Abstract
After undergoing several rounds of divisions normal human fibroblasts enter a terminally non-dividing state referred to as cellular or replicative senescence. We cloned MORF4 (mortality factor on human chromosome 4), as a cellular senescence inducing gene that caused immortal cells assigned to complementation group B for indefinite division to stop dividing. To facilitate analyses of this gene, which is toxic to cells at low levels, we obtained stable clones of HeLa cells expressing a tetracycline-induced MORF4 construct that could be induced by doxycycline in a dose-dependent manner. MORF4 induction resulted in reduced colony formation after 14 days of culture, as previously observed. We determined that MORF4 protein was unstable and that addition of the proteasome inhibitor MG132 resulted in accumulation of the protein. Following removal of MG132 the protein was rapidly degraded. Subcellular fractionation following MG132 treatment demonstrated that the protein accumulates primarily in the cytoplasm with some amounts present in the nucleus. It is therefore possible that MORF4 protein, which escapes degradation in the cytoplasm, is transported to the nucleus where it is functional. The results suggest that levels of MORF4 in cells must be tightly controlled and one mechanism involves stability of the protein.
Keywords: cellular senescence, protein stability, proteasome, ubiquitin, chromatin
INTRODUCTION
Normal human somatic cells do not divide indefinitely, cease proliferating after a certain number of divisions and enter a terminal loss of division termed replicative senescence [1, 2]. Although senescent cells do not divide, they remain metabolically active and exhibit specific characteristics, such as enlarged cell size, expression of senescent associated β-galactosidase (SA β-gal) activity, and changes in gene expression patterns [3–7]. Cell fusion studies between normal and tumor human cells showed that the senescent phenotype is dominant over immortalization and that cellular senescence is a potential tumor suppression mechanism [8]. Furthermore, 4 complementation groups for indefinite division were identified from cell fusion studies of over 40 human tumor cell lines [9]. Introduction of a normal human chromosome 4 induced senescence in tumor cell lines that assigned to complementation group B [10]. We cloned mortality factor on chromosome 4 (MORF4) as a senescence inducing factor specific to complementation group B cell lines [11]. The MORF4 protein has defined motifs such as a nuclear localization signal (NLS), helix-loop-helix (HLH) and leucine zipper (LZ) regions, which have been found in many transcriptional regulators. Our hypothesis is that MORF4 regulates transcription and modulates the expression of key genes to induce the senescent phenotype.
MORF4 is a member of a new gene family and two other genes, MORF4 related gene on chromosome 15 and X (MRG15 and MRGX), are ubiquitously expressed [11–13]. Interestingly, MRG15 and MRGX do not have the ability to induce senescence in tumor cells. MORF4 is a truncated version of MRG15 and MRGX, and has over 90% identity to these proteins at the amino acid level [14, 15]. It has become evident these family members share many common interacting protein partners and are present in many nuclear protein complexes. It is not known whether the complexes that contain the different protein family members have the same function or not.
We and others have shown that MRG15 and MRGX associate in complexes with histone acetyltransferase (HAT) Tip60 and histone deacetylases (HDAC) 1 and 2 and thereby regulate transcription [16–25]. MORF4 may also associate with similar complexes and since it is a truncated protein it has the potential to disrupt MRG15/MRGX interactions and induce the senescent phenotype through a key change(s) in gene expression. However, RNA expression levels of MORF4 are very low in all cell types analyzed and despite the production of a good polyclonal antibody to the protein we have had difficulty in detection by Western blot. To address these issues and enable a more detailed analysis of MORF4 we isolated a stable clone of HeLa cells expressing tetracycline (tet)-inducible MORF4 construct.
The results presented here demonstrate that MORF4 is a highly unstable protein and is rapidly degraded by the ubiquitin-proteasome pathway in cancer cells using this inducible cell culture system. Inhibition of the proteasome by a chemical inhibitor resulted in a rapid accumulation of MORF4 in the cells. The results provide a mechanistic basis for the role of MORF4 in cell growth control and an explanation of the need to maintain MORF4 protein at very low levels in cells.
MATERIALS AND METHODS
Plasmids
The fragment for a C-terminal FLAG-tagged MORF4 was amplified by PCR with Pfu Ultra DNA polymerase using genomic clone as a template. The amplified fragment was cloned into the BamHI and XbaI sites of pcDNA3.1(+) (Invitrogen) and pTRE2 (Clontech) vectors. C-terminal HA-tagged MORF4 vector was generated in a similar manner except the BamHI and XhoI sites for cloning into pcDNA3.1(+) vector were used. MORF4Flag ΔNLS was also constructed by PCR using pcDNA3.1(+)MORF4Flag as a template. The inserted fragments of all vectors were verified by DNA sequencing. The HA-tagged ubiquitin expression vector (MT123) was kindly provided by Dr. Dirk Bohmann (University of Rochester) [26].
Cell lines, culture conditions and reagents
HeLa (human cervical carcinoma cells) and the PT67 retrovirus packaging cell lines were maintained in Earle’s minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum and 0.1 mM non-essential amino acids at 5% CO2 and 37°C in saturated humidity. Puromycin, polybrene, doxycycline (dox), DAPI, phenylmethylsulfonyl fluoride (PMSF) and mouse anti-Flag (M2) monoclonal and rabbit anti-Flag antibodies were purchased from Sigma. Hygromycin B was purchased from Invitrogen. MG132 was purchased from Calbiochem, dissolved in DMSO at 10 mM, and kept at −20°C. Epoxomicin and mouse anti-mono-and polyubiquitinylated conjugate antibody (FK2) were purchased from Enzo Life Sciences. Ubiquitin E1 inhibitor UBEI-41 was purchased from Biogenova Corporation. Cycloheximide (CHX) was purchased from Calbiochem. Rabbit anti-HA (Y-11), rabbit anti-Sp1 (PEP2), and mouse anti-α-tubulin (B-7) antibodies were purchased from Santa Cruz Biotechnology. Rabbit antiserum against a MORF4 peptide (MRWAAPGKK, corresponding to 1–9 aa of MORF4) was produced for us by Cocalico Biologicals Inc. and the antibody was affinity-purified from antiserum using antigen column.
Production of retrovirus and retroviral infection
PT67 packaging cells (3 ×105 cells) were seeded into 35-mm tissue culture dishes and transfected with 1 µg of pLIB-rtTA-M2-TRSID-puro retroviral vector using Lipofectoamine PLUS reagent (Invitrogen) [27]. The next day, transfected cells were trypsinized, re-plated onto three 100-mm culture dishes and incubated for one more day. The transfected cells were selected by incubation with puromycin (1 µg/ml) containing medium for 2 weeks. The puromycin resistant colonies were trypsinized, combined and seeded into 35-mm tissue culture dishes (3 × 105 cells). After 48 h, the retrovirus-containing supernatant was collected, centrifuged at 10,000xg and kept at −80°C.
For retrovirus infection, HeLa cells (3 × 105 cells) were seeded in 35-mm tissue culture dishes and incubated overnight. The cells were incubated with the retrovirus-containing supernatant, with polybrene (final concentration 8 µg/ml), overnight. The cells were trypsinized, plated into three 100-mm culture dishes, and incubated. The next day, 1 µg/ml puromycin was added to the culture medium and puromycin-resistant clones selected after a 2 week-incubation. The puromycin-resistant clones were amplified from a single colony and maintained in complete medium containing 0.5 µg/ml of puromycin. Tet-responder clones were screened by transient transfection of the pTRE2-MORF4Flag expression vector following stimulation by dox at 1 µg/ml for 24 h. MORF4Flag expression was detected by Western blot using an anti-Flag (M2) monoclonal antibody. For isolation of tet-responder lines stably expressing MORF4Flag, the pTRE2-MORF4Flag expression vector was co-transfected with a pTK-Hyg vector, which encodes the hygromycin-resistant gene, at a 20:1 ratio in 60-mm culture dishes. The next day, the cells were trypsinized, plated into three 100-mm culture dishes, and incubated overnight. Hygromycin B (150 µg/ml) and puromycin (0.5 µg/ml) was added to the culture medium and the double-resistant clones selected after a 2 week-incubation. The double-resistant clones were amplified from single colony and maintained in complete media containing 100 µg/ml of hygromycin B and 0.5 µg/ml of puromycin. The clones were incubated with 1 µg/ml of dox for 24 h and total cell lysates prepared. MORF4Flag expression was detected by Western blot using rabbit anti-MORF4 or mouse anti-FLAG (M2) antibodies.
Analysis of growth properties
For the colony formation assay [28], 100 cells were seeded into 60-mm tissue culture dishes in medium EMEM+10% FBS supplemented with or without 1 µg/ml dox. Culture media were replaced ever 3 to 4 days because dox may be degraded during culture. The cells were incubated for 14 days, fixed with 1% glutaraldehyde, stained with 0.1% crystal violet and the number of cells per clone determined microscopically.
Transfection
For transient transfection experiments, we plated 6 × 105 cells in 60-mm tissue culture dishes and transfected them the next day using Lipofectamine Plus (Invitrogen). For protein stability experiments, the transfected cells were trypsinized 3 h after transfection and re-plated equally into 2 wells in 6 well plates to normalize for transfection efficiency. After 22 h incubation, MG132 or DMSO (vehicle control, less than 0.1%) was added to the culture medium and the cells incubated for 2 h. The cells were washed with PBS, lysed with lysis buffer (20 mM Tris-HCl [pH 7.5], 1% NP-40, 150 mM NaCl, 10% glycerol, and protease inhibitor cocktail set I [Calbiochem]), and total cell lysates prepared.
Western blot analysis and immunoprecipitation
Cells were lysed with 0.5 ml per 60-mm culture dish of lysis buffer, the lysates kept on ice for 30 min and centrifuged at 20,000xg for 15 min. For preparation of cytoplasmic and nuclear fractions, we used NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo) according to manufacturer’s instruction. Protein concentration of the supernatants was determined by the Bradford method (Bio-Rad). Total proteins (50 µg) were separated on 10% SDS-PAGE and transferred to nitrocellulose membrane. MRG15 was detected using a rabbit anti-MRG15 antibody raised by our laboratory. Mouse anti-β-actin antibody (abcam, AC-15) was used as a loading control. For nuclear and cytoplasmic fractionation, rabbit anti-Sp1 as nuclear marker and mouse anti-α-tubulin as cytoplasmic marker antibodies were used as a loading control.
For immunoprecipitation, the lysates were pre-cleared for 1 h by addition of ImmunoPure Immobilized Protein G or A Plus beads (30 µl, Thermo). The mouse anti-Flag (M2) or rabbit anti-Flag antibody (2 µg) was added to the pre-cleared lysates and kept at 4°C for 3 h. Protein G or A beads (30 µl) were added to each tube and kept at 4°C for another 1 h. Beads were washed four times with 0.5 ml of lysis buffer, and 1x loading buffer (25 mM Tris-HCl [pH 6.5], 5% glycerol, 1% SDS, 1% 2-mercaptoethanol and 0.05% BPB) was then added. Samples were boiled for 3 min and run on SDS-PAGE gels. The separated proteins were blotted on nitrocellulose membranes (Bio-Rad), which were probed with indicated antibodies and subsequently with horseradish peroxidase-conjugated second antibody. To visualize the proteins, SuperSignal West Pico Chemiluminescent Substrate was used (Thermo).
Immunofluorescence
HeLa cells were cultured onto coverslips and were transfected with 1 µg of the indicated plasmids. After 22 h incubation, cells were treated with 10 µM MG132 for 2 h, fixed with 4% paraformaldehyde in PBS for 20 min, and permeabilized with 0.2% Triton X-100 in PBS for 10 min. Monoclonal mouse anti-Flag (M2) antibody (10 µg/ml in 2% goat serum in PBS) was incubated for 1 h at room temperature. After washing, BODIPY FL goat anti-mouse IgG (1:1000 dilution, Molecular Probes) was incubated for 45 min at room temperature in the dark. Nuclei were counterstained with 1 µg/ml DAPI. Coverslips were washed with PBS and mounted on glass slides using GEL/MOUNT (biomeda). Cells were analyzed by Zeiss Axiovert 200M fluorescnce microscope using 63x objective lens.
RESULTS
Isolation of tet-inducible MORF4Flag clones
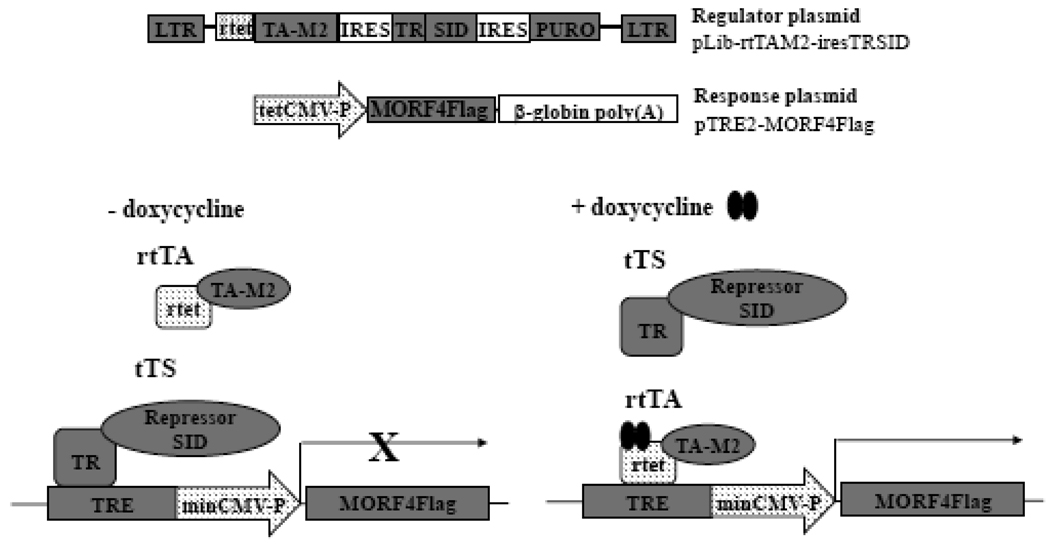
Transfection and overexpression of MORF4 in all cell types was extremely toxic. Therefore, to understand the mechanism of MORF4 function, we isolated tet-inducible MORF4Flag clones from HeLa a human carcinoma cell line, which assigns to complementation group B for indefinite division. We used the pLIB-rtTA-M2-TRSID-puro retroviral system for this purpose (Figure 1) [27]. This vector encodes a transcriptional repressor (TRSID) and tet-responsible activator (rtetTA-M2), which recognizes a tet-responsive element (TRE). A transcriptional repressor always binds to the TRE and inhibits transcription of the target gene in the absence of dox. When dox is added to culture media, the transcriptional activator replaces the repressor, and activates expression of the target gene. This system eliminates leakiness of expression of the target gene.
Figure 1. Schematic presentation of the inducible system for expression of MORF4Flag in cultured cells.
MORF4 expression will be completely shut down without dox because the TRSID repressor complex (tTS) will recruit a mSin3/HDAC complex to this promoter. When Dox is added to culture medium, tTS will be replaced by tetracycline-controlled transactivator (rtTA) and rtTA will activate the promoter. LTR, long terminal repeat; SID, mSin3 interaction domain; PURO, puromycin resistant gene; CMV-P, cytomegalovirus promoter; HH4-P, histone H4 promoter; NEO, neomycin resistant gene; TRE, tetracyclin responsive element.
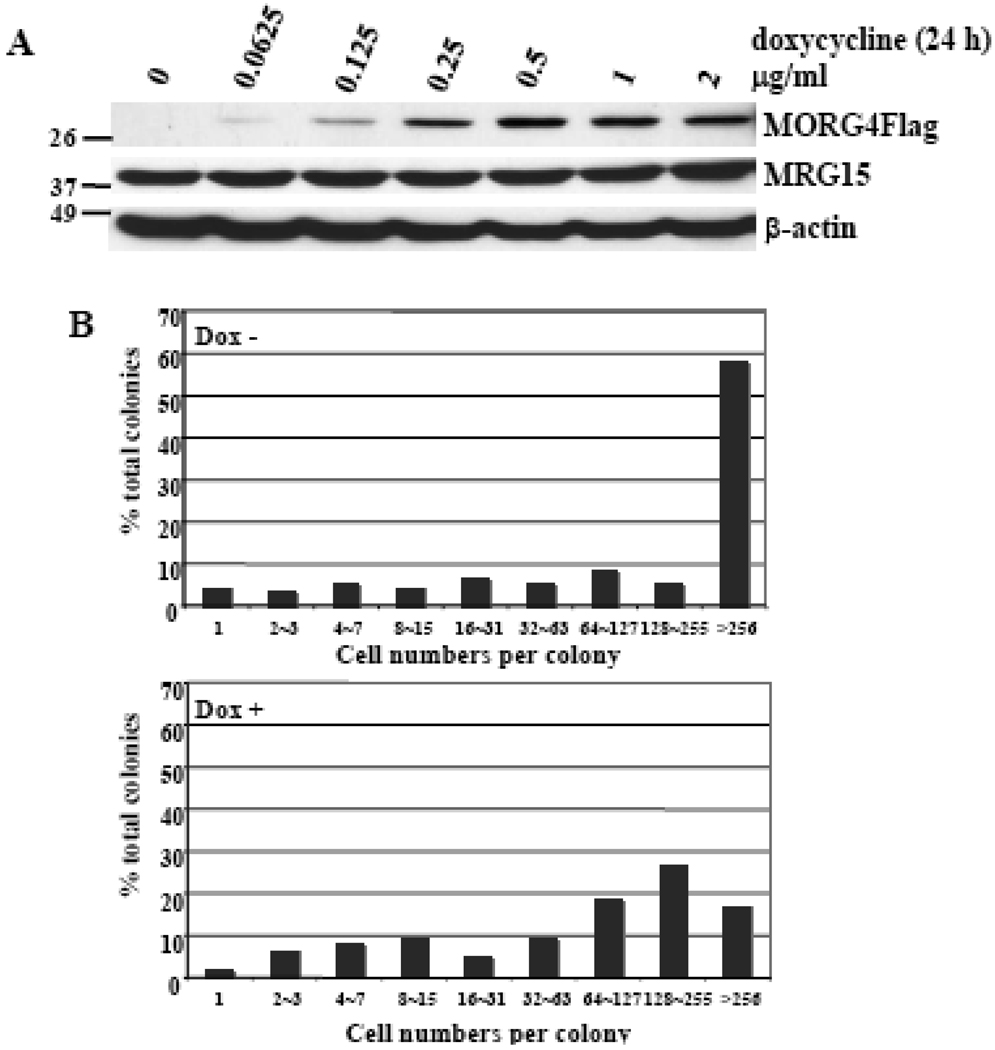
We isolated two independent clones in which MORF4Flag was induced by dox. Both clones gave the same results in initial dox dose dependent expression of MORF4 experiments. We therefore continued our studies with one clone designated HT-1. MORF4 expression (Figure 2A) was maximum between 0.5 and 1 µg/ml dox and 1 µg/ml dox was used in all subsequent experiments. We used a rabbit anti-MORF4 peptide antibody that we had generated for Western analyses and confirmed the results with a mouse anti-Flag (M2) antibody.
Figure 2. Studies of a tet-inducible MORF4Flag clone of HeLa cells (HT-1).
(A) Dox dose dependent induction of MORF4Flag protein in HT-1 cells. The location of molecular markers is indicated (kDa). (B) Colony size distribution (CSD) assay for HT-1 cells. HT-1 cells (100 cells) were seeded onto 60-mm tissue culture dishes and incubated with or without 1 µg/ml dox for 14 days. The cells were fixed and stained and cell numbers per colony scored.
MORF4 induction affects HeLa cell growth
MORF4 was originally isolated as a cellular senescence inducing gene in tumor cells assigned to complementation group B for indefinite division. To investigate the effect of MORF4Flag induction on the growth of a group B cell line, HeLa, we used the colony size distribution (CSD) assay [28] and plated 100 cells in 60-mm tissue culture dishes. Following a 2-week incubation the clones were fixed and stained and the number of cells per colony determined. The ratio of colonies with large cell number was high in untreated control cells, whereas the number of non-dividing colonies was much higher in dox-treated cells (Figure 2B). Although the dox treated group had large and actively dividing colonies, the ratio of such colonies to small colonies was reduced compared with the untreated cells. It is possible that these colonies may have lost dox responsiveness and MORF4Flag induction or that these clones could undergo additional cell divisions prior to becoming senescent.
MORF4 is an unstable protein
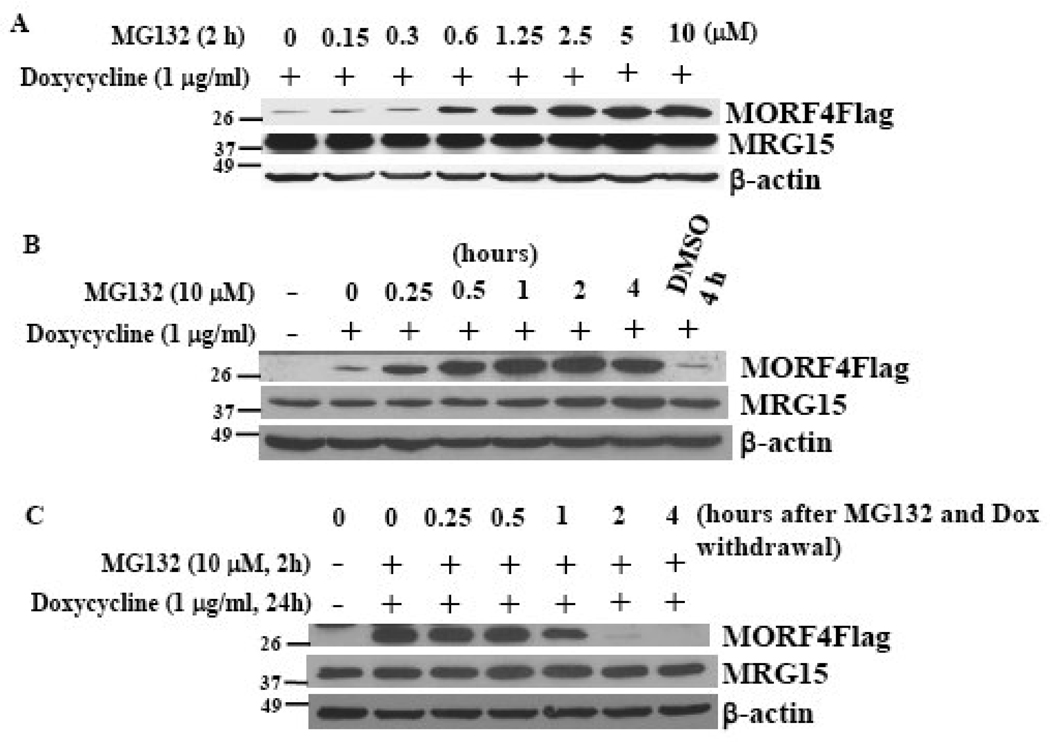
When compared with endogenous protein levels of MRG15 and β-actin in HeLa cells, MORF4Flag protein was expressed at low amounts even in the stable clone. Since levels of many proteins are often controlled by proteasomal degradation, we treated HT-1 cells with the proteasome inhibitor, MG132. MORF4Flag protein was stabilized and accumulated in the cells and the protein was stabilized at 15 min post treatment and reached a maximum at 2 h (Figure 3A). DMSO, which was used as the vehicle, had no effect on MORF4Flag protein accumulation. Interestingly, endogenous MRG15 protein levels were not affected by MG132 in this cell line (Figure 3A). Since the major difference in the two proteins is the additional N-terminal sequence in MRG15, this region may protect it from degradation. MORF4Flag protein accumulation was dependent on the concentration of MG132 and the effect was maximal at 2.5 µM for 2 h (Figure 3B). To measure how rapidly MORF4Flag is degraded in the cells, we determined protein amount after removal of MG132 and dox. The amount of MORF4Flag protein decreased after 1 h and was completely degraded at 2 h post removal of these reagents (Figure 3C). This may be an underestimation of the degradation of the protein because transcription of MORF4Flag may not cease immediately after removal of dox and translation may also continue. The results indicate that MORF4Flag is an unstable protein that is degraded by the proteasome.
Figure 3. Proteasomal degradation of MORF4 protein.
(A) Dose dependent MORF4Flag accumulation following treatment with the proteasome inhibitor, MG132. (B) Time course of MORF4Flag accumulation by 10 µM MG132 treatment. (C) Rapid disappearance of MORF4Flag after withdrawal of dox and MG132. The location of molecular markers is indicated (kDa).
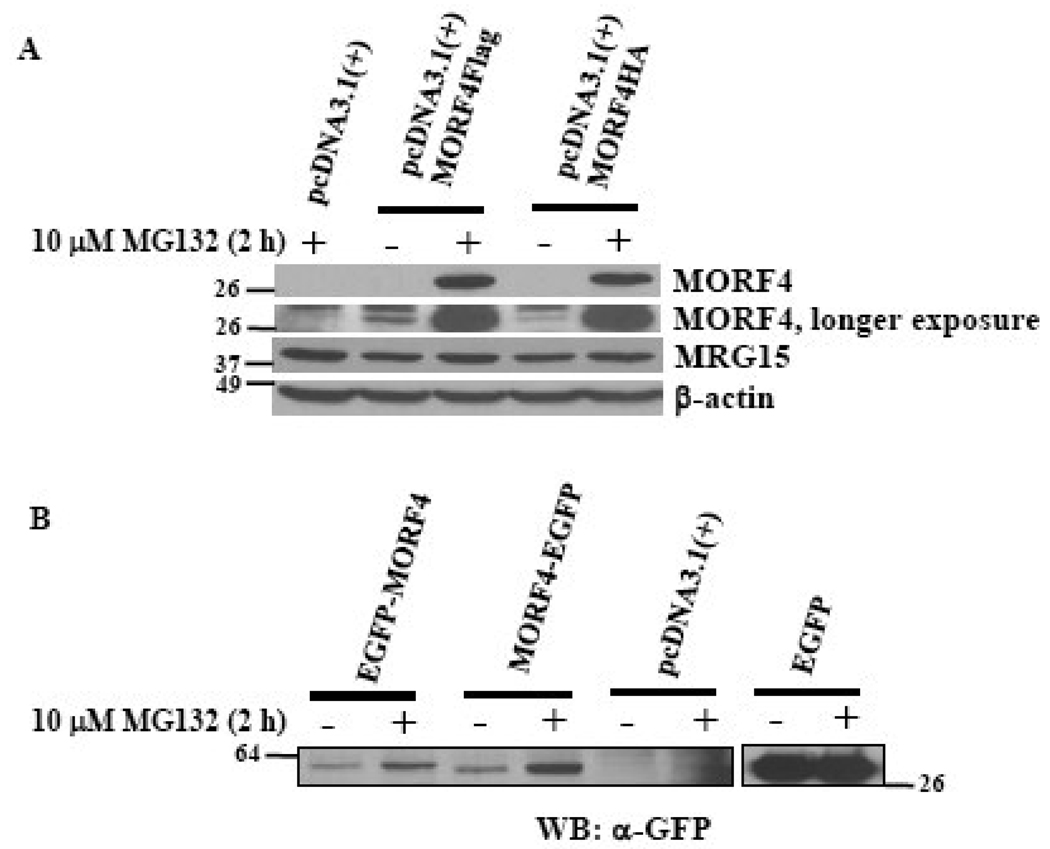
To eliminate the possibility that the tag and its location might affect MORF4 protein stability, MORF4Flag, MORF4HA, EGFP-MORF4 (N-terminal tag), and MORF4-EGFP (C-terminal tag) expression vectors were transiently transfected into HeLa cells. The transfected cells were equally divided into two plates to eliminate any differences in transfection efficiency versus the effect of MG132 treatment. Degradation of MORF4 protein expressed from all the constructs was protected by MG132 treatment, although it was slightly less effective for EGFP-tagged MORF4 (Figure 4A and 4B). Non-tagged MORF4, when transiently transfected into HeLa cells, was also protected from degradation by treatment with the proteasome inhibitor (data not shown).
Figure 4. Transient expression of various MORF4 constructs in HeLa cells.
(A) C-terminal Flag or HA tagged MORF4 constructs were transiently transfected into HeLa cells and the cells incubated overnight. Two hours after MG132 treatment, total cell lysates were prepared and MORF4 protein levels detected by Western blot. (B) N-terminal or C-terminal EGFP tagged MORF4 constructs were transiently transfected into HeLa cells and the cells incubated overnight. Two hours after MG132 treatment, total cell lysates were prepared and MORF4 protein levels detected by Western blot using a mouse anti-GFP antibody. The location of molecular markers is indicated (kDa).
MORF4 is degraded by ubiquitin/proteasome pathway
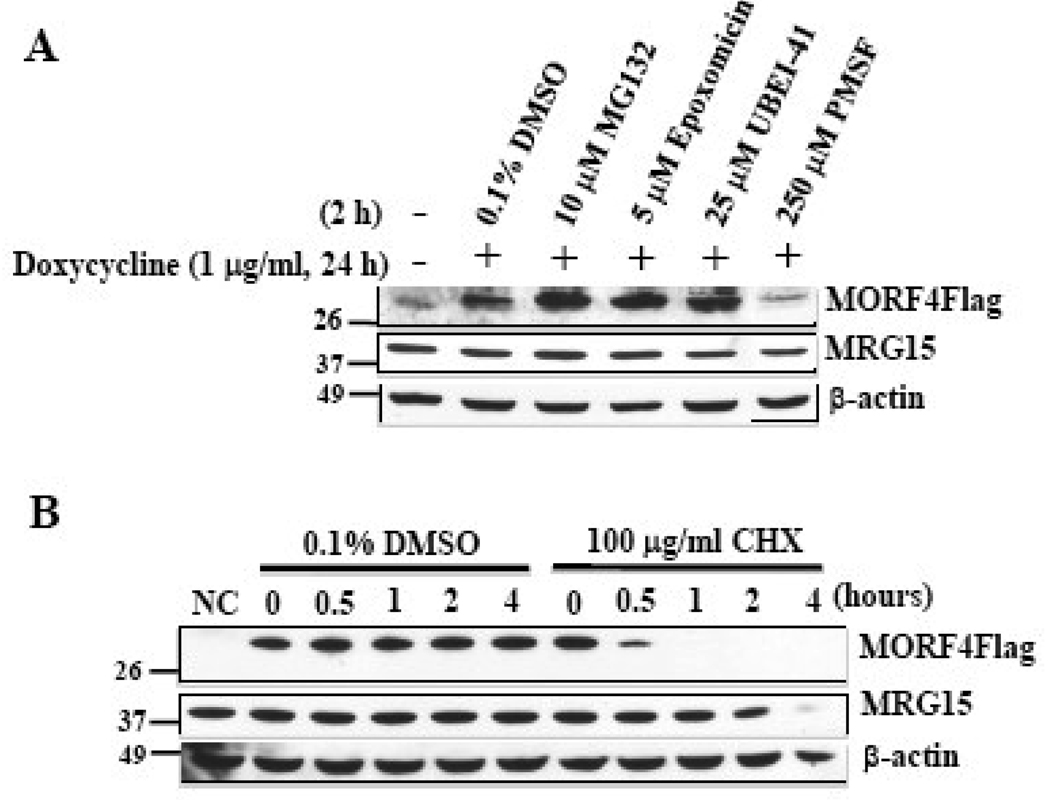
We determined whether MORF4 is degraded by the ubiquitin/proteasome pathway by treating HT-1 cells with various inhibitors (Figure 5A). Treatment with the proteasome inhibitors MG132 or Epoxomicin led to the stabilization of MORF4Flag protein. UBEI-41, also known as PYR-41, is a specific inhibitor of the E1 ubiquitin-activating enzyme, which acts at the first step of ubiquitination [29]. Treatment of UBEI-41 at 25 µM, at which 85% of E1 activity is inhibited [29, 30], also effectively stabilized MORF4Flag protein. However, the serine protease inhibitor PMSF had no effect on MORF4 protein stability at the effective concentration [31, 32]. All the inhibitors tested had no effect on MRG15 and β-actin protein levels. These results strongly suggest that MORF4 is degraded by ubiquitin/proteasome pathway.
Figure 5. MORF4 protein stability.
(A) Effect of various inhibitors on MORF4 stabilization. HT-1 cells were treated with or without Dox to induce MORF4 and then treated with various inhibitors at the indicated concentration for 2 h. Total cell lysates were prepared and MORF4, MRG15 and β-actin proteins detected by Western blot. (B) HeLa cells were transiently transfected with MORF4Flag expression or empty (NC) vector. After 3 h incubation, transfected cells were divided equally into 12 well plates to normalize for transfection efficiency. The cells were treated with CHX (100 µg/ml) or DMSO (0.1%, vehicle control) for indicated times. Total cell lysates were prepared and MORF4, MRG15 and β-actin proteins detected by Western blot. The location of molecular markers is indicated (kDa).
We next, examined MORF4 stability in HeLa cells following the addition of CHX, an inhibitor of translation. A half hour treatment with CHX resulted in a significant reduction of MORF4 protein levels and no protein was detected following 1 h of treatment (Figure 5B). MRG15 protein levels also significantly decreased 4 h after CHX treatment but β-actin protein levels did not change at this time. Vehicle control (0.1% DMSO) did not affect levels of any of the proteins.
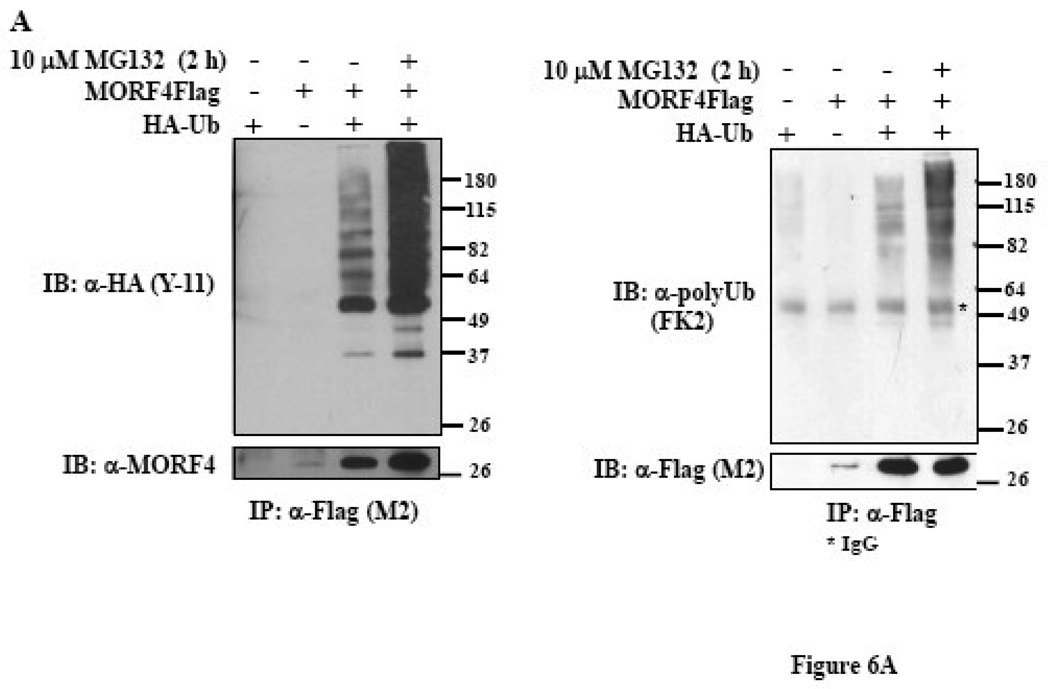
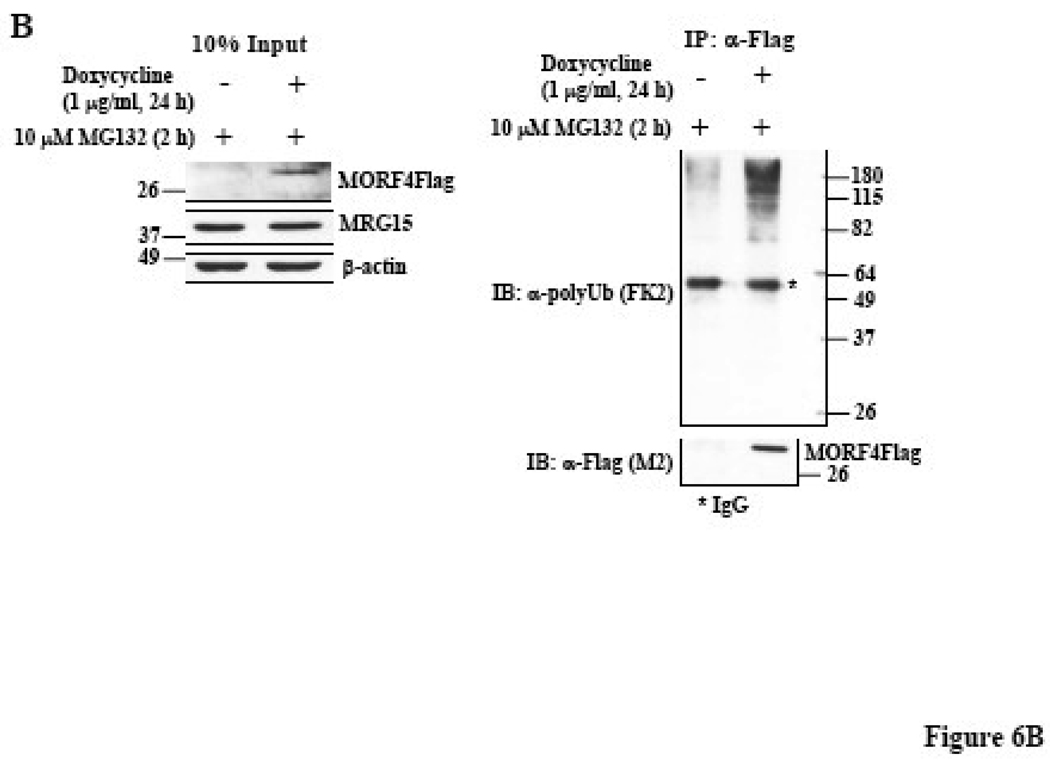
To further determine if MORF4 degradation by the proteasome is dependent on ubiquitination, the MORF4Flag construct was transiently co-transfected with or without a HA-tagged ubiquitin (HA-Ub) construct, into HeLa cells. MORF4Flag was immunoprecipitated from total cell lysates using mouse anti-Flag (M2) antibody (Figure 6A, left panel) or rabbit anti-Flag antibody (Figure 6A, right panel) and ubiquitinated proteins detected by Western blot using an anti-HA antibody or an anti-polyUb antibody. MORF4Flag was indeed polyubiquitinated and ubiquitinated MORF4Flag protein accumulated in the cells following MG132 treatment. We also confirmed polyubiquitination of MORF4 in HT-1 cells. MORF4Flag was immunoprecipitated from total cell lysates using rabbit anti-Flag antibody and ubiquitinated proteins detected by Western blot using an anti-polyUb antibody (Figure 6B). Ubiquitinated MORF4Flag was specifically detected in dox-induced lysate. This indicates that MORF4 protein is degraded in the cells by the ubiquitin-proteasome pathway.
Figure 6. Polyubiquitination of MORF4 protein in HeLa cells.
(A) MORF4Flag construct was transiently transfected into HeLa cells, with or without HA-tagged ubiquitin (HA-Ub). Twenty-four hours after transfection, total cell lysates were prepared and MORF4Flag protein was immunoprecipitated with a mouse anti-Flag (M2) (left panel) or a rabbit anti-Flag (right panel) antibody. Ubiquitinated MORF4 proteins were detected by Western blot with a rabbit anti-HA or a mouse anti-polyUb antibody and MORF4 protein was detected with a rabbit anti-MORF4 or a mouse anti-Flag (M2) antibody. The asterisk indicates IgG. (B) HT-1 cells were treated with or without Dox to induce MORF4 and then treated with MG132 at 10 µM for 2 h to allow accumulation of MORF4 protein in the cells. Total cell lysates were prepared, 10% of lysates was applied to SDS-PAGE, and MORF4, MRG15 and β-actin proteins detected by Western blot (left panel). The remaining lysates were used for immunoprecipitation/Western blot analyses. MORF4Flag protein was immunoprecipitated with a rabbit anti-Flag antibody. Ubiquitinated MORF4 proteins were detected by Western blot with a mouse anti-polyUb antibody and MORF4 protein was detected with a mouse anti-Flag (M2) antibody. The asterisk indicates IgG. The location of molecular markers is indicated (kDa).
MORF4 predominantly localizes to the cytoplasm
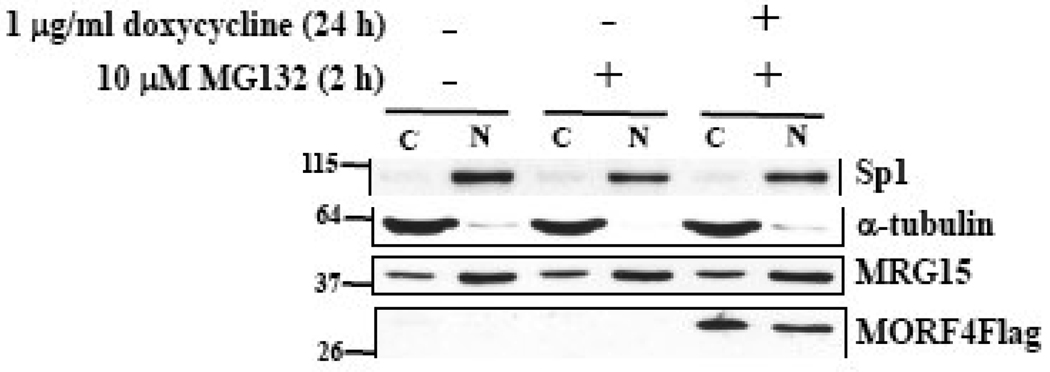
To examine the localization of MORF4Flag protein in cells, we separated cytoplasmic and nuclear proteins and determined that it was localized to both the nucleus and cytoplasm in HT-1 cells (Figure 7). In contrast, endogenous MRG15 predominantly localized to the nucleus in this cell line and induction of MORF4Flag did not affect MRG15 localization. Efficient separation of nuclear and cytoplasmic fractions was confirmed by Western blot for Sp1 and α-tubulin, respectively.
Figure 7. Cellular localization of MORF4 protein in HeLa cells.
HT-1 cells were treated with or without 1 µg/ml dox for 22 h and then treated with or without 10 µM MG132 for 2 h. Cytoplasmic and nuclear lysates were prepared and MORF4 and MRG15 proteins in the fraction detected by Western blot. Sp1 and α-tubulin were used as a loading control for nuclear and cytoplasmic fractions, respectively. The protein amount for loading was adjusted by cell number. The location of molecular markers is indicated (kDa). C; cyctoplasmic fraction and N; nuclear fraction.
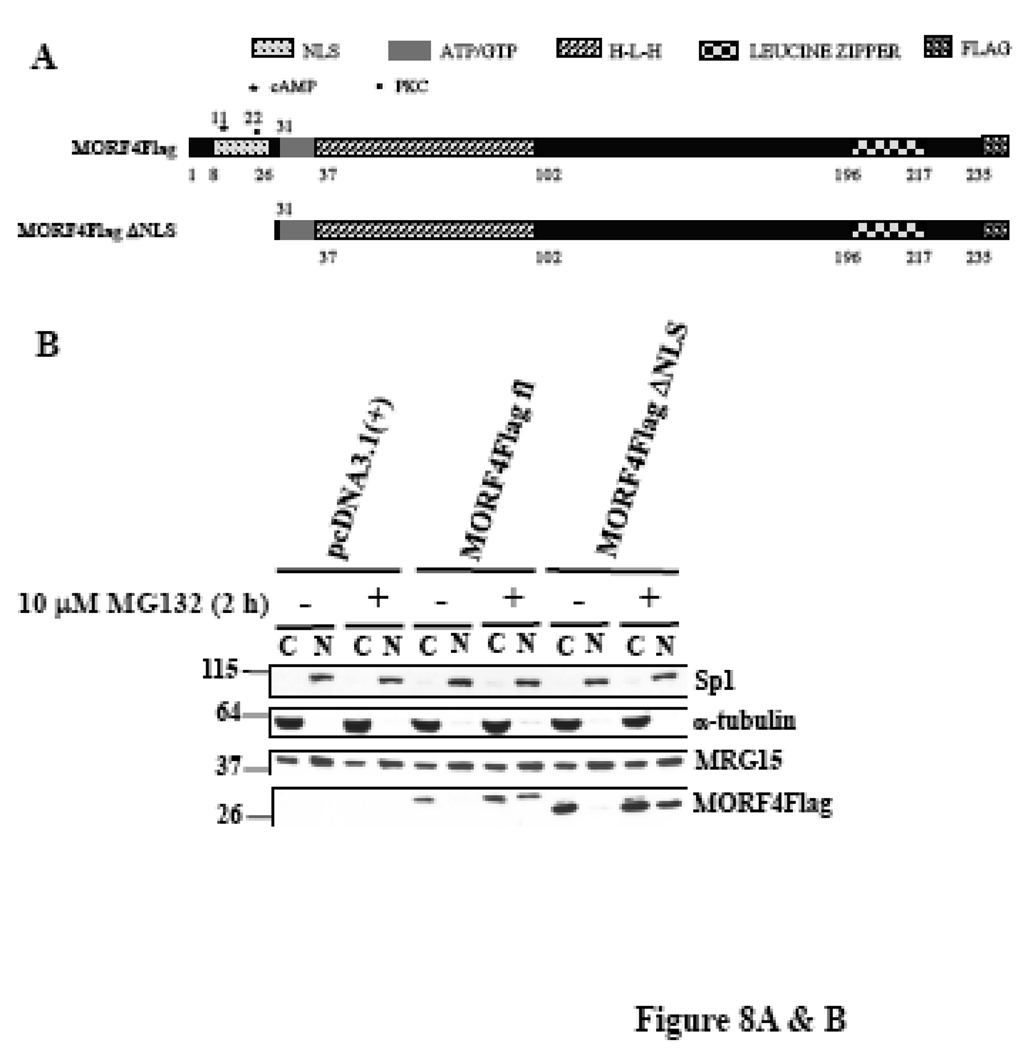
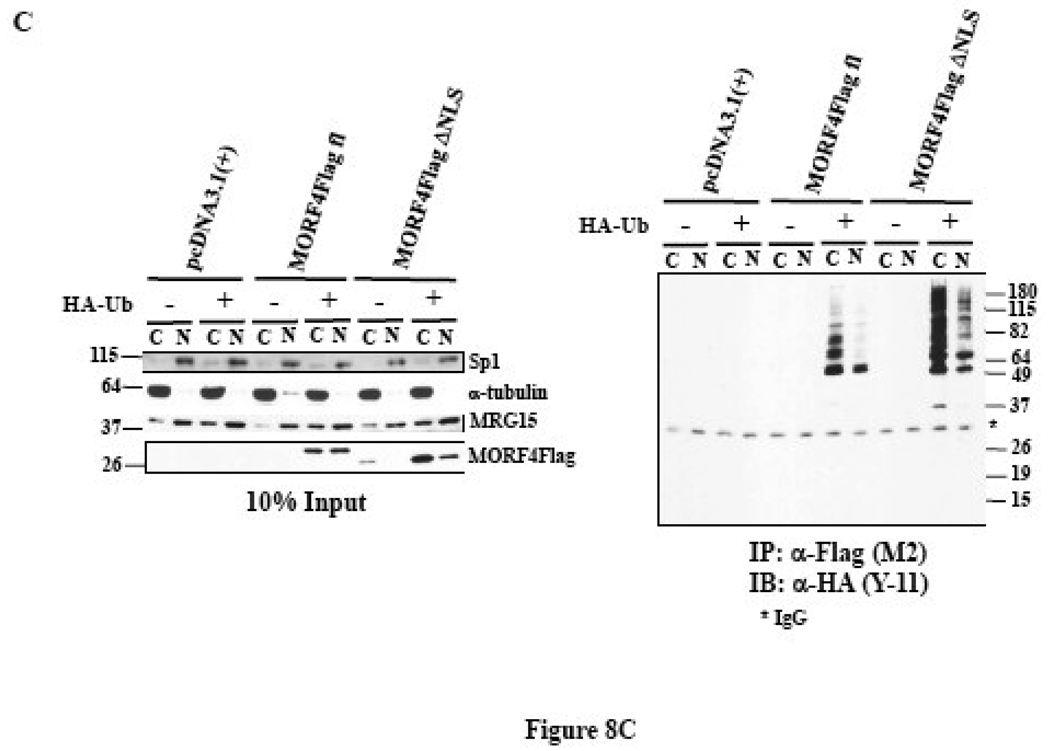
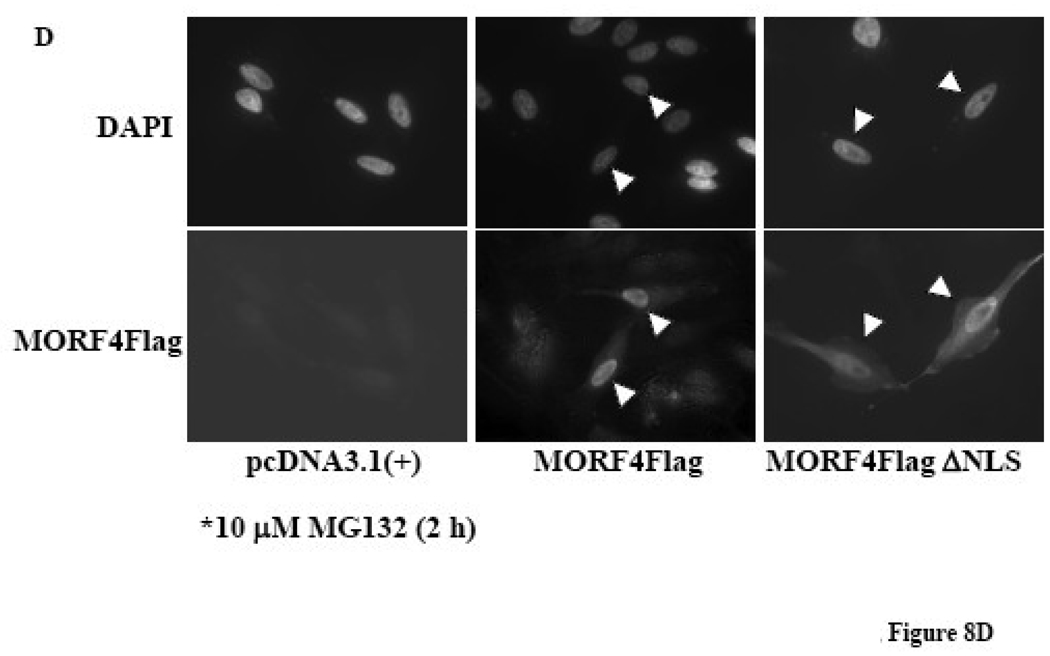
MORF4 has a putative bipartite NLS in the N-terminus (Figure 8A). We made a deleted NLS MORF4Flag construct (Δ2–26 amino acids in MORF4) in an expression vector, transfected it into HeLa cells, and 24 h later treated the cells with or without 10 µM MG132 for 2 h to allow for accumulation of protein in the cells. To examine the localization of MORF4Flag ΔNLS protein in cells, we separated cytoplasmic and nuclear proteins and determined that it was localized to both the nucleus and cytoplasm in transfected cells (Figure 8B). As expected, MORF4Flag ΔNLS tended to be more accumulated in cytoplasm compared with MORF4Flag. Next, we examined cellular localization of polyubiquitinated MORF4Flag by biochemical analysis using nuclear and cytoplasmic fractionation. Polyubiquitinated MORF4Flag was mainly present in the cytoplasmic fraction (Figure 8C). MORF4Flag ΔNLS was also polyubiquitinated in the cells and polyubiquitinated MORF4 Flag ΔNLS was more efficiently accumulated in the cytoplasm compared with wild-type MORF4Flag (Figure 8C). Immunocytochemistry using an anti-Flag (M2) antibody indicated that in the majority of the transfected cells, positive staining of full-length MORF4Flag was in the nucleus. In contrast, positive staining of MORF4Flag ΔNLS was observed to be stronger in the cytoplasmic fraction, although there was some localization to the nucleus (Figure 8D). These results suggest polyubiquitinated MORF4 is mainly degraded in the cytoplasm and non-ubiquitinated MORF4 can translocate to the nucleus and perform its functions.
Figure 8. Stability and ubiquitination of MORF4 mutant which has the NLS deletion in HeLa cells.
(A) Schematic representation of full length and NLS deleted MORF4 (MORF4Flag ΔNLS). (B) Stability of MORF4Flag ΔNLS mutant in HeLa cells. Full length MORF4Flag and MORF4Flag ΔNLS constructs were transiently transfected into HeLa cells and the cells incubated overnight. Two hours after MG132 treatment, total cell lysates were prepared and MORF4 protein levels were detected by Western blot using mouse anti-Flag (M2) antibody. The location of molecular markers is indicated (kDa). C; cyctoplasmic fraction and N; nuclear fraction. (C) Ubiquitinated MORF4 is predominantly localized in the cytoplasm in HeLa cells. MORF4Flag and MORF4Flag ΔNLS constructs were transiently transfected into HeLa cells with and without HA-Ub and the cells incubated overnight. Twenty-four hours after transfection, cytoplasmic and nuclear lysates were prepared and MORF4Flag and MORF4Flag ΔNLS protein in each fraction was immunoprecipitated with anti-Flag (M2) antibody. Ubiquitinated proteins were detected by Western blot with rabbit anti-HA antibody. Ten percent of each lysate was used as an Input to confirm fractionation. The location of molecular markers is indicated (kDa). C; cyctoplasmic fraction and N; nuclear fraction. (D) Localization of MORF4Flag ΔNLS protein in HeLa cells. Full length MORF4Flag and MORF4Flag ΔNLS constructs were transiently transfected into HeLa cells and the cells were incubated overnight. Two hours after MG132 treatment, MORF4Flag protein was detected by immunocytochemistry using anti-Flag (M2) antibody.
DISCUSSION
It has been shown that the control of protein degradation is important for the normal physiology of cells and individuals. Dysregulation of protein degradation not only affects maintenance of homeostasis of cells and individuals but also causes some diseases such as cancer and neurodegenerative disease [33]. In this study, we established tet-inducible MORF4 cell lines from HeLa cells to examine MORF4 functions in the cell and determined that MORF4 is rapidly degraded by the ubiquitin/proteasome pathway. Because MORF4 may be a general negative regulator for cell growth, it is necessary that the amount of MORF4 in cells is tightly controlled. Protein degradation is one mechanism that maintains control of MORF4 protein levels.
It is known that proteasome activity gradually decreases during aging in vivo and in vitro [34–36]. Loss of proteasome function has been reported in several aged tissues such as liver [37], heart [38], muscle [39, 40], retina [41] and spinal cord [42] from human and other mammals. Gene expression profiling by microarray analyses has shown that expression of genes involved in the ubiquitin/proteasome pathway of protein turnover decreases during aging in mouse gastrocnemius muscle [43]. Interestingly, caloric restriction, which is known as an intervention to retard aging in mammals, also retards the reduced expression of these genes. Furthermore, an age-dependent decline of proteasomal activity has been reported in human primary fibroblast culture undergoing senescence, and partial inhibition of proteasome activity in fibroblasts led to a shortened lifespan and induction of a senescent-like phenotype [44, 45]. Chondrogianni et al. has shown that senescent cells express lower levels of the catalytic subunits of the 20 S proteasome and subunits of the 19 S regulatory complex and this correlated with reduced proteasome activity and an accumulation of ubiquitinated or oxidized proteins [46].
We have previously reported that Mrg15 null embryos display growth retardation, Mrg15 null MEFs exhibit impaired proliferation in culture and the p21 protein is expressed at higher levels in early passage Mrg15 null MEFs compared to wild-type [47]. Early passage Mrg15 null MEFs also exhibit enlarged and flattened morphology when compared with wild-type cells and enter thenon-dividing state more rapidly. Other groups have also reported that the Tip60 complex, of which MRG15 is a component, is involved in regulation of expression of E2F dependent cell cycle related genes [48]. MRG15 is a positive regulator of cell growth whereas the major function of MORF4 is growth suppression. Therefore, MORF4 protein may need to be maintained at low levels by the proteasome degradation pathway. Our working hypothesis is that MORF4 protein that escapes degradation translocates to the nucleus and negatively affects MRG15 related complexes, resulting in changes in gene expression that cause entry into the non-proliferative state. We have observed disruption of the MRG15-associated factor complex (MAF2) following transfection with a chromodomain deleted MRG15 construct, which essentially mimics MORF4 [18]. However, we cannot rule out the possibility that MORF4 is either present in or interferes with the function of additional complexes in the nucleus, to induce senescence.
There are a number of ubiquitin ligases (E3) that determine the specificity and timing of ubiquitination of the target proteins. It is expected that 500 to 1000 different E3 ligases exist to maintain cell and tissue homeostasis [49]. Identification of the E3 that recognizes MORF4 is needed to elucidate the molecular mechanism of MORF4 action. Ubiquitination of p27, the cyclin-dependent kinase inhibitor (CKI), is regulated by the F-box protein, S-phase kinase-associated protein 2 (Skp2) and p27 CKI is stabilized by a decrease in Skp2 in senescent fibroblasts [50]. Another CKI, p21 which is important for replicative senescence [51, 52], can also be regulated by Skp2 dependent ubiquitination [53]. Another interesting E3 related to senescence is senescence evasion factor (SNEVPrp19/Pso4) [54]. SNEVPrp19/Pso4 expression is down-regulated in senescent endothelial cells and fibroblasts and over-expression of SNEVPrp19/Pso4 in endothelial cells increases the in vitro lifespan [55, 56]. Levels of the tumor suppressor protein p53, which plays a role in cellular senescence and apoptosis, are also regulated by the ubiquitin/proteasome pathway, and multiple E3s, such as Mdm2, MdmX, Pirh2, COP1, and ARF-BP1, can thereby modulate the various activities of p53 in cells [57]. It is possible that these E3 ligases, important for senescence, may also be involved in MORF4 ubiquitination.
Our efforts to determine the mechanism by which MORF4 specifically induces senescence in cell lines assigned to complementation group B have been hampered by a number of factors. These include low levels of expression at the RNA level (this has been independently demonstrated by another laboratory, in a study unrelated to senescence [58]), the toxicity caused by over-expression of the gene under other promoters, and the inability to generate an optimal antibody, because of the difficulty in identifying a reasonable stretch of amino acid sequence that is different from MRG15 and MRGX. We have produced an anti-peptide antibody to a small section of the protein but it is not useful for many studies as it has low affinity for MORF4. It is for these reasons that we generated the inducible cell system described here. It has defined a mechanism of regulation of MORF4 protein levels by the proteasome pathway and demonstrated how unstable the protein is in cells. It provides a tool to elucidate the precise molecular mechanism of MORF4 growth suppressive action in group B cell lines.
ACKNOWLEDGEMENTS
We thank Drs. Dirk Bohmann for providing the HA-tagged ubiquitin expression vector and Hai Rao and James R. Smith for helpful discussion. We also thank the members of Smith group at UTHSCSA for valuable comments and discussion. This work was supported by NIH grants R01AG032134 and the Ellison Medical Foundation (O.M.P.S.), and the American Federation for Aging Research and the American Heart Association grant 0765084Y (K.T.).
Abbreviations
- CHX
cycloheximide
- DMSO
dimethyl sulfoxide
- dox
doxycycline
- MORF4
mortality factor on chromosome 4
- MRG15
MORF4 related gene on chromosome 15
- MRGX
MORF4 related gene on chromosome X
- NLS
nuclear localization signal
- tet
tetracycline
- Ub
ubiquitin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 3.Dimri GP, Lee X, Basiel G, Acosta M, Scott G, Roskelley C, Medrano E, Linskens M, Rubelj I, Pereira-Smith OM, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 5.Tominaga K, Olgun A, Smith JR, Pereira-Smith OM. Genetics of cellular senescence. Mech. Ageing Dev. 2002;123:927–936. doi: 10.1016/s0047-6374(02)00030-1. [DOI] [PubMed] [Google Scholar]
- 6.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid Redox Signal. 2009;11:59–98. doi: 10.1089/ars.2008.2104. [DOI] [PubMed] [Google Scholar]
- 8.Pereira-Smith OM, Smith JR. Evidence for the recessive nature of cellular immortality. Science. 1983;221:964–966. doi: 10.1126/science.6879195. [DOI] [PubMed] [Google Scholar]
- 9.Pereira-Smith OM, Smith JR. Genetic analysis of indefinite division in human cells: identification of four complementation groups. Proc. Natl. Acad. Sci. USA. 1988;85:6042–6046. doi: 10.1073/pnas.85.16.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ning Y, Weber JL, Killary AM, Ledbetter DH, Smith JR, Pereira-Smith OM. Genetic analysis of indefinite division in human cells: evidence for a cell senescence-related gene(s) on human chromosome 4. Proc. Natl. Acad. Sci. USA. 1991;88:5635–5639. doi: 10.1073/pnas.88.13.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, Clark LJ, Parkinson EK, Barrett JC, Smith JR, Pereira-Smith OM. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tominaga K, Pereira-Smith OM. The genomic organization, promoter position and expression profile of the mouse MRG15 gene. Gene. 2002;294:215–224. doi: 10.1016/s0378-1119(02)00787-4. [DOI] [PubMed] [Google Scholar]
- 13.Tominaga K, Matzuk MM, Pereira-Smith OM. MrgX is not essential for cell growth and development in the mouse. Mol. Cell. Biol. 2005;25:4873–4880. doi: 10.1128/MCB.25.12.4873-4880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marin I, Baker BS. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 2000;17:1240–1250. doi: 10.1093/oxfordjournals.molbev.a026407. [DOI] [PubMed] [Google Scholar]
- 15.Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- 16.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 17.Leung JK, Berube NG, Venable A, Ahmed S, Timchenko N, Pereira-Smith OM. MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 2001;276:39171–39178. doi: 10.1074/jbc.M103435200. [DOI] [PubMed] [Google Scholar]
- 18.Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15 a novel chromodomain protein is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 2002;277:50860–50866. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 19.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol. Cell. Biol. 2002;22:7868–7876. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 21.Tominaga K, Leung JK, Rookard P, Echigo J, Smith JR, Pereira-Smith OM. MRGX: a novel transcriptional regulator that exhibits activation or repression of B-myb promoter in a cell type dependent manner. J. Biol. Chem. 2003;278:49618–49624. doi: 10.1074/jbc.M309192200. [DOI] [PubMed] [Google Scholar]
- 22.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyon Y, Cote J. The highly conserved and multifunctional NuA4 complex. Curr. Opin. Genet. Dev. 2004;14 doi: 10.1016/j.gde.2004.02.009. in press. [DOI] [PubMed] [Google Scholar]
- 24.Kusch T, Florens L, MacDonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 25.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. USA. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 27.Ausserlechner MJ, Obexer P, Deutschmann A, Geiger K, Kofler R. A retroviral expression system based on tetracycline-regulated tricistronic transactivator/repressor vectors for functional analyses of antiproliferative and toxic genes. Mol. Cancer Ther. 2006;5:1927–1934. doi: 10.1158/1535-7163.MCT-05-0500. [DOI] [PubMed] [Google Scholar]
- 28.Smith JR, Pereira-Smith OM, Schneider EL. Colony size distributions as a measure of in vivo and in vitro aging. Proc. Natl. Acad. Sci. USA. 1978;75:1353–1356. doi: 10.1073/pnas.75.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Kitagaki J, Dai R-M, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li C-CH, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 30.Satheshkumar PS, Anton LC, Sanz P, Moss B. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J. Virol. 2009;83:2469–2479. doi: 10.1128/JVI.01986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarin A, Adams DH, Henkart PA. Protease inhibitors selectively block T cell receptor-triggered programmed cell death in a murine T cell hybridoma and activated peripheral T cells. J. Exp. Med. 1993;178:1693–1700. doi: 10.1084/jem.178.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosslau A, Jao DL-E, Butler R, Liu AY-C, Chen KY. Thermal killing of human colon cancer cells is associated with the loss of eukaryotic initiation factor 5A. J. Cell. Physiol. 2009;219:485–493. doi: 10.1002/jcp.21696. [DOI] [PubMed] [Google Scholar]
- 33.Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. BioEssays. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- 34.Gaczynska M, Osmulski PA, Ward WF. Caretaker or undertaker? The role of the proteasome in aging. Mech. Ageing Dev. 2001;122:235–254. doi: 10.1016/s0047-6374(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 35.Chondrogianni N, Gonos ES. Proteasome dysfunction in mammalian aging: steps and factors involved. Exp. Gerontol. 2005;40:931–938. doi: 10.1016/j.exger.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Vernace VA, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging and regulated protein degradation: who has the UPPer hand? Aging Cell. 2007;6:599–606. doi: 10.1111/j.1474-9726.2007.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibatani T, Nazir M, Ward WF. Alteration of rat liver 20S proteasome activities by age and food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51:B316–B322. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- 38.Bulteau AL, Szweda LI, Friguet B. Age-dependent decline in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 39.Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 41.Louie JL, Kapphahn RJ, Ferrington DA. Proteasome function and protein oxidation in the aged retina. Exp. Eye Res. 2002;75:271–284. [PubMed] [Google Scholar]
- 42.Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000;98:149–156. doi: 10.1016/s0306-4522(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 43.Lee C-K, Klopp RG, Weindruch R, Prolla A. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 44.Torres C, Lewis L, Cristofalo VJ. Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J. Cell. Physiol. 2006;207:845–853. doi: 10.1002/jcp.20630. [DOI] [PubMed] [Google Scholar]
- 45.Chondrogianni N, Trougakos IP, Kletsas D, Chen QM, Gonos ES. Partial proteasome inhibition in human fibroblasts triggers accelerated M1 senescence or M2 crisis depending on p53 and Rb status. Aging Cell. 2008;7:717–732. doi: 10.1111/j.1474-9726.2008.00425.x. [DOI] [PubMed] [Google Scholar]
- 46.Chondrogianni N, Stratford FLL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts. J. Biol. Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 47.Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan H-M, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 50.Wagner M, Hampel B, Hutter E, Pfister G, Krek W, Zwerschke W, Jansen-Durr P. Metabolic stabilization of p27 in senescent fibroblasts correlates with reduced expression of the F-box protein Skp2. Exp. Gerontol. 2001;37:41–55. doi: 10.1016/s0531-5565(01)00165-6. [DOI] [PubMed] [Google Scholar]
- 51.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp. Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 52.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 53.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 54.Grillari J, Katinger H, Voglauer R. Aging and the ubiquitinome: traditional and non-traditional functions of ubiquitin in aging and tissues. Exp. Gerontol. 2006;41:1067–1079. doi: 10.1016/j.exger.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Grillari J, Hohenwarter O, Grabherr RM, Katinger H. Subtractive hybridization pf mRNA from early passage and senescent endothelial cells. Exp. Gerontol. 2000;35:187–197. doi: 10.1016/s0531-5565(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 56.Voglauer R, Chang M, Wieser M, Dampier B, Baumann K, Schreiber M, Katinger H, Grillari J. Overexpression of SNEV extends the replicative life span of human endothelial cells. Exp. Cell Res. 2006;312:746–759. doi: 10.1016/j.yexcr.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okamura K, Nakai K. Retrotransposition as a source of new promoters. Mol. Biol. Evol. 2008;25:1231–1238. doi: 10.1093/molbev/msn071. [DOI] [PMC free article] [PubMed] [Google Scholar]