Abstract
The replication of many viral and subviral pathogens as well as the amplification of certain cellular genes proceeds via a rolling circle mechanism. For potato spindle tuber (PSTVd) and related viroids, the possible role of a circular (−)strand RNA as a template for synthesis of (+)strand progeny is unclear. Infected plants appear to contain only multimeric linear (−)strand RNAs, and attempts to initiate infection with multimeric (−)PSTVd RNAs generally have failed. To examine critically the infectivity of monomeric (−)strand viroid RNAs, we have developed a ribozyme-based expression system for the production of precisely full length (−)strand RNAs whose termini are capable of undergoing facile circularization in vitro. Mechanical inoculation of tomato seedlings with electrophoretically purified (−)PSTVd RNA led to a small fraction of plants becoming infected whereas parallel assays with an analogous tomato planta macho viroid (−)RNA resulted in a much larger fraction of infected plants. Ribozyme-mediated production of (−)PSTVd RNA in transgenic plants led to the appearance of monomeric circular (−)PSTVd RNA and large amounts of (+)PSTVd progeny. No monomeric circular (−)PSTVd RNA could be detected in naturally infected plants by using either ribonuclease protection or electrophoresis under partially denaturing conditions. Although not a component of the normal replicative pathway, precisely full length (−)PSTVd RNA appears to contain all of the structural and regulatory elements necessary for initiation of viroid replication.
Keywords: RNA replication, circular RNAs, ribozymes
Viroids—small, single-stranded, circular RNAs containing 246–463 nucleotides arranged in a rod-like secondary structure—are the smallest pathogenic agents yet described. Unlike the many satellite or defective interfering RNAs associated with plant viruses, viroids replicate autonomously on inoculation of a susceptible host (1, 2). The absence of a protein capsid and of detectable messenger RNA activity implies that the information necessary for replication and pathogenesis resides within the unusual structure of the viroid genome.
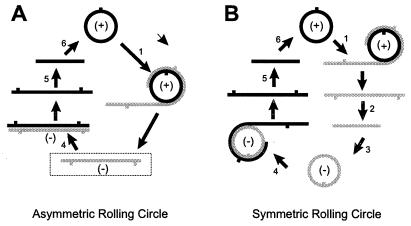
Viroid-infected tissue contains a variety of multimeric RNAs, and replication is thought to proceed via a “rolling circle” mechanism (see Fig. 6). Replication of avocado sunblotch viroid (ASBVd) uses a symmetric pathway in which circular forms of both (+) and (−)ASBVd RNA serve as a template for synthesis of complementary multimers that are cleaved into monomers and ligated to form circular progeny (3). Replication of potato spindle tuber (PSTVd) and related viroids, in contrast, appears to proceed via an asymmetric pathway in which multimeric linear (−)strands synthesized from the circular (+)PSTVd inoculum serve as a template for synthesis of (+)strand multimers from which the circular progeny are derived via cleavage and ligation (4). Although multimeric ASBVd RNAs of either polarity undergo ribozyme-mediated self-cleavage (5), one or more host-encoded nuclease activities appear to be necessary for the specific cleavage/ligation of multimeric (+)PSTVd RNAs (6–8). Synthesis of both (+) and (−)PSTVd RNAs is inhibited by low levels of α-amanitin (9), indicating the involvement of DNA-dependent RNA polymerase II.
Figure 6.
Initiation of PSTVd replication by circularized (−)PSTVd RNA. Viroid replication may involve as many as six separate steps: i.e., synthesis of multimeric (−)RNAs and (+)RNAs (steps 1 and 4), cleavage of these multimeric RNAs (steps 2 and 5), and circularization of the resulting linear (−)RNA and (+)RNA monomers (steps 3 and 6). During normal asymmetric replication (A), multimeric linear (−)PSTVd RNAs (boxed) serve as template for synthesis of (+)PSTVd progeny. In the symmetric rolling circle mechanism used by ASBVd and certain satellite RNAs (B), the corresponding multimeric (−)strand RNAs are cleaved and circularized before acting as template for (+)strand synthesis. Thus, the initial replicative steps after inoculation with precisely full length (−)PSTVd or (−)TPMVd RNA may resemble steps 3–6 of the symmetric mechanism.
Interest in their mode of replication has been heightened by the possibility that viroids may represent “relics” of a precellular RNA world (10). For example, their circular structure and the existence of efficient mechanisms for the precise cleavage of monomeric progeny from oligomeric replication intermediates may eliminate the need for discrete initiation or termination signals. Circular (+)PSTVd (i.e., viroid progeny) is always present in vast excess over any replicative intermediates, and available nucleic acid hybridization and fingerprinting data alone do not exclude a possible role for a circular (−)PSTVd RNA in PSTVd replication (4, 11). Similarly, interpretation of infectivity studies with multimeric (−)PSTVd RNAs is complicated by the relatively low specific infectivity of such RNAs (12).
To investigate the possible role of a circular (−)RNA in PSTVd replication, we have developed an expression system for the ribozyme-mediated production of a precisely full length (−)PSTVd RNA whose termini are capable of undergoing facile circularization. The infectivity of such an RNA appears to represent a previously undescribed pathway for initiating viroid replication, one that is distinctly different from the normal replicative pathway.
MATERIALS AND METHODS
Plasmid Constructs.
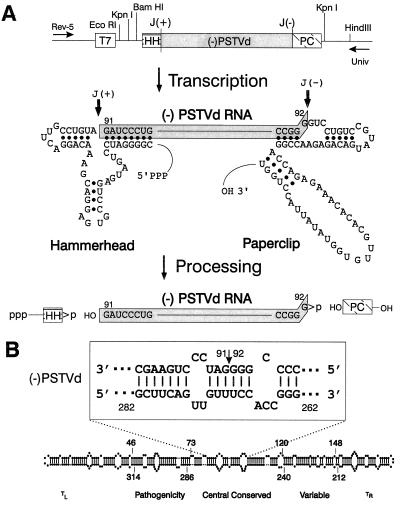
A paperclip ribozyme sequence designed to leave the sequence CCCCGGG>p at the 3′ end of either (−)PSTVd or (−)TPMVd RNA transcripts was generated from plasmid pT7(−)miniMn = 7 (13) in PCRs containing a “universal” M13 sequencing primer (5′-GTAAAACGACGGCCAGT-3′) plus an oligonucleotide designated “PSTVd PC(−)” (5′ GGGAGATCTAGCGCTTCAGGGTCCTGTCCGTATGACAGAGAACTGAACCAG-3′). After digestion with BglII and HindIII, this PCR product was cloned in pTZ18R Rz6′-1, a vector containing a hammerhead ribozyme and designed to leave the sequence HO-GATCC at the 5′ terminus of either (−)PSTVd or (−)TPMVd RNA transcripts. The resulting vector construct was designated pTZ18R Rz(−)4. After digestion with BamHI and XmaI, a double-stranded PSTVd cDNA was cloned into BglII + AvaI-digested pTZ18R Rz(−)4, and colonies containing full length inserts were used for synthesis of (−)PSTVd RNA. A slightly different vector was used for the (−)TPMVd construct, and, as a result, the hammerhead ribozyme formed a 10-bp stem with the (−)TPMVd RNA rather than the 8-bp stem shown in Fig. 1.
Figure 1.
Production of precisely full length (−)PSTVd RNA. (A) T7 expression cassette and processing pathway for (−)PSTVd RNA transcripts. T7, promoter for bacteriophage T7 RNA polymerase; (−)PSTVd, full length PSTVd cDNA; J(+) and J(−), cleavage sites for the hammerhead (HH) and paperclip (PC) ribozyme sequences from (+) and (−)sTRSV RNAs; Rev-5 and Univ, binding sites for M13 sequencing primers. (B) Location of the ribozyme cleavage sites (arrow) within the central conserved region. Note that the sequences in (−)PSTVd RNA are written in an unconventional orientation (i.e., 5′ on the right → 3′ on the left) and that individual nucleotides are numbered according to their positions in the corresponding (+)PSTVd RNA. The rod-like native structure of (+)PSTVd and the relative locations of the five proposed structural domains of (+)PTSVd (23) also are shown.
Transgenic Plants.
To obtain transgenic Nicotiana benthamiana L. plants constitutively expressing (−)PSTVd RNA, the PSTVd cDNA-ribozyme cassette was excised from pTZ18R Rz(−)4 by digestion with EcoRI and HindIII and recloned in plasmid pGEM-7Zf(+) (Promega). After conversion of the unique HindIII site in the resulting plasmid to an NheI site, the cassette was excised by digestion with NheI and XbaI and transferred to the partially filled HindIII site of pGA643 (14) downstream from the 35S promoter of cauliflower mosaic virus. The resulting recombinant plasmid was transformed directly into competent Agrobacterium tumefaciens (strain 5922) cells containing the helper plasmid pC2760 (15). Protocols for transformation of leaf tissue from N. benthamiana and identification of transformed plants by PCR have been described elsewhere (16, 17).
Biological Activity of Viroid-Complementary RNAs.
Plasmid DNAs were linearized with HindIII and transcribed with T7 RNA polymerase as suggested by the manufacturer (Promega) except that the Mg2+ concentration was increased to 15 mM. RNAs were recovered by phenol-chloroform extraction and ethanol precipitation, and monomeric linear (−)PSTVd or (−)TPMVd RNAs were purified by electrophoresis in 5% polyacrylamide gels containing 1 × TBE buffer [90 mM Tris, 90 mM boric acid, and 2.5 mM EDTA (pH 8.3)]/7 M urea.
After elution with 0.5 M ammonium acetate and ethanol precipitation (18), RNAs were dissolved in 20 mM sodium phosphate buffer (pH 7.0), and aliquots containing ≈30 ng/ml (−)PSTVd or (−)TPMVd RNA were rubbed on the carborundum-dusted cotyledons of week-old tomato seedlings. Inoculated plants were maintained in a greenhouse for 6–7 weeks under conditions suitable for viroid replication and symptom development, and tissue samples were assayed for the presence of viroid progeny by dot–blot hybridization with digoxygenin-labeled, viroid-specific RNA probes (19).
Characterization of PSTVd-Related RNAs.
32P-labeled PSTVd RNA transcripts synthesized in vitro were purified by electrophoresis in 5% polyacrylamide gels containing 1× TBE buffer/7 M urea and circularized by incubation with a cell-free wheat germ extract (Promega). After phenol-chloroform extraction and ethanol precipitation, the resulting mixtures of linear and circular RNAs were analyzed by temperature gradient gel electrophoresis in 5% polyacrylamide gels containing 0.2 × TBE buffer/5 mM NaCl (20).
Total RNA was extracted from samples of leaf tissue collected from individual transgenic plants (0.1 gm) by using the TRI reagent (Molecular Research Center, Cincinnati) and quantitated by UV spectrophotometry. PSTVd concentrations in individual RNA preparations were estimated by dot–blot hybridization as described above. For Northern analysis, aliquots (15 μl) containing 40–60 pg PSTVd-related RNA [either (+) or (−)strands] were mixed with a equal volume of loading buffer (90% formamide/10 mM EDTA/0.02% bromophenol blue/0.02% xylene cyanol), heated for 3 min at 94°, and chilled on ice before fractionation in 5% polyacrylamide gels containing 1 × TBE buffer/8 M urea. After electrophoresis, the RNAs were electrotransferred to Nytran Plus membranes (Schleicher & Schuell), UV-crosslinked, and hybridized with digoxygenin-labeled, strand-specific RNA probes.
Ribonuclease protection assays (RPAs) were carried out as described by the manufacturer (Ambion, Austin, TX) by using 32P-labeled probes synthesized from plasmids pST64-B5 and pST65-B5 (21). In both cases, the PSTVd cDNA termini were derived from the unique BamHI site at positions 87–92, and the respective probes contained either a 6-nt or an 11-nt sequence duplication. Each assay contained 30 pg (+)PSTVd or (−)PSTVd RNA (determined by dot–blot hybridization) and ≈1 × 105 cpm 32P-labeled probe. Protected fragments were fractionated on 6% nondenaturing polyacrylamide gels and visualized by autoradiography.
RESULTS
Ribozyme-Mediated Production of Viroid-Complementary RNAs.
As shown in Fig. 1A, appropriate modifications of noncatalytic sequences in ribozymes derived from the satellite RNA of tobacco ringspot virus (sTRSV RNA) (22) resulted in an expression cassette that can be used to synthesize precisely full length (−)PSTVd RNA. Spontaneous cleavage of the initial T7 transcript at both the 5′- and 3′-cleavage sites releases a precisely full length (−)PSTVd RNA plus the two flanking ribozyme sequences. Cleavage at either the hammerhead or paperclip site alone produces a mixture of longer RNAs containing both ribozyme- and PSTVd-specific sequences. As shown in Fig. 1B, the choice of positions 91–92 as the sites of ribozyme cleavage results in a linear (−)PSTVd RNA molecule whose termini are located within a helical portion of the central conserved domain (23).
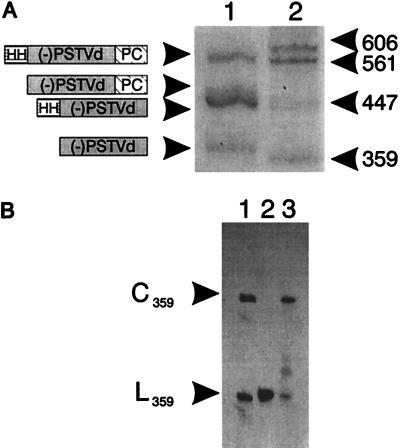
After overnight incubation with T7 RNA polymerase, transcription products from the (−)PSTVd RNA expression cassette were examined by PAGE under denaturing conditions. Data presented in Fig. 2A show that, in addition to a residual full length RNA transcript, each of the predicted single- and double-cleavage products also was present. Because (−)PSTVd RNA is a highly structured molecule (24), the relatively high yield of fully processed RNA suggests that ribozyme cleavage probably occurs before synthesis of the nascent transcript is complete. Furthermore, the 5′-hydroxyl and 2′,3′-cyclic phosphate termini generated by ribozyme cleavage permit efficient circularization of fully processed (−)PSTVd RNA by incubation with wheat germ ligase (compare lanes 2 and 3 of Fig. 2B). The initial product of ligation is a 2′-phosphomonoester, 3′,5′-phosphodiester linkage also known as a “Filipowicz junction” (25).
Figure 2.
Processing and circularization of (−)PSTVd RNA transcripts in vitro. (A) Electrophoretic fractionation of 32P-labeled processing products from overnight transcription reactions. Lanes: 1, (−)PSTVd RNA; 2, (−)sTRSV RNA markers. Sizes (in nucleotides) of individual cleavage products are indicated. (B) Circularization of (−)PSTVd RNA. Lanes: 1, circular (C359) and linear (L359) (−)sTRSV RNA markers; 2 and 3, (−)PSTVd RNA. The (−)PSTVd RNA sample in lane 3 was incubated with wheat germ extract before electrophoresis. After cleavage, the termini of full length (−)sTRSV RNA undergo spontaneous ligation to form both monomeric circles and dimeric linear molecules (22).
Infectivity of (−)PSTVd and (−)TPMVd RNAs.
The infectivity of precisely full length (−)PSTVd and (−)TPMVd RNAs released by ribozyme cleavage and purified by gel electrophoresis under denaturing conditions was examined in a series of bioassays on tomato seedlings. As shown in Table 1, only a small proportion of plants became infected after inoculation with 20–30 ng/ml (−)PSTVd RNA. A considerably higher proportion of infected plants was observed in parallel assays with (−)TPMVd RNA, but the specific infectivity of either RNA was still quite low—i.e., ≈10−4 that of a comparable preparation of precisely full length (+)PSTVd. (−)TPMVd RNA was chosen to confirm the results obtained with (−)PSTVd because no studies involving (+)TPMVd RNAs had been carried out recently in our laboratory or greenhouse facilities.
Table 1.
Infectivity of ribozyme-derived PSTVd and TPMVd RNAs
| Inoculum | Relative concentration
|
||||||
|---|---|---|---|---|---|---|---|
| 100 | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | |
| (+)PSTVd | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 4/10 | nd |
| 8/10 | 5/10 | 1/10 | |||||
| (−)PSTVd | 1/10 | ||||||
| 0/10 | |||||||
| 1/10 | |||||||
| (−)TPMVd | 5/20 | ||||||
| 15/18 | 5/18 | 3/18 | |||||
Initial inoculum concentration = 20–30 ng/μl. Each inoculum was tested in at least two independent infectivity trials. All data expressed as plants infected/plants inoculated. nd, not done.
Circularization of (−)PSTVd RNA in Vivo.
To further examine the possible role of monomeric (−)PSTVd RNA as template for PSTVd replication, we constructed several lines of transgenic N. benthamiana plants in which constitutive transcription of an expression cassette similar to that shown in Fig. 1 is driven by a cauliflower mosaic virus 35S promoter. In one group of plants, ribozyme-mediated cleavage of the initial RNA transcript was expected to release monomeric (−)PSTVd RNA; for the second group, the expected cleavage product was a monomeric (+)PSTVd RNA. Total RNA was prepared from leaf tissue collected from individual transformed plants, and the nature of the PSTVd-related RNAs present was assessed by a combination of Northern analysis and RPAs. Figs. 3 and 4 present the results of these analyses.
Figure 3.
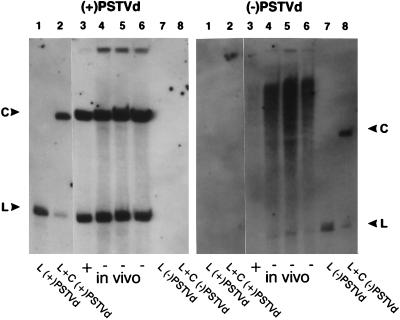
Expression of PSTVd-related RNAs in transgenic N. benthamiana. Total cellular RNA isolated from PSTVd-infected plants was fractionated by electrophoresis under denaturing conditions, and duplicate blots were hybridized with probes specific for either (+)PSTVd (Left) or (−)PSTVd (Right) RNAs. Samples isolated from transgenic plants (lanes 3–6) contained equal amounts (≈50 pg) of (+) or (−)PSTVd RNAs. The purified (+) and (−)PSTVd RNA transcripts analyzed in lanes 1, 2, 7, and 8 provide mobility standards for circular (C) and linear (L) molecules; samples in lanes 2 and 8 were incubated with wheat germ extract before analysis. Note that monomeric linear (−)PSTVd RNA (arrow) is only detectable in those plants in which it is expressed constitutively. The light area visible in lanes 4–6 (Right) opposite the circular standard in lane 8 is caused by the presence of large amount of circular (+)PSTVd progeny migrating at that position.
Figure 4.
Circularization of (−)PSTVd RNA in planta. Duplicate sets of RPAs using greater-than-full length probes specific for (+)PSTVd (Left) and (−)PSTVd RNA (Right) are shown. Lanes: 1–4, purified in vitro PSTVd RNA transcripts; 5–8, total RNA isolated from transgenic plants expressing either (+)PSTVd (lane 5) or (−)PSTVd RNA (lanes 6–8). All plants were PSTVd-infected, and samples 5–8 each contained 30 pg of (+)PSTVd or(−)PSTVd RNA. Positions of RNA:RNA duplexes containing circular (C) and linear (L) target molecules are marked (arrows). Strands shown in black and gray correspond to (+) and (−) PSTVd sequences, respectively, and the asterisks indicate the strand used as assay probe. Note that circularized (−)PSTVd RNA is detectable only in those plants that express the corresponding linear molecule. Exposure time for the (−)PSTVd RNA analysis was 10-fold longer than that for the (+)PSTVd analysis.
All 10 plants that constitutively expressed (+)PSTVd RNA became PSTVd-infected while still in tissue culture. Most (but not all) of the plants expressing (−)PSTVd RNA also became infected, and there were no obvious differences in PSTVd titer among the various infected plants (results not shown). Total RNA was extracted from four PSTVd-infected plants: three plants expressing (−)PSTVd RNA and one “positive control” that expressed (+)PSTVd. Aliquots containing equal amounts of (+) or (−)PSTVd RNAs were fractionated by electrophoresis under denaturing conditions and subjected to Northern analysis by using strand-specific probes.
As shown in Fig. 3, all four infected plants contained easily detectable levels of circular (+)PSTVd progeny (Fig. 3 Left, lanes 3–6). A parallel analysis using a probe specific for (−)PSTVd RNA revealed the presence of monomeric linear (−)PSTVd RNA only in those plants that constitutively expressed such a molecule (in Fig. 3 Right, compare lanes 4–6 with lane 3). Those three plants that expressed (−)PSTVd also appeared to contain higher relative concentrations of greater-than-unit-length (−)PSTVd. None of the four samples, however, yielded a discrete band corresponding to the circularized (−)PSTVd RNA marker (Fig. 3 Right, lane 8).
Total RNA preparations also were examined for the presence of circularized (−)PSTVd by using RPA. As described in Materials and Methods, the strand-specific probes used in these analyses contained short sequence duplications overlapping the potential ligation sites in their respective targets. Digestion of the resulting RNA:RNA hybrids with RNase would, therefore, be expected to produce small amounts of a nicked, circular (i.e., incompletely digested) duplex as well as the completely digested, linear, 359-bp duplex. Indeed, data presented in Fig. 4 show that, when circularized in vitro RNA transcripts were used as targets, easily detectable amounts of such a slowly migrating species were formed. Similar results were obtained when either (−)PSTVd or (+)PSTVd RNA was used as target (compare Fig. 4 Left, lane 2 with Fig. 4 Right, lane 4). Although all four transgenic plants contained circular (+)PSTVd progeny, circularized (−)PSTVd RNA was detected only in those plants that constitutively expressed (−)PSTVd RNA (Fig 4 Right, lanes 3–6).
Electrophoretic Separation of Circular (+) and (−)PSTVd RNAs.
Results of dot–blot hybridization assays carried out before the RPA analyses described above showed that transgenic plants constitutively expressing (−)PSTVd RNA contained ≈25-fold more (−)PSTVd RNA than comparable plants expressing (+)PSTVd RNA. To circumvent possible problems associated with probe dilution during the RPA assay, we sought to exploit differences in the structural properties of circularized (+)PSTVd and (−)PSTVd RNAs to separate these two molecules before analysis.
Optical melting studies and temperature gradient gel electrophoretic analysis of linear RNA transcripts have shown that (−)PSTVd and (+)PSTVd RNAs have quite different structural properties (24, 26). Denaturation of a monomeric (+)PSTVd RNA with termini derived from positions 281–282 was characterized by a single cooperative transition, whereas the profile of the corresponding (−)PSTVd RNA was biphasic, exhibiting two transitions of roughly equal magnitude. The ability of wheat germ RNA ligase to circularize the precisely full length (+) and (−)PSTVd RNAs produced by ribozyme cleavage in vitro allowed us to compare directly by temperature gradient gel electrophoresis the structural properties of the respective circular forms of (+) and (−)PSTVd RNAs.
As shown in Fig. 5, denaturation of circularized (−)PSTVd RNA also was biphasic. Note, in particular, that the first transition was virtually complete at temperatures where (+)PSTVd was just beginning to denature. By using appropriate combinations of buffer concentration and running temperature (e.g., 0.2 × TBE buffer/5 mM NaCl at 44° C), it was possible to separate circularized (−)PSTVd RNA from a vast excess of circular (+)PSTVd progeny by a more traditional one-dimensional PAGE protocol. The sensitivity of such an analysis carried out at 44° in the presence of 0.2 × TBE buffer/5 mM NaCl was sufficient to detect 5 pg of circularized (−)PSTVd RNA transcripts in the presence of 5000 pg of circularized (+)PSTVd RNA. The absence of a comparable band from an analysis of total RNA extracted from a transgenic plant constitutively expressing (+)PSTVd RNA and containing 10 ng (+)PSTVd indicates that the ratio of circular (+)PSTVd to circular (−)PSTVd RNA in such plants must have been >2000:1 (results not shown).
Figure 5.
Structural properties of circularized (−) and (+)PSTVd RNAs. (A) Samples of 32P-labeled PSTVd RNAs, each containing a mixture of circular (C) and linear (L) molecules, were applied successively to the central slot of a temperature gradient gel; the first sample contained (−)PSTVd RNAs, and the second contained (+)PSTVd RNAs. Direction of migration was from top to bottom, and positions of the denaturation profiles for the circularized molecules are indicated by arrows. (B) Tracings of the individual denaturation profiles. Note that both the linear and circularized forms of (−)PSTVd RNA began to denature at a lower temperature than the corresponding (+)PSTVd RNAs. Only the central portion of the gel is shown.
DISCUSSION
Previous infectivity studies with oligomeric (−)PSTVd RNAs synthesized in vitro have shown such RNAs to be, at best, weakly infectious (12). Complicating the interpretation of such data is the possibility of contamination by traces of infectious (+)strand RNA transcribed from the opposite strand of the DNA template (27). Three features of our double ribozyme expression strategy—the absence of any known RNA polymerase promoter sequences in the opposite strand of the DNA template, the inability of sequences complementary to the ribozymes to undergo self-cleavage, and the presence of only a 2-nt duplication in the (+)viroid sequence—make the presence of an infectious (+)PSTVd or (+)TPMVd contaminant in our experiments extremely unlikely.
After in vitro transcription, ribozyme-matured (−)viroid RNAs to be used as inocula were purified by electrophoresis. Such purification greatly reduced the possibility of retaining in the inoculum trace amounts of accidentally transcribed (+)viroid RNA. In the case of the transgenic plants designed to express (−)PSTVd RNA, insertion of the transgene in the vicinity of a plant promoter could drive production of (+)PSTVd RNA. Three observations make the presence of such a promoter an unlikely explanation for the infections observed. First, in experiments involving Agrobacterium-mediated insertions of promoterless genes, only 25% of insertion events were oriented properly near an endogenous promoter (28, 29). In our experiments, 60% of the transgenic lines became infected. Second, the cauliflower mosaic virus 35S promoter was 100-fold more active than the endogenous promoters found driving the promoterless genes (28). Third, the absence of ribozyme activity and the presence of only a 2-nt sequence severely limits the potential for (+)PSTVd RNA transcripts to mature to the correct size. For example, previous Agrobacterium-mediated inoculation studies have shown that a 35S promoter-driven (+)PSTVd cDNA construct containing a 4-nt sequence duplication is only weakly infectious (30). The question then arises: How are precisely full length, linear (−)strand viroid RNAs, molecules that normally are not detectable in vivo, able to initiate the infection process?
Fig. 6 contrasts an asymmetric, rolling circle mechanism of the type believed responsible for PSTVd replication with the symmetric mechanism used by ASBVd and certain viral satellite RNAs. Note that the template for synthesis of (+)strand progeny in the asymmetric mechanism is an uncleaved multimeric (−)strand RNA. If normal PSTVd replication were to proceed via a symmetric (rather than an asymmetric) mechanism, infected tissues should contain circular forms of (−)PSTVd as well as (+)PSTVd RNA.
Although an earlier analysis of partially double-stranded PSTVd replicative intermediates labeled in vivo with 32P failed to detect such molecules, the fingerprinting methods used would not have been able to detect relatively low levels of circular (−)PSTVd RNA (11). Even the 100-fold greater sensitivity of our methodologies, however, failed to reveal any evidence for replication via a symmetric pathway. Circularized (−)PSTVd RNA was detected only in transgenic plants that constitutively expressed the corresponding precisely full length linear RNA. Once the incoming (−)PSTVd RNA, as an inoculum either synthesized in vitro or expressed in planta, becomes circularized, the initial stages of the ensuing replication process in the host cell would resemble steps 3–6 of a symmetric, rolling circle mechanism (Fig. 6B). After the appearance of the first molecules of (+)PSTVd or (+)TPMVd progeny, replication then could continue via the normal asymmetric pathway shown in Fig. 6A.
Provided that precisely full length (−)PSTVd RNA is able to initiate infection, why should its specific infectivity be so much lower than that of the corresponding (+)strand RNA? Production of a large (i.e., 20- to 100-fold) excess of (+)RNA during the replication of most (+)strand RNA viruses implies that the viral replicase prefers (−)RNA as a template, and the transition from (−) to (+)strand synthesis appears to involve the recruitment of additional host factors by the viral replicase (31). The RNA replicase encoded by RNA 1 of the flock house virus (a member of the Nodaviridae) has been shown to support the in vivo replication of either (+) or (−)strand transcripts of RNA 2, but the presence of even two nonviral nucleotides at the 5′ end of the primary (−)strand transcript was sufficient to severely inhibit RNA 2 replication (32). As first pointed out by Diener (10), the existence of an efficient mechanism for the precise cleavage of monomeric viroid progeny from oligomeric (+)strand replicative intermediates may obviate the need for defined signals specifying initiation or termination by an RNA replicase on a circular template. Circularization would thus represent a key event in the infection process for either an incoming (+) or (−)strand viroid RNA.
Like the hepatitis delta virus, PSTVd and related viroids replicate in the nuclei of infected cells, where the enzyme involved appears to be the α-amanitin-sensitive, host-encoded RNA polymerase II (33, 34). The vast majority of both (+) and (−)strand PSTVd RNAs is located in the nucleolus (35), and in planta transcriptional assays independent of PSTVd replication recently have shown that monomeric PSTVd RNAs of either polarity contain promoter sequences recognized by cellular RNA polymerase(s) (36). Differences in the efficiency of transport from cytoplasm to nucleus after mechanical inoculation (possibly as a result of nuclease degradation associated with its less stable secondary structure; see Fig. 5) could explain the relatively low specific infectivity of (−)PSTVd RNA. Studies have shown that both (+) and (−)strand monomeric hepatitis delta virus RNAs are infectious (37, 38).
The relationship between the secondary/tertiary structure of viroids and their various biological properties remains poorly understood. For PSTVd and related viroids, pathogenicity and/or replication is modulated by at least three discrete sequence or structural elements (39), and within the so-called pathogenicity domain of PSTVd, there is a strong correlation between symptom severity and the relative orientation of three short helical regions within the overall rod-like native structure (40). Formation of other critical structural elements, especially those involved in replication-related events, appears to require disruption of the familiar rod-like native structure.
Thermal denaturation in vitro causes the native structure of PSTVd and related viroids to undergo a highly cooperative transition to a branched structure containing 2–3 alternative interactions known as “secondary hairpins” (41). Formation of secondary hairpin II appears to be essential for infectivity (21, 42), but it is not yet clear whether this interaction is operating at the level of the (+) or (−)strand RNA. Mutational analysis suggests that formation of secondary hairpin II is essential for the ability of multimeric (−)PSTVd RNA to act as template for the synthesis of (+)strand progeny, possibly by regulating the competition between the energetically favored, rod-like native structure and various metastable structures (43). In this view, the primary function of the native structure may be to ensure the long-term survival of the unencapsidated viroid RNA within the host plant.
At the present time, mutational analysis of viroid function is heavily dependent on infectivity assays carried out in vivo. Studies of the structural properties of the various (+) and (−)strand RNAs found in infected cells have, in contrast, been carried out almost exclusively in vitro. The ability to conveniently prepare large amounts of infectious (−)strand PSTVd RNA in vitro means that, for the first time, it is now possible to examine directly the biological and structural properties of all components of the replicative pathway. Studies comparing the ability of (+)strand and (−)strand PSTVd RNAs to enter the nucleus of potential host cells and move from cell to cell via the plasmodesmata are currently in progress.
Acknowledgments
We thank J.N. Culver, R.W. Hammond, J.M. Kaper, and E.V. Podleckis for helpful discussions and critical review of the manuscript. P.A.F. and Y.H. were supported by Grant 91–373303-6649 from the United States Department of Agriculture/National Research Initiative Competitive Grants Program.
ABBREVIATIONS
- ASBVd
avocado sunblotch viroid
- PSTVd
potato spindle tuber viroid
- RPA
ribonuclease protection assay
- sTRSV RNA
satellite RNA of tobacco ringspot virus
- TPMVd
tomato planta macho viroid
References
- 1.Diener T O, editor. The Viroids. New York: Plenum; 1987. [Google Scholar]
- 2.Semancik J S, editor. Viroids and Viroid-Like Pathogens. Boca Raton, FL: CRC; 1987. [Google Scholar]
- 3.Daros J A, Marcos J F, Hernandez C, Flores R. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branch A D, Robertson H D. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 5.Hutchins C J, Rathjen P D, Forster A C, Symons R H. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steger G, Baumstark T, Mörchen M, Tabler M, Tsagris M, Sänger HL, Riesner D. J Mol Biol. 1992;227:719–737. doi: 10.1016/0022-2836(92)90220-e. [DOI] [PubMed] [Google Scholar]
- 7.Baumstark T, Riesner D. Nucleic Acids Res. 1995;23:4246–4254. doi: 10.1093/nar/23.21.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumstark T, Schröder ARW, Riesner D. EMBO J. 1997;16:599–610. doi: 10.1093/emboj/16.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindler IM, Mühlbach HP. Plant Sci. 1992;8:221–229. [Google Scholar]
- 10.Diener TO. Proc Natl Acad Sci USA. 1989;86:9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branch AD, Benenfeld BJ, Robertson HD. Proc Natl Acad Sci USA. 1988;85:9128–9132. doi: 10.1073/pnas.85.23.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabler M, Sänger HL. EMBO J. 1985;4:2191–2199. doi: 10.1002/j.1460-2075.1985.tb03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldstein PA, Bruening G. Nucleic Acids Res. 1993;21:1991–1998. doi: 10.1093/nar/21.8.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An G, Ebert PR, Nitra A, Ha SB. In: Plant Molecular Biology Manual. Gelvin SB, Schilperoort RA, editors. Dordrecht, Netherlands: Kluwer; 1988. pp. A3/1–13. [Google Scholar]
- 15.Gallie DR, Zaitlin D, Perry KL, Kado C. J Bacteriol. 1984;157:739–745. doi: 10.1128/jb.157.3.739-745.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond J, Kamo K. Mol Plant–Microbe Interact. 1995;8:674–682. doi: 10.1094/mpmi-8-0674. [DOI] [PubMed] [Google Scholar]
- 17.McGarvey P, Kaper JM. BioTechniques. 1991;11:428–432. [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Podleckis EV, Hammond RW, Hurtt SS, Hadidi A. J Virol Methods. 1993;43:147–158. doi: 10.1016/0166-0934(93)90072-y. [DOI] [PubMed] [Google Scholar]
- 20.Owens RA, Chen W, Hu Y, Hsu Y-H. Virology. 1995;208:554–564. doi: 10.1006/viro.1995.1186. [DOI] [PubMed] [Google Scholar]
- 21.Owens R A, Hammond R W, Gardner R C, Kiefer MC, Thompson S M, Cress DE. Plant Mol Biol. 1986;6:179–192. doi: 10.1007/BF00021487. [DOI] [PubMed] [Google Scholar]
- 22.Bruening G. Semin Virol. 1990;1:127–134. [Google Scholar]
- 23.Keese P, Symons RH. Proc Natl Acad Sci USA. 1985;82:4582–4586. doi: 10.1073/pnas.82.14.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecker R, Wang Z, Steger G, Riesner D. Gene. 1988;72:59–74. doi: 10.1016/0378-1119(88)90128-x. [DOI] [PubMed] [Google Scholar]
- 25.Konarska M, Filipowicz W, Domdey H, Gross HJ. Nature (London) 1981;293:112–116. doi: 10.1038/293112a0. [DOI] [PubMed] [Google Scholar]
- 26.Steger G, Tabler M, Brüggemann W, Colpan M, Klotz G, Sänger HL, Riesner D. Nucleic Acids Res. 1986;14:9613–9630. doi: 10.1093/nar/14.24.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewelt A, Prinsen E, Schell J, Van Onckelen H, Schmulling T. Plant J. 1994;6:879–891. doi: 10.1046/j.1365-313x.1994.6060879.x. [DOI] [PubMed] [Google Scholar]
- 29.Koncz C, Schell J. Mol Gen Genet. 1989;204:383–396. doi: 10.1007/BF00261151. [DOI] [PubMed] [Google Scholar]
- 30.Gardner RC, Chonoles KR, Owens RA. Plant Mol Biol. 1986;6:221–228. doi: 10.1007/BF00015228. [DOI] [PubMed] [Google Scholar]
- 31.Pogue, G.P, Huntley, C.C. & Hall, T.C. (1994) Arch. Virol.9, Suppl., 181–194. [DOI] [PubMed]
- 32.Ball LA. Proc Natl Acad Sci USA. 1994;91:12443–12447. doi: 10.1073/pnas.91.26.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai M. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 34.Flores R, Di Serio F, Hernandez C. Semin Virol. 1997;8:65–73. [Google Scholar]
- 35.Harders J, Lukács N, Robert-Nicoud M, Jovin TM, Riesner D. EMBO J. 1989;8:3941–3949. doi: 10.1002/j.1460-2075.1989.tb08577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maroon CJM. Ph.D. dissertation. College Park: University of Maryland; 1997. [Google Scholar]
- 37.Glenn JS, Taylor JM, White JM. J Virol. 1990;64:3104–3107. doi: 10.1128/jvi.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang SB, Jeng KS, Lai MMC. In: The Unique Hepatitis Delta Virus. Dinter-Gottlieb G, editor. Austin, Texas: R.G. Landes; 1995. pp. 95–108. [Google Scholar]
- 39.Sano T, Candresse T, Hammond RW, Diener TO, Owens RA. Proc Natl Acad Sci USA. 1992;89:10104–10108. doi: 10.1073/pnas.89.21.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owens RA, Steger G, Hu Y, Fels A, Hammond RW, Riesner D. Virology. 1996;222:144–158. doi: 10.1006/viro.1996.0405. [DOI] [PubMed] [Google Scholar]
- 41.Riesner D. Semin Virol. 1990;1:83–99. [Google Scholar]
- 42.Loss P, Schmitz M, Steger G, Riesner D. EMBO J. 1991;10:719–727. doi: 10.1002/j.1460-2075.1991.tb08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu F, Heinrich C, Loss P, Steger G, Tien P, Riesner D. EMBO J. 1993;12:2129–2139. doi: 10.1002/j.1460-2075.1993.tb05861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]