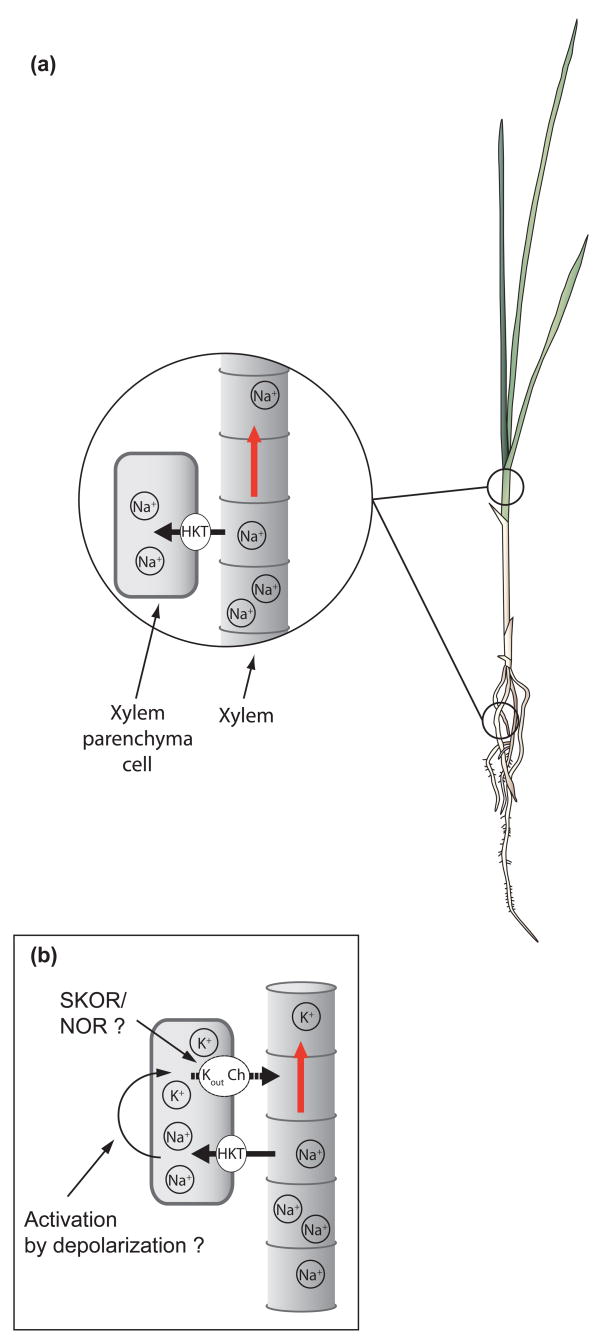
The mechanism of action of AtHKT1;1 has been characterized further. AtHKT1;1 was found to localize at the plasma membrane of xylem parenchyma cells [26]. Analyses of xylem sap emerging from roots and in stems showed that AtHKT1;1 mediates removal of Na+ from root xylem vessels during salinity stress [26]. These findings led to an alternative model for the function of the AtHKT1;1 transporter in protecting leaves from salinity stress by mediating removal of Na+ from the xylem sap via Na+ uptake into xylem parenchyma cells [26,27,55] (Figure 2a). In support of this model, enhancer trap-mediated AtHKT1;1 overexpression in the root pericycle, which includes xylem parenchyma cells, causes enhanced inward Na+ transport in the targeted cells, and results in improved salt tolerance in targeted AtHKT1;1-overexpressing lines compared with wild-type plants [60].
Figure 2.
A model for AtHKT1;1 and OsHKT1;5 functions in mediating Na+ exclusion from leaves by removing Na+ from the xylem sap during salinity stress. (a) AtHKT1;1 and OsHKT1;5 mediate Na+ unloading from xylem vessels at the plasma membrane of xylem parenchyma cells [25,26]. The Na+ uptake function of AtHKT1;1 and OsHKT1;5 prevents Na+ overaccumulation in shoots and leaves, thus protecting vital salt-sensitive photosynthetic processes from salinity stress [11,25–27]. (b) Working hypothesis for the coupling of Na+ unloading from the xylem by class 1 HKT transporters with K+ loading into xylem vessels via depolarization-activated K+ channels. HKT-mediated Na+ uptake from xylem vessels would induce membrane depolarization of xylem parenchyma cells, which in turn could activate outward-conducting K+ channels (i.e. depolarization-activated outward-rectifying K+ channels). This mechanism would result in promoting the concomitantly observed K+ accumulation in the xylem and leaves, which contributes to protection of leaves from salinity stress [25,26].
Independent analyses of the quantitative trait locus (QTL) in rice, named SKC1 (shoot K+ content), resulted in an identical model for the function of the rice gene OsHKT1;5 [25]. The SKC1 QTL was caused by point mutations in OsHKT1;5 that replace several amino acid residues in the salt-tolerant cultivar Nona Bokra [25]. These point mutations enhance the overall Na+ transport activity of OsHKT1;5, based on electrophysiological activity analyses of OsHKT1;5 expressed in Xenopus oocytes [25].
The OsHKT1;5 transporter was found to encode a Na+-selective transporter in Xenopus oocytes [25]. OsHKT1;5 promoter-beta-glucuronidase (GUS) reporter analyses show expression in rice xylem parenchyma cells and OsHKT1;5 reduces Na+ levels in the xylem sap [25]. These findings are similar to the characterized functions of the close orthologs, OsHKT1;5 and AtHKT1;1, which are members of the class 1 sub-family of HKT transporters [25,26,58] (Figure 1). Interestingly, the Mesembryanthemum crystallinum McHKT1;1 class 1 HKT transporter was also targeted to xylem parenchyma cells [48].
The findings that OsHKT1;5 and AtHKT1;1 mediate Na+ exclusion from leaves via Na+ removal from the xylem sap, do not exclude a role for AtHKT1;1 in Na+ loading into phloem vessels [24]. Research using the identical methods reported for phloem sap extraction [24] reproduced the significant reduction in Na+ accumulation in phloem extracts of athkt1;1 null-mutant plants under salinity stress [26]. By contrast, physiological experiments using wild-type and athkt1;1 mutant plants that were loaded with radioactive tracer 22Na+ in the presence of 50 mM NaCl indicated that shoot Na+ contents of both wild-type and mutant plants were relatively unaffected over 2 days after Na+ loading, which led to the conclusion that the phloem sap Na+ recirculation model [24] is unlikely [55,61,62]. This controversy regarding the phloem Na+ recirculation model [24,55,62] highlights an important question, namely: where does the Na+ that is removed from the xylem sap end up?
An intracellular compartment that can accommodate substantial Na+ concentrations is the vacuole, where Na+ sequestration occurs via AtNHX Na+/cation-H+ antiporters [8]. Sodium efflux from roots mediated by the SOS1 Na+-H+ antiporter is another candidate mechanism for eliminating Na+ ions from the cytoplasm, as Na+ exclusion activity by SOS1 from the epidermal layer of the mature root zone [63] and the root tip region [23] have been shown. Further research is needed to address whether vacuolar Na+ sequestration is the only mechanism used by plants to deal with Na+ toxicity in their aerial tissues after HKT-mediated removal of Na+ from the xylem sap. Apart from these questions, present findings in Arabidopsis and rice agree with the model that a major role for AtHKT1;1 and OsHKT1;5 during salinity stress can be defined as ‘Na+ exclusion from shoots’ via Na+ removal from the xylem sap [25–27,64], which thus protects leaves from Na+ toxicity including vital photosynthetic reactions (Figure 2a).
A role for xylem parenchyma-HKT transporters in increasing K+ levels in leaves during salinity stress
Potassium protects plant cells from Na+ stress. Mutations in the rice and Arabidopsis Na+ transporters have been also found to reduce K+ accumulation in shoots during salt exposure, which is likely to further enhance salinity stress [25,26]. The reduced K+ accumulation in the xylem sap, shoots and roots of hkt mutants was inverted to enhanced Na+ accumulation in the same tissues. Specifically, enhanced Na+ levels in the xylem sap of athkt1;1 mutants was accompanied by reduced xylem sap K+ levels [25,26]. As K+ counteracts Na+ toxicity, the reduced K+ accumulation in leaves of hkt mutants further compounds Na+ toxicity. However, the effect of athkt1;1 disruption and point mutations found in OsHKT1;5 proteins on K+ accumulation is smaller than the inverse effect on Na+ accumulation [25,26]. These results suggest that AtHKT1;1 and OsHKT1;5 indirectly propel K+ release from xylem parenchyma cells into the xylem vessel.
A probable driving force for xylem parenchyma K+ release in combination with the Na+ removal function of AtHKT1;1 and OsHKT1;5 should be membrane depolarization. Na+ absorption into xylem parenchyma cells via HKT transporters would depolarize xylem parenchyma cells, which could activate ion channels that can mediate K+ efflux [65–67]. Two independent depolarization-activated outward K+ rectifying systems have been identified by patch clamping analyses using xylem parenchyma cells from barley (Hordeum vulgare) roots: (i) the K+ outward-rectifying (KOR) channel; and (ii) the nonselective outward-rectifying (NOR) channel [66,67]. The KOR channel was found to be largely K+ selective, whereas the NOR channel was shown to be predominantly permeable to cations including K+ and Ca2+ [67]. These reports lead to the hypothesis that KOR and/or NOR channel activity are coupled to the AtHKT1;1-mediated depolarization at xylem parenchyma cells in Arabidopsis (Figure 2b). The cDNA encoding the shaker type outward-rectifying K+ channel (SKOR) was identified in Arabidopsis [68]. Characterizations of the AtSKOR channel in Xenopus oocytes and loss-of-function atskor mutant plants have shown that this channel mediates K+ release into the xylem vessels from xylem parenchyma cells [68]. Whether the NOR-type channel activity also contributes to the K+ release from xylem parenchyma cells, will require identification of the NOR-encoding gene(s). Patch clamp analyses in combination with GFP fluorescence-labeled xylem parenchyma cells could enable further characterization of electrogenic K+ efflux-mediating transporters in xylem parenchyma cells.
Findings on the inverted control of Na+ and K+ levels in the xylem sap of hkt1 mutant plants, suggest that AtHKT1;1 and OsHKT1;5 provide two essential mechanisms towards mediating salt tolerance: (i) prevention of Na+ overaccumulation in leaves; and (ii) promotion of K+ accumulation in leaves [25,26] (Figure 2b).
Major salinity tolerance QTL in wheat are encoded by class 1 HKT genes
An important salinity tolerance locus Kna1 in T. aestivum controls the selectivity of Na+ and K+ transport to shoots, resulting in a high K+:Na+ ratio in leaves [69–72]. The underlying Kna1 gene has been long sought [69–72] and has been mapped to the distal region of the long arm of chromosome 4D [72], but identifying the underlying gene was complicated by the polyploid nature of the wheat genome. Nax1 and Nax2 loci, which contribute to salt tolerance of wheat plants, were identified by QTL analyses using durum wheat [73,74]. Analogous to the findings in Arabidopsis and rice discussed earlier [11,25,26], the Nax1 and Nax2 loci were shown to reduce Na+ transport from roots to shoots and maintain low Na+ concentrations in the leaf blade by excluding Na+ from the xylem [75]. The Nax1 locus has been further shown to have a leaf-located function in exclusion of Na+ from leaves, such that Nax1 enhances removal of Na+ from the xylem of the leaf sheath (the base of leaves), preventing Na+ entry into the leaf blade [75]. Furthermore, the presence of Nax1 and Nax2 was shown to enhance K+ accumulation in leaf blades and sheaths, suggesting that Nax1 and Nax2 mediate K+ loading into the xylem [75]. The Nax2 chromosomal region of a salt-tolerant durum wheat, Line149, was found to correspond to the Kna1 region of T. aestivum, suggesting that Kna1 and Nax2 are orthologs [29]. Interestingly, sequencing analyses of known Na+ transporter genes, including HKTs [39,40], NHXs [8,9,76] and SOS1 [23], within the rough mapping regions of Kna1, Nax1 and Nax2 have suggested that all three of these major salinity tolerance QTL are attributable to polymorphisms in copies of wheat HKT genes, TmHKT1;4-A1 and TmHKT1;4-A2 (also named TmHKT7-A1 and -A2) for Nax1 and TmHKT1;5 and TaHKT1;5 for Nax2 and Kna1 [28,29]. Indeed, the wheat TmHKT1;4 and HKT1;5 genes encode class 1 HKT transporters that are orthologs of the transporters encoded by AtHKT1;1 and OsHKT1;5 [28,29] (Figure 1). These findings in wheat implicate that class 1 HKT Na+ transporters mediate Na+ exclusion from leaves and leaf Na+ tolerance for all three of these major Na+ tolerance QTL.
The Kna1, Nax1 and Nax2 QTL in wheat control Na+ accumulation in leaf sheaths and in Na+ removal from the xylem sap, suggesting that they share analogous or similar functions with AtHKT1;1 in Arabidopsis and OsHKT1;5 and OsHKT1;4 (previously named OsHKT7) in rice (Figure 2) [25,26]. Together, the above studies suggest that class 1 HKT transporters (Figure 1) in Arabidopsis, rice and wheat have major functions in mediating Na+ tolerance and Na+ exclusion from leaves, by removing Na+ from the xylem sap [25–29,75]. This mechanism protects leaves and the photosynthetic machinery from Na+ overaccumulation in dicots and monocots (Figure 2a) and could provide a mechanism for engineering leaf Na+ exclusion and thus enhanced Na+ tolerance in plants [25,26]. Targeted overexpression of AtHKT1;1 in the pericycle of Arabidopsis enhances salinity resistance, supporting this model [60]. Moreover, this recent AtHKT1;1 overexpression study [60] and athtk1;1 T-DNA knockout analyses [27] both could not confirm a previous study reporting that AtHKT1;1 enhances salt sensitivity [77]. Further research is needed to illuminate specialized functions and regulation mechanisms of these salinity tolerance-mediating HKT transporters.
Class 2 HKT transporters
The leaf Na+ exclusion mechanism in plants discussed above is mediated by class 1 HKT transporter sub-family members (Figure 1) [11,24–27,54,55,64]. A second clade of HKT transporters exists in plants, the HKT2 family [39,44] for which in planta functions are only now beginning to be revealed. TaHKT2;1 was the first HKT transporter identified in plants, which belongs to the class 2 subfamily [39] (Figure 1). The transcript level of several HKT2-type transporter genes has been shown to increase by K+ starvation in wheat, rice and barley [44,49,78]. As summarized below, several members of the HKT2 class of transporters are more permeable to K+ compared with the class 1 HKTs based on chimeric studies and biophysical transport analyses in Xenopus oocytes and yeast [39–41,44,57,59,79]. However, the in vivo physiological functions of this HKT2 transporter class remain unknown in plants, with the exception of the recently characterized function of OsHKT2;1 in rice [51], which has been demonstrated to be a unique member of class 2 HKT transporters in terms of its relative selectivity for Na+ over K+ [44,49,57].
Among the class 2 HKT transporters, the OsHKT2;1 transporter is distinct because it has a serine residue in the first putative selectivity pore-forming region (‘loop’) [80], which represents one of the important factors determining Na+ selectivity in HKT transporters [57], with some exceptions [43,46,48]. Other presently known members of the HKT2 class have a glycine residue at the corresponding position instead of a serine residue [39,40,44,49,79]. A different model has suggested that an unusual translation start at a non-ATG start site is responsible for the different ion selectivities of HKT transporters [50], but more recent work revised this hypothesis and concluded that differences in heterologous expression levels of HKT cDNA mutated in their 5′ UTRs were responsible for the reported differential extracellular cation removal activities [81]. Consistent with the K+ permeability of class 2 HKT transporters, in vivo analyses of all four pore-forming loops in the bacterial HKT2 homolog, KtrB, showed that these glycine residues are required for K+ transport activity in vivo [82]. The transport selectivity of Na+ and K+ of HKT transporters depends on the ionic conditions and HKT2 transporters can mediate selective Na+ influx at high Na+ concentrations [40,41,44,47,57,83]. Furthermore, studies have shown that the relative cation selectivities of HKT transporters depend on the ionic conditions [39–41,44,83], which is typical for ion channels that show multiple occupancy by more than one cation at a time [79,84]. The importance of the glycine residue for K+ permeability at the filter position did not apply to some class 1 HKT transporters [43,48] and the ion selectivity of OsHKT2;1 reported differed among studies from relatively more Na+ selective [44,49] to showing a slightly larger relative K+ permeability [59] and no selectivity among mono-valent cations in heterologous expression systems [46]. Further research is needed in which HKT transporters are expressed in plant cells to determine the in vivo cation selectivities of HKT2-class transporters with four glycines in their pore loop domains, as presently all cation selectivity studies on this class of plant transporters have been conducted in heterologous systems.
OsHKT2;1 transporter-mediated nutritional Na+ uptake in K+-starved rice roots
Salt toxicity arises mainly owing to Na+ influx into plant roots, which is probably mediated by more than one type of Na+ transporter [6,13,34,85–89]. No mutation that diminishes Na+ influx into plant roots had been isolated until recently. The OsHKT2;1 transporter was characterized as a Na+ transporter in heterologous expression systems [44,49,57], with evidence for additional K+ transport activity [46,59]. Analyses of three rice oshkt2;1 disruption mutant alleles revealed a major role for OsHKT2;1 in mediating Na+ influx into K+-starved rice roots [51]. Plant physiological experiments showed that Na+ enhances growth of many different plant species [90,91]. Findings from oshkt2;1 analyses provide genetic evidence that Na+ acts as a substitute nutrient for K+ in K+-starved rice plants under moderate Na+ concentrations [51], supporting this long-standing hypothesis [90,91]. Thus, oshkt2;1 mutant alleles represent a first mutant gene that diminish Na+ influx into plant roots [51].
Given that many transporters are expected to contribute to Na+ influx into plant roots, it is not surprising that single gene mutants with greatly reduced Na+ influx have not been previously described [6,86,87]. Nevertheless, the OsHKT2;1-mediated short-term Na+ influx rates into K+-starved rice roots are as high as Na+ influx rates found for low-affinity Na+ influx transporters in plants [92,93], indicating that OsHKT2;1 is a primary Na+ transporter in rice roots when extracellular K+ is limiting. Given that OsHKT2;1 is conditionally induced by K+ starvation, the question arises whether this transporter mediates measurable K+ influx into intact rice roots. However, no clear differences in Rb+ (a K+ analog in transport experiments) influx rates were found among wild-type and oshkt2;1 mutant alleles [51], consistent with data showing relative Na+ over K+ selectivity of OsHKT2;1 in heterologous expression systems [44,49].
OsHKT2;1-dependent Na+ influx is highly regulated and activated under K+ starvation conditions, but is rapidly downregulated by protein kinase inhibitors and also by salinity stress with a half-time of < 1.5 hr [51], suggesting that Na+ transport activity of OsHKT2;1 is post-translationally regulated by as yet unknown cellular signaling pathways. Analyses of OsHKT2;1 regulation mechanisms will provide insights into molecular mechanisms by which a plant root Na+ influx transporter can be controlled.
In spite of the lower K+ permeability of OsHKT2;1 compared with other HKT2 class transporters [44], many of the presently characterized HKT2 class genes are also induced by K+ starvation, including those in wheat, barley and rice [44,49,78]. Furthermore, anti-sense repression of the wheat gene TaHKT2;1 (previously named HKT1) was shown to reduce Na+ influx into wheat roots [47]. Together with its root cortex expression pattern [39], induction by K+ starvation [78], and K+-starvation-induced Na+ influx currents in wheat cortex cells [88], TaHKT2;1 might have a similar function in root Na+ influx to OsHKT2;1 in rice. Thus, it is interesting that class 2 HKT transporters can shift from a Na+–K+ co-transport mode to Na+ transport, depending on the ionic conditions [40,41,83]. Na+ influx into plant roots has been suggested to be dependent on multiple pathways and Na+ influx to toxic levels is independent of the status of K+ availability [87,89,94], consistent with findings that OsHKT2;1 is rapidly downregulated by salinity stress and does not contribute to salinity stress [51].
K+ starvation-induced Na+ influx is likely to be mediated by more than one transporter family in plants. The findings that the TaHKT2;1 transporter mediates Na+–K+ uptake [40,41], indicated that other types of high-affinity K+ uptake transporter should exist in plants. Arabidopsis genomic sequencing led to identification of the KUP–HAK–KT family, which is closely related to bacterial KUP K+ uptake transporters [95–99]. The findings that Na+–K+ uptake is a major high-affinity K+ uptake mechanism in aquatic plants [100,101], but not in land plants [102] further make clear that non-HKT transporters should mediate high-affinity K+ uptake. KUP/HAK/KT transporters function in high-affinity K+ uptake [103,104], and have also have been reported to enable Na+ permeation [95,98]. Members of the KUP/HAK/KT transporter family are strongly induced by K+ starvation [96,98,104,105] and thus these transporters might also contribute to K+-starvation-induced Na+ influx into plant roots, depending on the plant species. Furthermore, some plants, such as Arabidopsis, do not have class 2 HKT Na+ transporters [13,42], and therefore it would be trivial to conclude that no other Na+ influx transporter classes exist in plant roots. Future research is likely to shed molecular insights into these classes of additional Na+ influx transporters [85–89,94,106].
Important future questions for HKT transporter-mediated salinity tolerance
As described above, research on Arabidopsis and rice has led to the uncovering of molecular genetic and mechanistic functions for a major salinity tolerance mechanism and class of HKT transporters that mediate leaf Na+ exclusion in dicot and monocot reference plants [11,24–27,54,55]. In spite of this important advance, there are many new questions. For example, no protein interactor and regulator of HKT transporters is currently known in plants. Recent research indicates roles for post-translational modification of HKT-dependent transport activity in rice [51]. Similarly, the abundance of a relatively Na+ selective transporter in the plasma membrane of xylem parenchyma cells (Figure 2) [26,48] calls for transport activity regulation mechanisms. Such HKT transporters would probably have damaging effects on xylem parenchyma membrane potential, if they were not downregulated when not needed at low extracellular Na+ levels. HKT-mediated membrane potential changes could cause hyperpolarized resting potentials of xylem parenchyma cells in the absence of Na+ stress [39,41], assuming baseline Na+ concentrations in the cytoplasm. Such HKT-mediated effects on membrane potential could be debilitating on physiological xylem transport of many other ions and nutrients.
Another question that remains is whether the physiological functions of these HKT1 transporters are fully understood. The answer is currently no, given the debate regarding whether AtHKT1;1 affects phloem Na+ levels (see above and Refs[24,55]). Other open questions include: where does Na+ go after being removed from the xylem? It is hard to imagine that all Na+ removed from xylem vessels remains in xylem parenchyma cells even though some Na+ will be sequestered in vacuoles [8,19]; independent of whether Na+ re-circulation via the phloem occurs, what is the identity of the Na+ transporters that mediate Na+ efflux from xylem parenchyma cells into the neighboring apoplastic space? Does the AtHKT1;1-type transporter also mediate outward Na+ transport in vivo, as proposed in the Na+ re-circulation model [24]? Alternatively, a SOS1-type Na+-H+ exchanger [23] in the plasma membrane of phloem companion cells would be suited for mediating Na+ efflux.
Several studies have suggested that HKT transporters function as ion channels [41,57,79,107], but to test this hypothesis directly, in vivo electrophysiological analyses are needed to determine whether reversal potentials of HKT transporters follow the electrochemical equilibrium (Nernst) potential for imposed ion gradients. Research on rice and the rice genome sequencing efforts showed that monocots have multiple HKT transporters [49]. In a japonica rice cultivar Nipponbare, five functional class 1 HKT transporters including OsHKT1;5 have been found to be expressed [49] (Figure 1). Recently, detailed ion selectivity analyses of OsHKT1;1 and OsHKT1;3 have been reported [59]. OsHKT1;1 and OsHKT1;3, which are two closely related class 1 transporters, have been demonstrated to show Na+ selective transport [59]. In situ hybridization experiments showed that the tissue specificity of the expression of OsHKT1;1, OsHKT1;3 and OsHKT2;1 overlap in root periphery and vascular tissues of both roots and shoots, also showing some gene-specific expression patterns. These OsHKT mRNAs were also detected in osmocontractile leaf bulliform cells that are thought to function in leaf folding. These results suggest that Na+ transport via OsHKT transporters have roles in several different physiological aspects of rice [59]. Thus far, no gene knockout data on the monocot class 1 HKT transporters exist and only one HKT2 class transporter knockout has been reported [51]. In wheat, where HKT1-linked salinity resistance QTL have been characterized [28,29], this is complicated by the fact that wheat includes multiple copies of HKT1;4 and HKT1;5 genes with similar proposed functions as the Arabidopsis AtHKT1;1 gene. No knockout mutants in the rice orthologs HKT1;4 and HKT1;5 [25] are available yet. Recent advances in creating transposon and T-DNA insertional mutagenesis lines in rice [108–111], rice transformation [112], and rice genome sequencing [113–115] render rice a suitable crop plant model system to characterize the functions of these HKT transporters. Further evidence from loss-of-function mutants of these genes will be needed to elucidate fully the roles of HKT1;4 and HKT1;5 in plant salinity tolerance. Another question that arises is whether these findings can be translated into engineering improved Na+ tolerance of crops, which is likely to require an in-depth understanding of the multiple HKT isoforms found in crop plants, their cellular targeting and their regulators and protein interactors. Nevertheless present studies suggest that manipulation of HKT-mediated transport provides an approach for engineering Na+ exclusion and protection of leaves from salinity stress.
Conclusions
Recent research points to a major function of specific class 1 HKT transporters in mediating salt tolerance and leaf Na+ exclusion in dicots and monocots, via reducing Na+ content in the xylem sap and thus protecting plant leaves from Na+ overaccumulation. This research provides an example that basic findings in Arabidopsis are relevant to crop plants. However, important questions remain. Elucidating a complete picture of how class 1 HKT transporters contribute to salinity tolerance in plants, together with uncovering the downregulation mechanisms of HKT2 transporter-mediated Na+ influx into roots under high Na+ concentrations, will be important steps in determining distinct and relevant salt tolerance mechanisms in plants. Further research will contribute to understanding mechanisms by which leaf metabolic processes and photosynthesis can be protected during salt stress, which will become increasingly important for engineering plants for food and renewable biomass production in light of globally increasing salinization of agricultural utilized lands.
Acknowledgments
This research was support by DOE grant DOE-DE-FG02-03ER15449 and NIEHS grant 1 P42 ESI0337 (to J.I.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tarczynski MC, et al. Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science. 1993;259:508–510. doi: 10.1126/science.259.5094.508. [DOI] [PubMed] [Google Scholar]
- 2.Rains DW, Epstein E. Transport of sodium in plant tissue. Science. 1965;148:1611. doi: 10.1126/science.148.3677.1611. [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman JM. Mechanisms of salinity tolerance in plants. Plant Physiol. 1988;87:547–550. doi: 10.1104/pp.87.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkla BJ, Blumwald E. Identification of a 170-kDa protein associated with the vacuolar Na+/H+ antiport of Beta vulgaris. Proc Natl Acad Sci U S A. 1991;88:11177–11181. doi: 10.1073/pnas.88.24.11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munns R. Physiological processes limiting plant growth in saline soils: Some dogmas and hypotheses. Plant Cell Environ. 1993;16:15–24. [Google Scholar]
- 6.Schroeder JI, et al. Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: Biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct. 1994;23:441–471. doi: 10.1146/annurev.bb.23.060194.002301. [DOI] [PubMed] [Google Scholar]
- 7.Murguía JR, et al. A salt-sensitive 3′ (2′), 5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 8.Apse MP, et al. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 9.Gaxiola RA, et al. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification. Proc Natl Acad Sci U S A. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson DE, et al. Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol. 1999;119:165–172. doi: 10.1104/pp.119.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mäser P, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002a;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- 12.Ward JM, et al. Plants pass the salt. Trends Plant Sci. 2003;8:200–201. doi: 10.1016/S1360-1385(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 13.Horie T, Schroeder JI. Sodium transporters in plants. Diverse genes and physiological functions. Plant Physiol. 2004;136:2457–2462. doi: 10.1104/pp.104.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song CP, et al. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:10211–10216. doi: 10.1073/pnas.0403709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenway H, Munns RA. Mechanisms of salt tolerance in non-halophytes. Annu Rev Plant Physiol. 1980;31:149–190. [Google Scholar]
- 16.Flowers TJ. Salinisation and horticultural production. Sci Hort. 1999;78:1–4. [Google Scholar]
- 17.Tsugane K, et al. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell. 1999;11:1195–1206. doi: 10.1105/tpc.11.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munns R. Prophylactively parking sodium in the plant. New Phytol. 2007;176:501–504. doi: 10.1111/j.1469-8137.2007.02249.x. [DOI] [PubMed] [Google Scholar]
- 19.Blumwald E, Poole R. Na+/H+-antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 1985;78:163–167. doi: 10.1104/pp.78.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blumwald E, Poole RJ. Salt tolerance in suspension cultures of sugar beet : induction of Na+/H+ antiport activity at the tonoplast by growth in salt. Plant Physiol. 1987;83:884–887. doi: 10.1104/pp.83.4.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, et al. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/K+ antiporter. Proc Natl Acad Sci U S A. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H, et al. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 23.Shi H, et al. The putative plasma membrane Na+-H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthomieu P, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren ZH, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 26.Sunarpi et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005;44:928–938. doi: 10.1111/j.1365-313X.2005.02595.x. [DOI] [PubMed] [Google Scholar]
- 27.Horie T, et al. Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 2006;47:622–633. doi: 10.1093/pcp/pcj029. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, et al. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006;142:1718–1727. doi: 10.1104/pp.106.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrt CS, et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007;143:1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shubert S, Läuchli A. Sodium exclusion mechanisms at the root surfaces of two maize cultivars. Plant Soil. 1990;123:205–209. [Google Scholar]
- 31.Gaxiola R, et al. A novel and conserved salt-induced protein is an important determinant of salt tolerance in yeast. EMBO J. 1992;11:3155–3164. doi: 10.1002/j.1460-2075.1992.tb05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu SJ, et al. Sos1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu JK, et al. Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demidchik V, et al. Nonselective cation channels in plants. Annu Rev Plant Biol. 2002;53:67–107. doi: 10.1146/annurev.arplant.53.091901.161540. [DOI] [PubMed] [Google Scholar]
- 35.Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000;12:431–434. doi: 10.1016/s0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apse MP, Blumwald E. Na+ transport in plants. FEBS Lett. 2007;581:2247–2254. doi: 10.1016/j.febslet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Schachtman DP, Schroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370:655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- 40.Rubio F, et al. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 41.Gassmann W, et al. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 1996;10:869–882. doi: 10.1046/j.1365-313x.1996.10050869.x. [DOI] [PubMed] [Google Scholar]
- 42.Uozumi N, et al. The Arabidopsis HKT1 gene homologue mediates inward Na+ currents in Xenopus oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;121:1249–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fairbairn DJ, et al. Characterization of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol Biol. 2000;43:515–525. doi: 10.1023/a:1006496402463. [DOI] [PubMed] [Google Scholar]
- 44.Horie T, et al. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- 45.Rus A, et al. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci U S A. 2001;98:14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golldack D, et al. Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J. 2002;34:1–14. doi: 10.1046/j.1365-313x.2002.01374.x. [DOI] [PubMed] [Google Scholar]
- 47.Laurie S, et al. A role for HKT1 in sodium uptake by wheat roots. Plant J. 2002;32:139–149. doi: 10.1046/j.1365-313x.2002.01410.x. [DOI] [PubMed] [Google Scholar]
- 48.Su H, et al. Expression of the cation transporter McHKT1 in a halophyte. Plant Mol Biol. 2003;52:967–980. doi: 10.1023/a:1025445612244. [DOI] [PubMed] [Google Scholar]
- 49.Garciadeblás B, et al. Sodium transport and HKT transporters: the rice model. Plant J. 2003;34:788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- 50.Haro R, et al. HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol. 2005;139:1495–1506. doi: 10.1104/pp.105.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horie T, et al. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi R, et al. Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J Exp Bot. 2007;58:4387–4395. doi: 10.1093/jxb/erm306. [DOI] [PubMed] [Google Scholar]
- 53.Huang S, et al. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J Exp Bot. 2008;59:927–937. doi: 10.1093/jxb/ern033. [DOI] [PubMed] [Google Scholar]
- 54.Gong JI, et al. Microarray- based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004:101. doi: 10.1073/pnas.0404780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davenport RJ, et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007;30:497–507. doi: 10.1111/j.1365-3040.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 56.Horie T, et al. Functions of HKT transporters in sodium transport in roots and in protecting leaves from salinity stress. Plant Biotech. 2008;25:233–239. [Google Scholar]
- 57.Mäser P, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci U S A. 2002b;99:6428–6433. doi: 10.1073/pnas.082123799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Platten JD, et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006;11:372–374. doi: 10.1016/j.tplants.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Jabnoune M, et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. doi: 10.1104/pp.109.138008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Møller IS, et al. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell. doi: 10.1105/tpc.108.064568. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Møller IS, Tester M. Salinity tolerance of Arabidopsis: a good model for cereals? Trends Plant Sci. 2007;12:534–540. doi: 10.1016/j.tplants.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 63.Shabala L, et al. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta. 2005;222:1041–1050. doi: 10.1007/s00425-005-0074-2. [DOI] [PubMed] [Google Scholar]
- 64.Rus A, et al. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006;2:e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder JI, et al. Voltage dependence of K channels in guard-cell protoplasts. Proc Natl Acad Sci U S A. 1987;84:4108–4112. doi: 10.1073/pnas.84.12.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wegner LH, Raschke K. Ion channels in the xylem parenchyma of barley roots. Plant Physiol. 1994;105:799–813. doi: 10.1104/pp.105.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wegner LH, De Boer AH. Properties of two outward-rectifying channels in root xylem parenchyma cells suggest a role in K+ homeostasis and long-distance signaling. Plant Physiol. 1997;115:1707–1719. doi: 10.1104/pp.115.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaymard F, et al. Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell. 1998;94:647–655. doi: 10.1016/s0092-8674(00)81606-2. [DOI] [PubMed] [Google Scholar]
- 69.Gorham J, et al. Chromosomal location of a K/Na discriminating character in the D genome of wheat. Theor Appl Genet. 1987;74:584–588. doi: 10.1007/BF00288856. [DOI] [PubMed] [Google Scholar]
- 70.Gorham J, et al. Partial characterization of the trait for enhanced K+-Na+ discrimination in the D genome of wheat. Planta. 1990;180:590–597. doi: 10.1007/BF02411458. [DOI] [PubMed] [Google Scholar]
- 71.Luo MC, et al. Engineering of interstitial foreign chromosome segments containing the K+/Na+ selectivity gene Kna1 by sequential homoeologous recombination in durum wheat. Theor Appl Genet. 1996;93:1180–1184. doi: 10.1007/BF00230144. [DOI] [PubMed] [Google Scholar]
- 72.Dubcovsky J, et al. Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet. 1996;92:448–454. doi: 10.1007/BF00223692. [DOI] [PubMed] [Google Scholar]
- 73.Munns R, et al. Genetic control of sodium exclusion in durum wheat. Aust J Agri Res. 2003;54:627–635. [Google Scholar]
- 74.Lindsay MP, et al. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol. 2004;31:1105–1114. doi: 10.1071/FP04111. [DOI] [PubMed] [Google Scholar]
- 75.James RA, et al. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006;142:1537–1547. doi: 10.1104/pp.106.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yokoi S, et al. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313x.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- 77.Rus A, et al. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 2004;136:2500–2511. doi: 10.1104/pp.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang TB, et al. Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 1998;118:651–659. doi: 10.1104/pp.118.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corratge C, et al. Molecular and functional characterization of a Na+-K+ transporter from the Trk family in the ectomycorrhizal fungus Hebeloma cylindrosporum. J Biol Chem. 2007;282:26057–26066. doi: 10.1074/jbc.M611613200. [DOI] [PubMed] [Google Scholar]
- 80.Kato Y, et al. Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc Natl Acad Sci U S A. 2001;98:6488–6493. doi: 10.1073/pnas.101556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bañuelos MA, et al. Effects of polylinker uATGs on the function of grass HKT1 transporters expressed in yeast cells. Plant Cell Physiol. 2008;49:1128–1132. doi: 10.1093/pcp/pcn088. [DOI] [PubMed] [Google Scholar]
- 82.Tholema N, et al. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J Biol Chem. 2005;280:41146–41154. doi: 10.1074/jbc.M507647200. [DOI] [PubMed] [Google Scholar]
- 83.Rubio F, et al. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J Biol Chem. 1999;274:6839–6847. doi: 10.1074/jbc.274.11.6839. [DOI] [PubMed] [Google Scholar]
- 84.LeMasurier M, et al. KcsA: it’s a potassium channel. J Gen Physiol. 2001;118:303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amtmann A, et al. Multiple inward channels provide flexibility in K+/Na+ discrimination at the plasma membrane of barley suspension culture cells. J Exp Bot. 1997;48:431–440. doi: 10.1093/jxb/48.Special_Issue.481. [DOI] [PubMed] [Google Scholar]
- 86.Roberts S, Tester M. A patch clamp study of Na+ transport in maize roots. J Exp Bot. 1997;48:431–440. doi: 10.1093/jxb/48.Special_Issue.431. [DOI] [PubMed] [Google Scholar]
- 87.Tyerman S, et al. Pathways for the permeation of Na+ and Cl− into protoplasts derived from the cortex of wheat roots. J Exp Bot. 1997;48:459–480. doi: 10.1093/jxb/48.Special_Issue.459. [DOI] [PubMed] [Google Scholar]
- 88.Buschmann PH, et al. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000;122:1387–1397. doi: 10.1104/pp.122.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maathuis FJ, Sanders D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001;127:1617–1625. [PMC free article] [PubMed] [Google Scholar]
- 90.Mengel K, Kirkby EA. Principles of Plant Nutrition. 3. International Potash Institute; 1982. [Google Scholar]
- 91.Flowers TJ, Läuchli A. Sodium versus potassium: Substitution and compartmentation. Inorg Plant Nutr. 1983;15b:651–681. [Google Scholar]
- 92.Rains DW, Epstein E. Sodium absorption by barley roots: role of the dual mechanisms of alkali cation transport. Plant Physiol. 1967;42:314–318. doi: 10.1104/pp.42.3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Essah PA, et al. Sodium influx and accumulation in Arabidopsis. Plant Physiol. 2003;133:307–318. doi: 10.1104/pp.103.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davenport RJ, Tester M. A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol. 2000;122:823–834. doi: 10.1104/pp.122.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fu HH, Luan S. AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim EJ, et al. AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintero FJ, Blatt MR. A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett. 1997;415:206–211. doi: 10.1016/s0014-5793(97)01125-3. [DOI] [PubMed] [Google Scholar]
- 98.Santa-Maria GE, et al. The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 1997;9:2281–2289. doi: 10.1105/tpc.9.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gierth M, Mäser P. Potassium transporters in plants-involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett. 2007;581:2348–2356. doi: 10.1016/j.febslet.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 100.Smith FA, Walker NA. Transport of potassium in Chara australis. I A symport with sodium. J Membr Biol. 1989;108:125–137. doi: 10.1007/BF01869452. [DOI] [PubMed] [Google Scholar]
- 101.Walker NA, Sanders D. Sodium-coupled solute transport I charophyte algae: a general mechanism for transport energization in plant cells? Planta. 1991;185:443–445. doi: 10.1007/BF00201070. [DOI] [PubMed] [Google Scholar]
- 102.Maathuis F, et al. The physiological relevance of Na+-coupled K+-transport. Plant Physiol. 1996;112:1609–1616. doi: 10.1104/pp.112.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rigas S, et al. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gierth M, et al. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005;137:1105–1114. doi: 10.1104/pp.104.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nieves-Cordones M, et al. A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant Mol Biol. 2008;68:521–532. doi: 10.1007/s11103-008-9388-3. [DOI] [PubMed] [Google Scholar]
- 106.Volkov V, Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J. 2006;48:342–353. doi: 10.1111/j.1365-313X.2006.02876.x. [DOI] [PubMed] [Google Scholar]
- 107.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 108.Hirochika H, et al. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci U S A. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hirochika H. Retrotransposons of rice: Their regulation and use for genome analysis. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- 110.Jeon JS, et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 111.Miyao A, et al. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hiei Y, et al. Efficient transformation of rice (Oryza sative L.) mediated by Agrobacterium and sequence analysis of the boundaries of the DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 113.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp Japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 114.Sakata K, et al. An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 2002;30:98–102. doi: 10.1093/nar/30.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp Indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 116.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 117.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]