Abstract
Low penetration ability of Small Interfering RNA (siRNA) through the cellular plasma membrane combined with its limited stability in blood, limits the effectiveness of the systemic delivery of siRNA. In order to overcome such difficulties, we constructed a nanocarrier-based delivery system by taking advantage of the lessons learned from the problems in the delivery of DNA. In the present study, siRNA nanoparticles were first formulated with Poly(Propyleneimine) (PPI) dendrimers. To provide lateral and steric stability to withstand the aggressive environment in the blood stream, the formed siRNA nanoparticles were caged with a dithiol containing cross-linker molecules followed by coating them with Poly(Ethylene Glycol) (PEG) polymer. A synthetic analog of Luteinizing Hormone-Releasing Hormone (LHRH) peptide was conjugated to the distal end of PEG polymer to direct the siRNA nanoparticles specifically to the cancer cells. Our results demonstrated that this layer-by-layer modification and targeting approach confers the siRNA nanoparticles stability in plasma and intracellular bioavailability, provides for their specific uptake by tumor cells, accumulation of siRNA in the cytoplasm of cancer cells, and efficient gene silencing. In addition, in vivo body distribution data confirmed high specificity of the proposed targeting delivery approach which created the basis for the prevention of adverse side effects of the treatment on healthy organs.
Keywords: Poly(propyleneimine) dendrimer, siRNA targeted to BCL2 mRNA, LHRH peptide, tumor targeting, cytotoxicity, in vivo imaging
Introduction
RNA interference (RNAi), a post-transcriptional gene silencing method, mediated by small interfering RNA (siRNA) of 19–23 base pairs (bp) [1–5] has attracted attention for the development of novel highly effective therapeutics. However, like the other gene therapy strategies, the main obstacle to the success of siRNA therapeutics is its poor ability to penetrate across the cell membrane into the cytoplasm where it can guide sequence-specific mRNA degradation [2, 6]. Viruses have the ability to effectively deliver nucleic acids into cells; however, the immune response elicited by viral proteins possesses a major challenge for using viral vectors [7]. Therefore, a considerable interest is being devoted to the development of non-viral gene delivery vehicles [1, 3–5, 8–10].
Attempts to develop effective non-viral vectors for in vivo delivery of nucleic acids to cancer cells through a systemic route are hampered by difficulties of combining high extracellular stability in the blood stream with ready availability of the nucleic acids following entry into the cells. Extracellular stability is essential for resistance of the delivery system against the aggressive biological environment en-route to the target tumor site, while availability of the nucleic acids is required to permit efficient therapeutic effects within the cancer cells. It has been recognized that the complexation of the nucleic acid to nanoparticles of 100 – 200 nm size can form the basis for the effective transport of DNA through the cellular plasma membrane, even though the optimized size of nanoparticles is still in debate [11]. Currently, several modifications that increase extracellular stability and intracellular internalization were proposed. Caging of the nanoparticles with a dithiol containing cross-linker provides lateral stabilization, preventing unfavorable dissociation of the nanoparticles before entering the cytoplasm of target cells through the interaction with negatively charged biomacromolecules in the entity [12]. PEGylation of the caged nanoparticles stabilizes them against aggregation induced by salts and proteins existed in the serum [9]. The combined PEGylation and caging modifications of the DNA complexes led to the extended systemic circulation after intravenous administration to mice [12]. Furthermore, PEG coating allows for the attachment of specific targeting groups to its distal ends which can forward an entire system toward specific cells potentially eliminating nonspecific delivery to the healthy organs [13]. It was found that after entering the cellular cytoplasm, the disulfide bonds of coated and caged nanoparticles were reduced due to the reductive environment and the nucleic acid was released [14]. However, it was also reported that the gene transfection efficiency was largely decreased and even inhibited by increasing the number of disulfide linkages in caging due to over stabilization of the polyplex, and, consequently, less stable disulfide linkage was proposed [15]. It was found that the observed poor transfection efficiency was due to the difficulties in endosome-to-cytoplasm transfer of the disulfide caged complexes [12]. On the other hand, it was shown [16] that the balance between cationic charge and disulfide cross-linking densities of the polyplexes is crucial in order for the disulfide cross-linked polypolexes to achieve the intracellular release of entrapped plasmid DNA (pDNA) in the cytoplasm while maintaining its high stability in an extracellular environment.
The unique combination of reversible lateral and steric stabilization has not been exploited in siRNA therapeutic strategies. In spite of the similarities, pDNA and siRNA are very different in molecular weight and molecular topography with potentially important consequences. Typical DNA used in gene therapy often contains several thousands base pairs and possesses a molecular topography, which allows it to be packed into small, nanometer size particles when complexed with a cationic agent and be protected from enzymatic or physical degradation [17]. However, the persistent length of double-stranded RNA is around 70 nm [18], which is about 260 bp [19]. Therefore, siRNAs with 21 bp would essentially behave like rigid rods and are not likely to be further complexed. Consequently, their relatively uncontrolled interactions with cationic agents could result in undesirably large complexes or incomplete encapsulation of siRNA molecules, which exposes siRNA to potential enzymatic or physical degradation prior to the delivery to the targeted cells [17].
The overall goal of this work is an application and study of the surface modification approach for siRNA containing nanoparticles with an intention to answer the following questions: (1) whether siRNA nanoparticles formulated in the presence of cationic polymers are stable during the surface modification processes; (2) if caging with dithiol containing cross-linkers and PEGylation of siRNA nanoparticles are capable of preventing their dissociation and aggregation; (3) whether targeting of surface modified siRNA to specific cells can enhance their ability to effectively penetrate the plasma membrane of these cells and eliminate nonspecific delivery to healthy organs; (4) upon internalization into the cells, can siRNA nanoparticles escape from the 2 endosome to cytoplasm, and the siRNA can be effectively released from the modified nanoparticles with their bioactivity retained; and (5) are surface-modified siRNA tumor-targeted nanoparticles capable to preferentially accumulate in the tumor creating prerequisites for decreasing adverse side effects of the treatment.
Materials and methods
Materials
Dimethyl-3-3′-Dithiobispropionimidate-HCl (DTBP) was obtained from Pierce (Rockford, IL). Polypropylenimine tetrahexacontaamine Dendrimer Generation 5 (PPI G5), 2,4,6-trinitrobenzenesulphonic acid (TNBSA), Poly(methacrylic acid, sodium salt) solution (PMAA), and O-[2-(N-succinimidyloxycarbonyl)-ethyl]-O’-methylpolyethylene glycol 5000 (NHS-PEG(5000)OCH3) were obtained from Sigma-Aldrich (Milwaukee, WI). Ethidium Bromide (EtBr) solution was obtained from Promega (Madison, WI) and Glutathione Reduced from AMRESCO (Solon, OH). α-maleimide-ω-N-hydroxysuccinimide ester poly(ethylene glycol) (MAL-PEG-NHS, MW 5000 Da) was purchased from NOF Corporation (White Plains, NY). The sequence of siRNA targeted to BCL2 mRNA custom synthesized by Ambion (Austin, TX), was: sense strand, 5′-GUG AAG UCA ACA UGC CUG C-dTdT-3′; antisense strand, 5′-GCA GGC AUG UUG ACU UCA C-dTdT-3′. 6-FAM (siGLO Green) and DY-547 (siGLO Red) labeled siRNA was obtained from Applied Biosystems (Ambion, Inc., Foster City, CA). Cy5.5 NHS ester was purchased from GE Healthcare Life Sciences (Piscataway, NJ). siRNA used as a negative control (sense strand, 5′-CCU CGG GCU GUG CUC UUU U-dTdT-3′; antisense strand, 5′-AAA AGA GCA CAG CCC GAG G -dTdT-3′) was received from Dharmacon Inc. (Lafayette, CO). Synthetic analog of Luteinizing Hormone-Releasing Hormone (LHRH) decapeptide (Gln-His-Trp-Ser-Tyr-DLys(D-Cys)-Leu-Arg-Pro) was synthesized according to our design by American Peptide Company, Inc. (Sunnyvale, CA).
Preparation of PPI G5-siRNA complexes
The siRNA complexes were prepared at an amine/phosphate (N/P) ratio of 2.4 in 5 mM HEPES buffer (pH 8.0) by adding stock solution of PPI G5 dendrimer (typically, 500 μM) into a pre-prepared siRNA solution (Fig. 1A). The charge ratio was calculated by relating the number of positively charged primary amine groups on dendrimer with the number of negatively charged phosphate groups of siRNA. The final concentrations of siRNA and PPI G5 dendrimer in the solution were 4.0 μM and 6.3 μM, respectively. The samples were vortexed briefly, and the solutions were then incubated at room temperature for 30 min to ensure complex formation.
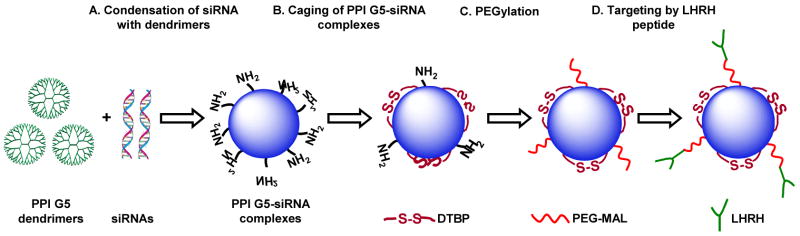
Fig. 1.
Engineering approach for the preparation of tumor-targeted, stable nanoparticles for the siRNA delivery.
Modification of PPI G5-siRNA complexes
In order to cross-link individual complexes, DTBP dissolved in 5 mM HEPES buffer (2.5 mg/mL, pH 8.0) was added to the above-formulated PPI G5-siRNA complexes solution at various concentrations depending on the desired cross-linking ratio. For example DTBP: NH2 = 3.2 indicates that PPI G5-siRNA complex was cross-linked with DTBP using molar ratio of 3.2 between DTBP and total amino groups of PPI G5 available after being complexed with siRNAs. After 3 h of reaction, pH of the solution was adjusted to 7.2, followed by the addition of MAL-PEG-NHS (35mg/mL). The NHS groups on the distal end of PEG reacted with amine groups on the periphery of PPI G5-siRNA complex. After the reaction within 1 h at room temperature, 12.5 mg/mL of LHRH peptide were added to the solution to covalently conjugate the peptide on the distal end of the PEG layer through the maleimide groups on the PEG and the thiol groups in LHRH (Fig. 1D). After 12 h of the reaction, the modified PPI G5-siRNA complexes were then purified by dialysis (MW cut-off 10 kDa) against water for 24 h and used for further studies. The percentage of amino groups available on the peripheral of the formulated PPI G5-siRNA complexes for cross-linking and PEGylation as well as the decrease in their concentration after the surface modification was determined by a modified spectrophotometric TNBSA assay [20]. The size and shape of the prepared complexes were determined by Atomic Force Microscopy (AFM) and Dynamic light scattering (DLS) (details please see in supplementary material).
Cell lines
Three types of human cancer cell lines were used: LHRH receptor-negative SKOV-3 and LHRH receptor-positive A2780 human ovarian and LHRH receptor-positive A549 human lung cancer cell lines. A2780 cells were obtained from Dr. T. C. Hamilton (Fox Chase Cancer Center, Philadelphia, PA), A549 and SKOV-3 cells were obtained from the ATTC (Manassas, VA). In addition, luciferase-transfected bioluminescent A549 cells were purchased from Xenogen (Alameda, CA). Cells were cultured in RPMI 1640 medium (Sigma Chemical Co., Louis, MO) supplemented with 10% fetal bovine serum (Fisher Chemicals, Fairlawn, NJ). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 (v/v) in air. All of the experiments were performed on the cells in exponential growth phase.
Cellular internalization
Cellular internalization of 6-FAM labeled siRNA complexes was analyzed by fluorescence (Olympus America Inc., Melville, NY) and laser scanning spectral confocal (Leica Microsystems Inc., Bannockburn, IL) microscopes as previously described [21–24]. Prior the visualization A549, A2780 and SKOV-3 cells were plated (20,000 cells/well) in 6-well tissue culture plates and treated with siRNA samples for 24 h. The concentration of siRNA was 0.25 μM. After 24 h of treatment cells were washed three times with DPBS, 1 mL of fresh medium was added to each well and photographed by a fluorescent or confocal microscope. To assess intracellular distribution of the substances, six optical sections, known as a z-series, were scanned sequentially along the vertical (z) axis from the top to the bottom of the cell.
Gene expression and In Vitro Cytotoxicity
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) and modified MMT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay were used for the analysis of expression of genes encoding BCL2 and β2-microglobulin (β2-m, internal standard) proteins and the evaluation of cellular cytotoxicity of all studied formulations as previously described [21–23], respectively (details please see in supplementary materials).
siRNA serum stability
Serum stability of both free siRNA and modified siRNA complexes (equivalent to 4.0 μM of siRNA) was investigated by incubating each formulations at 37 °C with equal volume of human serum to give 50% serum concentration. At each predetermined time interval, (0, 5, 15, 30min, 1, 2, 3, 4, 5, 6, 7, 24 and 48 h) 50 μL of the mixture were removed and stored at −20°C until gel electrophoresis was performed. In order to release siRNA from the complexes for gel electrophoresis, each sample was treated with 25 mM of reduced glutathione and 100 μM of PMAA. The aliquots from different incubation time periods were loaded onto 4% NuSieve 3:1 Reliant agarose gels in 1×TBE buffer (0.089 M Tris/Borate, 0.002 M EDTA, pH 8.3; Research Organic Inc., Cleveland, OH) and subjected to submarine electrophoresis. The gels were stained with EtBr, digitally photographed, and scanned using Gel Documentation System 920 (NucleoTech, San Mateo, CA).
In vivo body distribution
Experiments were carried out on nude mice bearing subcutaneous rapidly grown xenografts of human lung cancer cells as previously described [24]. According to the approved institutional animal use protocol, the tumor was measured by a caliper every day and its volume was calculated as d2×D/2 where d and D are the shortest and longest diameter of the tumor in mm, respectively. After tumor size reached about 200–300 mm3, LHRH targeted and non-targeted PPI G5-siRNA complexes were injected systemically into the mice. Animals were anesthetized with isoflurane using the XGI-8 Gas Anesthesia System (Xenogen, Alameda, CA). Bioluminescence of transfected with luciferase cancer cells after intraperitoneal injection of luciferin (10 min before visualization) and fluorescence of labeled with Cy5.5 dendrimer (supplementary material) and with DY-547 siRNA were visualized using IVIS Xenogen (Alameda, CA) imaging system 72 h after the injection. Visible light, bioluminescence, and fluorescence images were taken and overlaid using the manufacturer’s software to obtain composite images. The distribution of fluorescence was analyzed using original software developed in our laboratory.
Statistical analysis
Data were analyzed using descriptive statistics, single-factor analysis of variance (ANOVA), and presented as mean values ± standard deviation (SD) from four to eight independent measurements. The comparison among groups was performed by the independent sample student’s t-test. The difference between variants was considered significant if P < 0.05.
Results
Stabilization of siRNA nanoparticles by caging with dithiol containing cross-linkers and PEGylation; release of the siRNA triggered by intracellular reductive reagents
PPI G5 dendrimers were able to effectively package siRNA into discrete nanoparticles (Fig. 1A), probably through a zipping packaging pathway, similar to that responsible for the complexation of single stranded antisense oligonucleotides with PPI dendrimers [25].
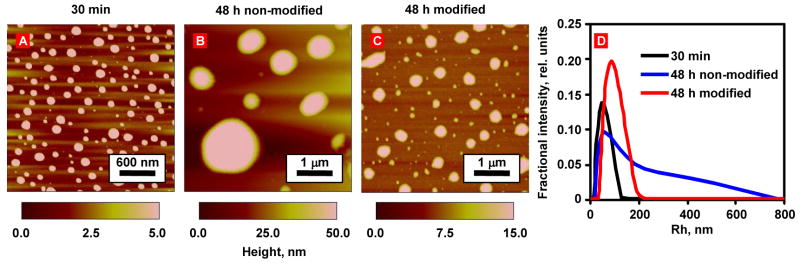
The nanoparticles appeared to be spherical after 30 minutes of complexation with an average diameter of 150.1 ± 22.2 nm and a height of 5.4 ± 0.9 nm (Fig. 2, AFM measurements). Additionally, DLS measurements demonstrated that the average diameter of nanoparticles was around 101.2 ± 43.0 nm with a relatively narrow size distribution (Fig. 2D). Note that the size of the nanoparticles determined by DLS was ~1.5 times smaller than that obtained by AFM diameter measurements, which possibly could be attributed to flattering of the nanoparticles on the mica surface during the drying process [26]. However, the formulated non-modified nanoparticles did not remain stable over a long period of time under the storage conditions. According to the AFM studies (Fig. 2B), 48 h after incubation in PBS buffer, the nanoparticles significantly (P < 0.05) increased the average size from 150.1 ± 22.2 nm to 612.7 ± 451.1 nm (Fig. 2A and 2B). Furthermore, DLS measurements also demonstrated the same trend, indicating the increase in the nanoparticles averaged diameter and very broad size of its distribution (262.0 ± 260.0 nm) after 48 h (Fig. 2D). These results showed that the siRNA nanoparticles experienced a serious aggregation in PBS buffer (containing sodium salts) during this period of time and it can be an obstacle for their in vivo application, since aggregation can lead to rapid clearance of the entire delivery complexes by phagocytic cells and the reticuloendothelial system [27]. Furthermore, some negatively charged proteins and polyanions in serum and blood can slowly penetrate and further destabilize the siRNA nanoparticles by replacing the siRNA molecules before reaching the targeted cancer cells [12].
Fig. 2.
Influence of the storage in PBS buffer at room temperature on the PPI G5-siRNA nanoparticles. Representative AFM images of modified and non-modified PPI G5-siRNA complexes. (A) Non-modified nanoparticles after 30 min of condensation. (B) non-modified siRNA nanoparticles stored for 48 h at room temperature. (C) siRNA complexes modified with DTBP, NHS-PEG-maleimide and LHRH and stored for 48 h at room temperature. (D) Representative curves of DLS size measurements of siRNA complexes presented on A–C.
In order to provide lateral stabilization and inhibit the possible replacement of siRNA by serum polyanions, the formulated siRNA nanoparticles were modified with cleavable dithiol containing cross linker, such as DTBP (Fig. 1B). It was estimated using TNBSA assays [28] that 26.5% of free amine groups on the siRNA nanoparticles were cross-linked. To prevent the siRNA nanoparticles aggregation and extend the circulation time of the siRNA nanoparticles in the blood stream, we further modified the caged siRNA nanoparticles with NHS functionalized PEG for steric stabilization (Fig. 1C). The stability of the siRNA nanoparticles with covalent stabilization by DTBP and the PEG layer was first evaluated by AFM and DLS. The data obtained revealed the fact that in comparison with non-modified particles, the modified nanoparticles became slightly larger (from 150.1 ± 22.2 nm to 256.8 ± 123.8 nm for non-modified and modified nanoparticles, respectively, based on AFM measurements, and 101.2 ± 43.0 to 182.4 ± 67.4 nm, based on DLS measurements). However, after 48 h in PBS buffer, the size of the particles retained almost the same with relatively narrow distribution (Fig. 2C and D). The increase in the size of the prepared nanoparticles could be attributed to the modification of their surfaces by PEG and the targeting peptide layer [29].
The stability of the siRNA nanoparticles with or without covalent caging by DTBP, with or without the PEG layer was further studied by analyzing the stability of the siRNA nanoparticles against polyanion disruption. This method has been widely used to study the stability of DNA nanoparticles in vitro by mimicking in vivo polyanion-induced DNA complexes disruption [30]. The experiments were performed by measuring the ability of a polyanion, PMAA to restore siRNA access to EtBr binding. To evaluate the effectiveness the siRNA release after the cytoplasmic delivery of PPI G5-siRNA complexes, glutathione, a reductive agent present in cytoplasm [12], was added to the cross-linked samples with and without PEGylation.
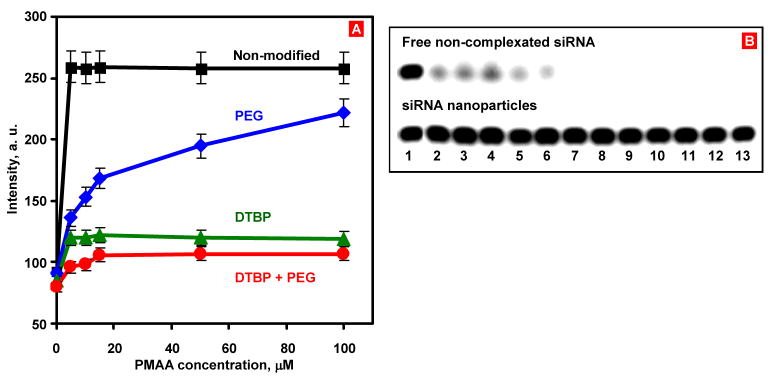
As expected, the EtBr fluorescence was dramatically increased upon its incubation with free siRNA (data not shown). Complexation of the siRNA with PPI G5 dendrimers decreased the intensity of the EtBr fluorescence to the level of free dye. Cross-linking the siRNA nanoparticles with DTBP did not introduce any significant change in fluorescence (data not shown), indicating that the siRNA nanoparticles remain compacted during the cross-linking process. As the concentration of PMAA, added to the siRNA nanoparticles solution, was increased, higher fluorescence readings were observed (Fig. 3A). This increase demonstrated that siRNA release from non-modified complexes or at least local de-complexation of the siRNA nanoparticles took place. For the siRNA nanoparticles without DTBP and PEG layer, 5 μM PMAA was able to release 85 % of the siRNA which then was intercalated by EtBr substantially increasing its fluorescence (Fig. 3A, black curve). For the siRNA nanoparticles caged with DTBP, 5 μM PMAA released only 14% of siRNA. Addition of PMAA up to 100 μM did not cause further release of siRNA, indicating that siRNA nanoparticles caged by DTBP were stable against PMAA disruption (Figure 3A, green curve). When 25 mM glutathione was added to the solution to promote reduction of the intramolecular disulfide bond in DTBP, the stabilization of siRNA was compromised and about 85% of siRNA was released from the nanoparticles (estimated from the EtBr fluorescence increase, data not shown). For the PEG-protected siRNA nanoparticles, with PMAA concentration increase, the fluorescence gradually reached 70% of the level observed for the free siRNA (Fig. 3A, blue curve). These data show that PEG layer alone could not effectively protect siRNA nanoparticles from the polyanions disruption. The addition of 5 μM PMAA to the nanoparticles with the combination of the DTBP cross-linking and PEGylation on the siRNA nanoparticles led to the release of 10% of siRNA and caused no further release at higher PMAA concentrations tested (Figure 3A, red curve). Addition of 25 mM glutathione into the solution, led to the release of about 80% of siRNA from the modified nanoparticles. The results of this series of the experiments demonstrated the efficiency of layer-by-layer modification approach to prevent the disruption of siRNA complexes in the presence of competing polyanions, which are widely abundant in extracellular environment. Moreover, the availability of disulfide bond in the structure of the cross-linking agent for the destruction by glutathione can provide a triggered release of siRNA in the cytoplasm of the targeted cell.
Fig. 3.
EtBr replacement assay to study the stability of siRNA nanoparticles against PMAA disruption. (A) EtBr fluorescence from interactions with different siRNA nanoparticles packaged by PPI G5 dendrimers (nonmodified, caged with DTBP cross-linker, PEGylated, and modified by the combination of DTBP caging and PEGylation) in the presence of PMAA. (B) The human serum stability of free siRNA (upper panel) and the engineered PPI G5-siRNA nanoparticles (lower panel). 1- 0 min; 2 – 5 min; 3 – 15 min; 4–30 min; 5-1 h; 6- 2 h; 7- 3 h; 8- 4 h; 9- 5 h; 10- 6 h; 11- 7 h; 12–24 h; 13–48 h, respectively.
PEGylation prevents the siRNA nanoparticles from aggregation and limits nonspecific delivery of the non-targeted siRNA nanoparticles inside cells
To demonstrate whether the PEG layer can prevent siRNA nanoparticles from aggregation in extracellular environments and non-specific uptake by cells, we have compared the ability of non-targeted siRNA nanoparticles with different modifications to undergo cellular internalization. In these experiments the cells were treated with the 6-FAM-labeled siRNA-dendrimer nanoparticles fabricated from PPI G5 dendrimer alone, and the PPI G5-siRNA nanoparticles caged by DTBP and PEG with different molar ratio between DTBP and the amine groups in the PPI G5 (DTBP: NH2=3.2:1 and 15.9:1). Nonmodified siRNA nanoparticles showed serious aggregation in the cell medium (Fig. 4A). In case of the modified nanoparticles, there is no significant aggregation of the siRNA nanoparticles (Fig. 4B, C), indicating that the PEG layer can remarkably prevent aggregation of siRNA-nanoparticle complexes induced by physiological salts and serum proteins. Furthermore, the modified non-targeted siRNA nanoparticles at the lower ratio of DTBP: NH2 (3.2:1) in the conjugation solution (Fig. 4C), showed significantly lower internalization by the cells, compared to the nanoparticles with the higher ratio (DTBP: NH2=15.9:1, Fig. 4B). The internalization efficiency of the siRNA complexes with different modification ratios was quantitatively evaluated by measuring the fluorescence intensity of 6-FAM labeled siRNA internalized by single A549 cancer cell under the same experimental conditions. The obtained data revealed that the complexes with higher degrees of modification (fluorescence intensity is 19.2×103 a.u. per cell) resulted in ~ 1.5 times stronger siRNA cellular internalization when compared to the nanoparticles with lower modification degree (12.6×103 a.u per cell).
Fig. 4.
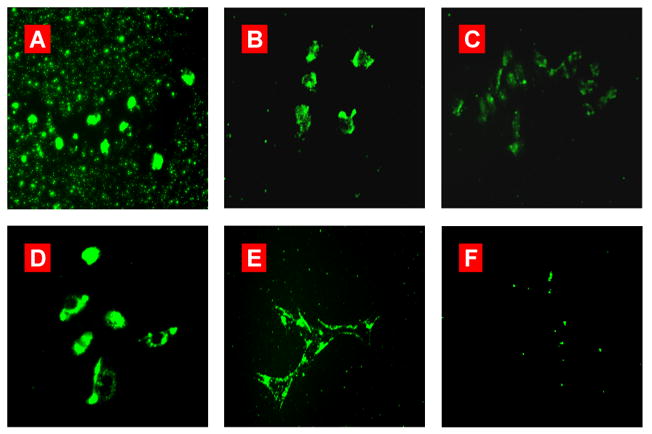
Cellular internalization of different PPI G5-siRNA complexes in various cell lines. Representative fluorescence microscopy images of (A) siRNA-PPI G5 complexes; (B) PEG-DTBP-PPI G5-siRNA complexes (DTBP:NH2=15.9:1); (C) PEG-DTBP-PPI G5-siRNA complexes (DTBP:NH2=3.2:1) and (D-F) LHRH-PEG-DTBP-PPI G5-siRNA complexes after 24 h of incubation with LHRH-positive A549 (D), A2780 (E), and negative SKOV-3 (F) cancer cells. siRNA was labeled with 6-FAM Green.
Incorporation of siRNA into caged and PEGylated nanoparticles prevents its degradation in serum
The stabilization of siRNA in serum against the nuclease degradation is essential for in vivo siRNA delivery. Therefore, to examine siRNA protection in the engineered siRNA nanoparticles from nuclease degradation, the complexes were incubated in the presence of 50% human serum at 37°C. Gel electrophoresis experiment demonstrated that the degradation of free siRNA was observed 5 min after the incubation, while 2 h after the beginning of the experiment, particles were fully digested (Fig. 3B, upper panel). On the other hand, the surface engineering approach effectively stabilized the siRNA nanoparticles in human serum for the entire duration of the experiment (48 h, Fig. 3B, bottom panel).
Bioavailability of complexated siRNA and suppression of targeted BCL2 mRNA
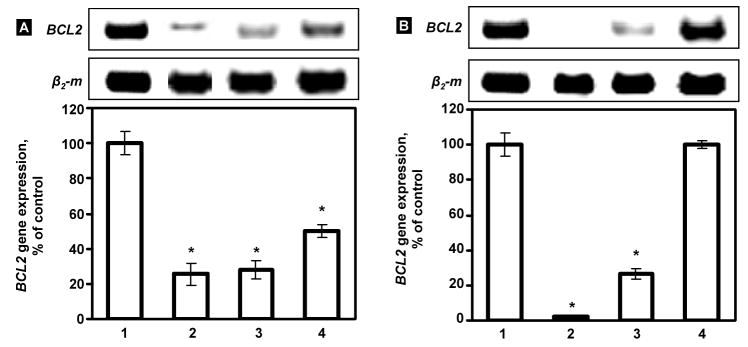
To demonstrate whether the siRNA can be released from the caged nanoparticles after internalization in cytoplasm and whether the bioactivity of the siRNA is retained after the modification of nanoparticles, the ability of the modified siRNA nanoparticles to silence their target mRNA was studied using RT-PCR. The sequence of siRNA was designed to target to BCL2 mRNA and can be used to suppress antiapoptotic cellular defense (nonpump drug resistance) [5, 21]. As shown in Figure 7A, caged nanoparticles with higher ratio of DTBP (DTBP: NH2=15.9:1, which result in lower level of nanoparticles PEGylation) induced the average suppression of the BCL2 gene on 72.8 % (down to 27.2 % of control level). Non-modified nanoparticles demonstrated similar effect decreasing the expression of targeted mRNA on 74.5 % (down to 25.5 % of control level). The nanoparticles with lower caging ratio (DTBP: NH2=3.2:1) showed significantly (P < 0.05) lower level of the suppression (on 53.2 %), which is in agreement with their lower cellular internalization. These results demonstrated that the siRNA can be released from the modified nanoparticles in the cytoplasm of cells with preserved biological activity.
Fig. 7.
Suppression of BCL2 mRNA by different nanoparticles containing BCL2 targeted siRNA. Typical images of RT-PCR products and average expression of the BCL2 gene determined by RT-PCR. Panel A: (1) control (fresh media), (2) PPI G5-siRNA nanoparticles (3) PEG-DTBP-PPI G5-siRNA nanoparticles (DTBP: NH2=15.9:1) and (4) PEG-DTBP-siRNA-PPI G5 nanoparticles (DTBP: NH2=3.2:1) incubated with A549 cancer cells. Panel B: (1) control (fresh media), A549 cancer cells; (2–4) engineered targeted siRNA nanoparticles (LHRH-PEG-DTBP-siRNA-PPI G5) incubated with LHRH-positive A2780 (2) and A549 cancer cells (3) and LHRH-negative SKOV-3 cancer cells (4). Messenger RNA levels are expressed as the percentage of the β2-m (internal standard) level. The expression of the BCL2 gene in control conditions was set at 100%. Means ± SD are shown. *P < 0.05 when compared with control.
Targeting of nanoparticles specifically to cancer cells by LHRH peptide
To prevent the non-specific accumulation of siRNA nanoparticles in healthy tissues and enhance their internalization specifically by cancer cells we used a synthetic analog LHRH peptide as a ligand to corresponding LHRH receptors that are overexpressed in the plasma membrane of many types of cancer cells [13, 24]. To incorporate LHRH peptide as a targeting moiety into designed siRNA delivery system, we employed a heterobifunctional PEG (5 kDa) molecule, MAL-PEG-NHS to modify the DTBP caged siRNA nanoparticles. It contains an amine reactive NHS ester and MAL reactive group on the opposite sides and allows for a further modification siRNA nanoparticles in a layer-by-layer fashion. We followed the well-documented coupling and separation procedures [31] to covalently link PEG with amine groups on the caged siRNA nanoparticles (DTBP:NH2=3.2:1), and then conjugated the LHRH peptides to the distal end of PEG layer through MAL group and the cysteine in the LHRH peptides. Since post-modification approach was employed in the study, one could expect that LHRH peptide is only conjugated on the surface of PPI G5-siRNA complexes. The presence of LHRH peptide on the complex surface was detected by Bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific Inc., Rockford, IL) according to manufacture protocol (Fig. S1, supplementary material).
Fluorescent microscopic study demonstrated that the LHRH-modified siRNA complexes can successfully be internalized by LHRH-positive A549 and A2780 cancer cells that are overexpressed LHRH receptors but not by LHRH-negative SKOV-3 cells which do not show a detectable level of LHRH receptors (Fig. 4D–F). The ability of the LHRH–modified complexes to suppress the level of target BCL2 mRNA was studied by RT-PCR. The results demonstrated that the siRNA delivered by the targeted nanoparticles effectively knocked down their target gene in cancer cells overexpressed LHRH receptors (Fig. 7B, bars 2 and 3). In contrast, the same siRNA complexes were unable to significantly suppress targeted mRNA in SKOV-3 cells with low expression of the receptors (Fig. 7B, bar 4). These data are in agreement with lower cellular internalization of targeted nanoparticles in LHRH-negative SKOV-3 cell line when compared with cells that are overexpressed LHRH receptors (compare Fig. 4, D–F). In order to demonstrate the specificity of siRNA, a negative control siRNA duplex (mock siRNA with a sequence having no substantial matching to any known mRNA) was complexated with similar targeted PPI dendrimer. The influence of these complexes on the expression of the targeted BCL2 mRNA was investigated. No significant inhibition of BCL2 mRNA was found after the incubation of the cells with these negative control PPI G5-siRNA nanoparticles (data not shown).
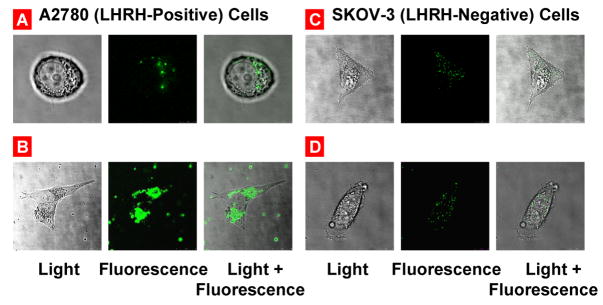
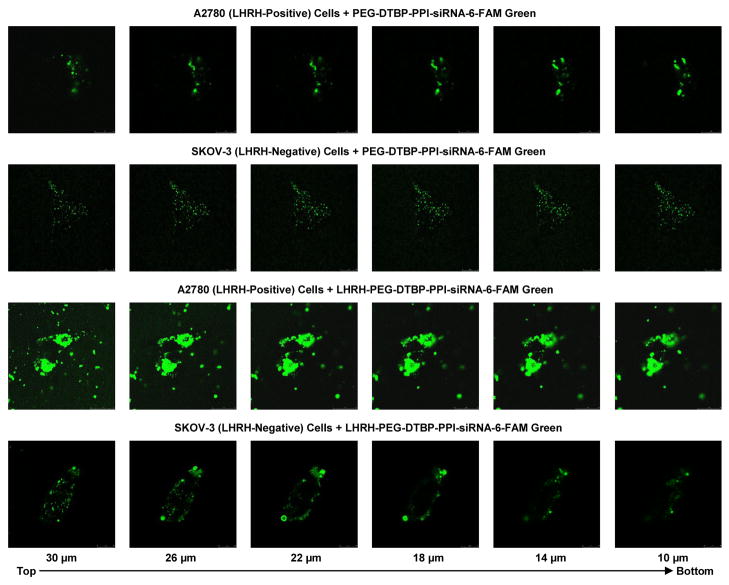
Confocal microscope study (Fig. 5 and Fig. 6) supports data obtained by fluorescent microscopy (Fig. 4) and shows that siRNA delivered by non-targeted nanoparticles possesses relatively low penetration ability in human ovarian cancer cells which express (A2780) and do not express (SKOV-3) LHRH receptors. Internalization of siRNA delivered by nanoparticles targeted to cancer cells by LHRH peptide was substantially higher in the cells overexpressed LHRH receptors (A2780) when compared with non-targeted nanoparticles. In contrast, the degrees of internalization of both non-targeted and LHRH-targeted nanoparticles by SKOV-3 ovarian cancer cells that do not express LHRH receptors were similar.
Fig. 5.
Confocal microscope images of LHRH receptor-positive A2780 (A, B) and LHRH receptor-negative SKOV-3 (C, D) human ovarian cancer cells incubated with fluorescent labeled PEG-DTBP-PPI G5-siRNA-6-FAM Green (A, C) and LHRH-PEG-DTBP-PPI G5-siRNA-6-FAM Green (B, D) nanoparticles.
Fig. 6.
Confocal microscopy fluorescent images of LHRH receptor-positive (A2780) and LHRH receptor-negative (SKOV-3) ovarian cancer cells incubated for 24 h with non-targeted (PEG-DTBP-PPI-siRNA-6-FAM Green) and targeted (LHRH-PEG-DTBP-PPI-siRNA-6-FAM Green) dendrimers (z-series, from the top of the cell to the bottom).
Intracellular distribution of the delivered siRNA
We applied confocal fluorescent microscopy to analyze the distribution of 6-FAM-labeled siRNAs in different cellular compartments (Fig. 8A–C) and various layers from the upper to the lower of fixed cells (z-sections, Fig. 8D). Fluorescence was observed in the cytoplasm but not in nuclear and the distribution was very similar for the most part of the different cellular layers. The maximal accumulation of delivered siRNA was found in the cytoplasm in the perinuclear region and near the plasma membrane.
Fig. 8.
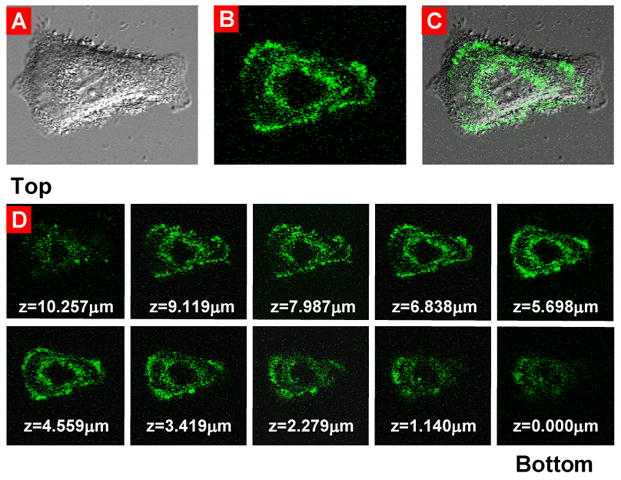
Intracellular localization of fluorescently labeled siRNA delivered by the targeted by LHRH modified PPI dendrimers complexes. Typical confocal microscopy images: (A) light; (B) fluorescent; (C) superimposed light and fluorescent of human A549 lung cancer cells incubated for 24 h with modified siRNA complexes. (D) z-series, from the top of the cell to the bottom.
In vitro cytotoxicity of the engineered siRNA nanoparticles
The cytotoxicity of the engineered nanoparticles was examined on A549 human lung cells using free PPI G5 dendrimer and the modified PPI G5 dendrimer with siRNA targeted to BCL2 mRNA (Fig. S2, supplementary material). The small decrease (~20 %) in cellular viability was observed in cells exposed to the modified nanoparticles with siRNA compare to free PPI G5 dendrimer under concentrations that were substantially higher than the working concentration of dendrimers (0.4 μM) used in the present study. The observed modest cellular toxicity of dendrimer-siRNA complexes is most likely attributed to the suppression by the siRNA targeted to the BCL2 mRNA of the antiapoptotic function of BCL2 protein. As we previously showed, such suppression leads to the slight cell death induction [5, 21].
In vivo body distribution of targeted and non-targeted dendrimer-siRNA complexes
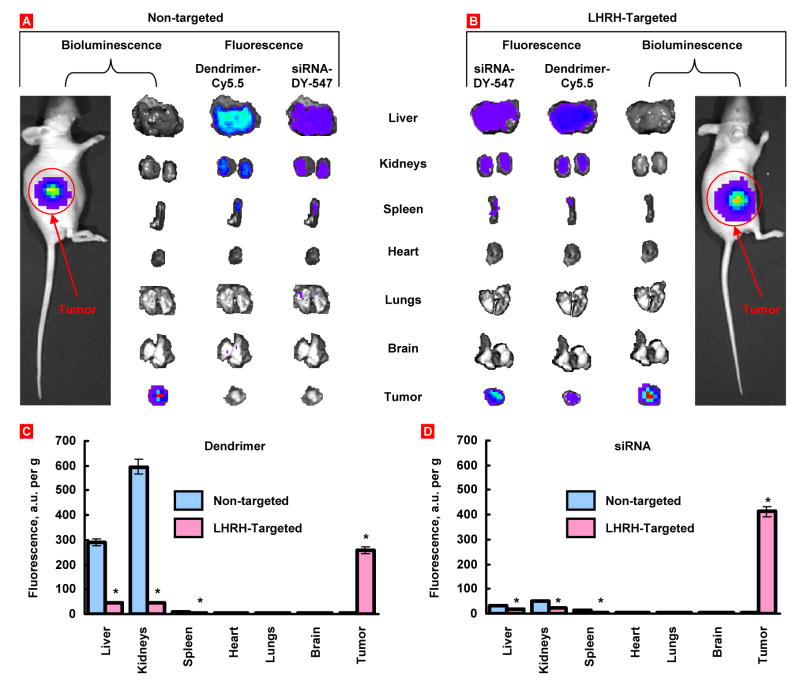
Body distribution of non-targeted and tumor-targeted nanoparticles and encapsulated siRNA was studied in vivo after their systemic administration. Nanocarriers were labeled with Cy5.5 near infrared dye; siRNA was labeled with DY-547 (siGLO red). In addition, A549 human cancer cells were transfected with luciferase prior to the inoculation to the flanks of nude mice. The bioluminescence of the transfected cells as well as fluorescence of dendrimers and siRNA was analyzed using IVIS imaging system (Xenogen, Alameda, CA). Animals were anesthetized with isoflurane and photographed 72 h after the injection of nanoparticles. The intensity of fluorescence was represented on composite light/fluorescent images by different colors with blue color reflecting the lowest fluorescence and red color – the highest intensity (Fig. 9A, B). After photographing, animals were euthanized; tumor and organs were excised, photographed and processed similarly to the images of an entire animal. Total fluorescence in the tumor and organs were calculated (Fig. 9C, D). The data showed that tumor targeting by LHRH peptide significantly changed body distribution of dendrimers and delivered siRNA. Non-targeted dendrimer and delivered by this carrier siRNA were found mainly in the liver and kidney 72 h after the injection. Only trace amounts of dendrimer and siRNA were found in the tumor. In contrast, targeted dendrimer and siRNA delivered by this carrier were found predominately in the tumors.
Fig. 9.
Organ and tumor content of non-targeted and LHRH- targeted PPI dendrimers and siRNA. Human A549 cancer cells were transfected with luciferace and injected subcutaneously into the flanks of nude mice. Dendrimers and siRNA were labeled with near infrared Cy5.5 dye and DY-547 (siGLO red) respectively. PPI G5-siRNA nanoparticles were injected into the mice. The distribution of labeled dendrimers and siRNA was analyzed in live anesthetized animals 72 h after injection using IVIS Xenogen imaging system. The intensity of bioluminescence (tumor cells) and fluorescence (dendrimer and siRNA) are expressed by different colors with blue color reflecting the lowest intensity and red color reflecting the highest intensity. After measuring the distribution of bioluminescence and fluorescence in the entire animal, tumor and organs were excised and their bioluminescence and fluorescence were registered and processed by the imaging system. Representative images are shown (A, B). The bottom panel demonstrates average concentration per gram of organ weight of labeled dendrimers (C) or siRNA (D). Means ± SD are shown. *P < 0.05 when compared with non-targeted complex.
Discussion
The proposed modified siRNA-PPI dendrimer complexes have been designed (1) to prevent the aggregation of nanoparticles; (2) to increase the stability of the nanoparticles against degradation en route to the targeted site of action; (3) to provide for a targeted delivery specifically to the tumor cells and prevent adverse side effects on healthy organs; (4) to release siRNA in the cytoplasm of targeted cancer cells; (5) to preserve the activity of packaged siRNA; (6) to ensure relatively low cellular toxicity of the developed nanoparticles; (7) to provide for a preferential accumulation of targeted siRNA complexes in the tumor. The characterization and testing of the developed nanoparticles showed that all aforementioned requirements have been achieved.
We packaged siRNA into nanoparticles with the average diameter of about 100 nm using PPI generation 5 dendrimer. The similar dendrimers have previously been efficiently used for the delivery of plasmid DNA and antisense oligonucleotides [32]. However, according to our experimental data, this dendrimer without further modification cannot provide for an effective intracellular delivery of siRNA because of aggregation of resulting nanoparticles, low stability in saline, and blood plasma and low penetration ability into cancer cells. To improve the efficiency of the systemic delivery of siRNA to targeted tumor cells, we have employed a layer-by-layer surface modification strategy which can stabilize the gene delivery vectors during the delivery phase and can be triggered to release DNA at the target sites. The data obtained indicate that siRNA nanoparticles are stabilized in terms of dissociation in the presence of a competing polyanion after DTBP cross-linking and PEGylation. Gel electrophoresis experiment demonstrated that the modified siRNA nanoparticles were stable in human serum for at least 48 h, while free siRNA was digested in 2 h. These results are in a good corroboration with the data obtained by Kim and co-workers [33] for the PEGylated siRNA delivery system based on polyethylenimine (PEI), which maintained the extended stability of siRNA against nuclease-mediated degradation in 50% serum. In contrast, the non-modified chitosan-siRNA nanoparticles in the same serum concentration kept siRNA intact up to 7 h and fully degraded after 48 h of incubation [34]. Therefore, the prolonged protection observed in our engineered nanoparticles was likely due to the presence of PEG chains in the outer shell layer that sterically hindered the access of nucleases into PPI G5-siRNA internal core. The DTBP cross-linking of the prepared nanoparticles could also contribute to overall serum stability through the prevention of siRNA release from the formed particles.
The second positive effect of the proposed surface modification includes the prevention of non-specific cellular internalization of nanoparticles. The present experimental data showed that caged and PEGylated non-targeted nanoparticles containing siRNA were unable to efficiently penetrate inside cells. The cross-linked siRNA nanoparticles increase the stability against polyelectrolyte exchange, with the remaining positive surface charges on the siRNA nanoparticle surface, which are essential for non-specific internalization of nucleic acid nanoparticles. It was reported that modification of the DNA nanoparticles with hydrophilic polymers such as PEG can stabilize the polyplexes against salt, and protein-induced aggregation. The increased stability against aggregation was attributed to the steric effects of the PEG layer that lead to decreased particle–particle and particle–protein interactions. At the same time, PEGylation has been shown to reduce internalization of non-targeted nanoparticles [9]. The effect depends on the molecular weight of the PEG and the grafting density on the particles. The higher molecular ratio (DTBP: NH2=15.9:1) during the caging process resulted in decreasing of NH2 groups on the siRNA particles surface for PEG conjugation. Even though the PEG layer prevents the aggregation of the siRNA nanoparticles, a large amount of siRNA was still internalized by the cells. On the other hand, with lower molecular ratio (DTBP: NH2=3.2:1) during the caging process, more NH2 groups left on the siRNA particles surface for PEG conjugation, so the density of the positive charges on the siRNA nanoparticle surfaces was largely decreased after further PEGylation of the DTBP caged siRNA nanoparticles limiting the cellular internalization of the siRNA nanoparticles.
Consequently, we used a special tumor-targeting moiety in order to increase the internalization of the modified nanoparticles specifically by cancer cells and further prevent their non-specific uptake by healthy cells in order to limit the adverse side effects of chemotherapy. Recently, we have successfully used a modified peptide synthetic analog of Luteinizing Hormone-Releasing Hormone decapeptide as a targeting moiety to tumors overexpressing LHRH receptors [13, 24, 35]. Data obtained in the present study support our previous findings and show that the use of this peptide prevented an accumulation of nanoparticles and siRNA in healthy organs, and enhanced both drug accumulation in tumors and its internalization by cancer cells. Most importantly, despite the protective effects of cross-linking, upon internalization into the targeted cancer cells, siRNA escaped from endosomes to cytoplasm and released from the nanoparticles to efficiently silence the targeted mRNA. In addition to enhancing the uptake by cancer cells, the use of LHRH peptide as a targeting moiety provided for a tumor-specific delivery of siRNA and prevented its accumulation in other healthy organs. This, in turn, created the basis for the prevention of severe adverse side effects of the treatment. Observed high targeting efficiency of used in the present study siRNA complexes can be explained by exceptional specificity of LHRH peptide to the cancer cells with overexpression of LHRH receptors. Previously, we found that LHRH receptors overexpressed in several types of cancer (including ovarian, prostate, lung, breast and colon). In contrast, we did not find detectable expression of these receptors in healthy human visceral organs (lung, heart, liver, kidney, spleen, etc.) [13, 23, 24, 35–37]. This reversible stability of the siRNA nanoparticles combined with the effective targeted delivery specifically to cancer tumor cells has the potential to improve siRNA therapeutic effects and minimize adverse side effects on healthy cells caused by the non-specific uptake.
Data obtained showed that siRNA delivered by the proposed modified targeted nanoparticles localized inside the cytoplasm of cancer cells relatively homogenously within the cell from the top to the bottom. It is known that short antisense oligonucleotides delivered to the cell cytosol can penetrate into the nucleus after escaping from endosome [38]. The cytoplasmic localization and distribution of siRNA is different depending on the type and the concentration of the used delivery agent [1, 3–5, 17]. Some reports suggest that the activity of the siRNA is closely related to their subcellular localization [39]. Being delivered by cationic liposomes, most of the siRNA localized near the perinuclear regions and did not enter the nucleus even after extended periods of time [1, 3, 5, 17]. In contrast, delivered by modified polyamidoamine (PAMAM) dendrimers siRNA was found at both the perinucleus and nucleolus regions [4, 39, 40]. It is found that such shift in siRNA localization pattern was associated with their lower RNAi activity, suggesting that sub-cellular localization of siRNA was important for siRNA function [39]. In the present study, siRNA delivered by the targeted modified nanoparticles localized in the cellular cytoplasm did not penetrate into the nuclei. At the same time, the higher efficiency of delivered siRNA was registered in terms of the suppression of targeted mRNA. Taken together, these data have shown that proposed tumor targeted modified nanoparticles were able to increase the stability siRNA in the serum, effectively deliver it into the cytoplasm of LHRH-positive cancer cells, and into the tumor while preserving its gene silencing ability.
Conclusion
The combination of serum resistance, the increased stability in biological liquids, tumor-specific targeting, effective penetration into cancer cells, accumulation of delivered siRNA in the cytoplasm and preferential accumulation in the tumor conferred by the caging, PEGylation, and targeting by LHRH peptide makes the proposed nanoparticles almost ideal nanoscale-based siRNA delivery vehicle. Consequently, one can conclude that this approach can be used for the in vivo systemic delivery of siRNA for efficient cancer therapy.
Supplementary Material
Acknowledgments
The research was supported in part by NIH CA100098, CA111766 and Charles & Johanna Bush Biomedical Grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Betigeri S, Pakunlu RI, Wang Y, Khandare JJ, Minko T. JNK1 as a molecular target to limit cellular mortality under hypoxia. Mol Pharm. 2006;3(4):424–430. doi: 10.1021/mp060014x. [DOI] [PubMed] [Google Scholar]
- 2.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with Disease: A Progress Report on siRNA-based Therapeutics. Nat Rev Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbuzenko OB, Saad M, Betigeri S, Zhang M, Vetcher AA, Soldatenkov VA, Reimer DC, Pozharov VP, Minko T. Intratracheal Versus Intravenous Liposomal Delivery of siRNA, Antisense Oligonucleotides and Anticancer Drug. Pharm Res. 2009;26(2):382–394. doi: 10.1007/s11095-008-9755-4. [DOI] [PubMed] [Google Scholar]
- 4.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19(7):1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 5.Saad M, Garbuzenko OB, Minko T. Co-delivery of siRNA and an anticancer drug for treatment of multidrug-resistant cancer. Nanomed. 2008;3(6):761–776. doi: 10.2217/17435889.3.6.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a Correction between siRNA Localization, Cellular Uptake, and RNAi in Living Cells. Chem & Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Bessis N, GarciaCozar FJ, Boissier MC. Immune Responses to Gene Therapy Vectors: Influence on Vector Function and Effector mechanisms. Gene Ther. 2004;11:S10–17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 8.Kim WJ, Kim SW. Efficient siRNA Delivery with Non-viral Polymeric Vehicles. Pharm Res. 2009;26(3):657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 9.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and Development of Polymers for Gene Delivery. Nat Rev Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 10.Shim MS, Kwon YJ. Controlled cytoplasmic and nuclear localization of plasmid DNA and siRNA by differentially tailored polyethylenimine. J Control Release. 2009;133(3):206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Blessing T, Remy JS, Behr JP. Monomolecular Collapse of Plasmid DNA into Stable Virus-like Particles. Proc Natl Acad Sci, USA. 1998;95:1427–1431. doi: 10.1073/pnas.95.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oupicky D, Carlisle RC, Seymour LW. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001;8(9):713–724. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 13.Dharap SS, Wang Y, Chandna P, Khandare JJ, Qiu B, Gunaseelan S, Sinko PJ, Stein S, Farmanfarmaian A, Minko T. Tumor-specific targeting of an anticancer drug delivery system by LHRH peptide. Proc Natl Acad Sci U S A. 2005;102(36):12962–12967. doi: 10.1073/pnas.0504274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooya T, Lee J, Park K. Hydrotropic dendrimers of generations 4 and 5: synthesis, characterization, and hydrotropic solubilization of paclitaxel. Bioconjug Chem. 2004;15(6):1221–1229. doi: 10.1021/bc049814l. [DOI] [PubMed] [Google Scholar]
- 15.Trubetskoy VS, Loomis A, Slattum PM, Hagstrom JE, Budker VG, Wolff JA. Caged DNA does not aggregate in high ionic strength solutions. Bioconjug Chem. 1999;10(4):624–628. doi: 10.1021/bc9801530. [DOI] [PubMed] [Google Scholar]
- 16.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, Kataoka K. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J Am Chem Soc. 2004;126(8):2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 17.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43(42):13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 18.Hagerman PJ. Investigation of the flexibility of DNA using transient electric birefringence. Biopolymers. 1981;20(7):1503–1535. doi: 10.1002/bip.1981.360200710. [DOI] [PubMed] [Google Scholar]
- 19.Shah SA, Brunger AT. The 1.8 A crystal structure of a statically disordered 17 base-pair RNA duplex: principles of RNA crystal packing and its effect on nucleic acid structure. J Mol Biol. 1999;285(4):1577–1588. doi: 10.1006/jmbi.1998.2385. [DOI] [PubMed] [Google Scholar]
- 20.Snyder SL, Sobocinski PZ. An improved 2,4,6-trinitrobenzenesulfonic acid method for the determination of amines. Anal Biochem. 1975;64(1):284–288. doi: 10.1016/0003-2697(75)90431-5. [DOI] [PubMed] [Google Scholar]
- 21.Pakunlu RI, Wang Y, Tsao W, Pozharov V, Cook TJ, Minko T. Enhancement of the efficacy of chemotherapy for lung cancer by simultaneous suppression of multidrug resistance and antiapoptotic cellular defense: novel multicomponent delivery system. Cancer Res. 2004;64(17):6214–6224. doi: 10.1158/0008-5472.CAN-04-0001. [DOI] [PubMed] [Google Scholar]
- 22.Jayant S, Khandare JJ, Wang Y, Singh AP, Vorsa N, Minko T. Targeted sialic acid-doxorubicin prodrugs for intracellular delivery and cancer treatment. Pharm Res. 2007;24(11):2120–2130. doi: 10.1007/s11095-007-9406-1. [DOI] [PubMed] [Google Scholar]
- 23.Khandare JJ, Chandna P, Wang Y, Pozharov VP, Minko T. Novel polymeric prodrug with multivalent components for cancer therapy. J Pharmacol Exp Ther. 2006;317(3):929–937. doi: 10.1124/jpet.105.098855. [DOI] [PubMed] [Google Scholar]
- 24.Saad M, Garbuzenko OB, Ber E, Chandna P, Khandare JJ, Pozharov VP, Minko T. Receptor targeted polymers, dendrimers, liposomes: Which nanocarrier is the most efficient for tumor-specific treatment and imaging? J Control Release. 2008;130(2):107–114. doi: 10.1016/j.jconrel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen AM, Santhakumaran LM, Nair SK, Amenta PS, Thomas T, He H, Thomas TJ. Oligonucleotide nanostructure formation in the presence of polypropyleneimine dendrimers and their uptake in breast cancer cells. Nanotechnology. 2006;17:5449–5460. [Google Scholar]
- 26.Oh JK, Tang C, Gao H, Tsarevsky NV, Matyjaszewski K. Inverse miniemulsion ATRP: a new method for synthesis and functionalization of well-defined water-soluble/cross-linked polymeric particles. J Am Chem Soc. 2006;128(16):5578–5584. doi: 10.1021/ja060586a. [DOI] [PubMed] [Google Scholar]
- 27.Dash PR, Read ML, Barrett LB, Wolfert MA, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999;6(4):643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- 28.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 29.van Steenis JH, van Maarseveen EM, Verbaan FJ, Verrijk R, Crommelin DJ, Storm G, Hennink WE. Preparation and characterization of folate-targeted pEG-coated pDMAEMA-based polyplexes. J Control Release. 2003;87(1–3):167–176. doi: 10.1016/s0168-3659(02)00361-9. [DOI] [PubMed] [Google Scholar]
- 30.Gosselin MA, Guo W, Lee RJ. Incorporation of reversibly cross-linked polyplexes into LPDII vectors for gene delivery. Bioconjug Chem. 2002;13(5):1044–1053. doi: 10.1021/bc025512c. [DOI] [PubMed] [Google Scholar]
- 31.Kleemann E, Neu M, Jekel N, Fink L, Schmehl T, Gessler T, Seeger W, Kissel T. Nano-carriers for DNA delivery to the lung based upon a TAT-derived peptide covalently coupled to PEG-PEI. J Control Release. 2005;109(1–3):299–316. doi: 10.1016/j.jconrel.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 32.Kim TI, Baek JU, Zhe Bai C, Park JS. Arginine-conjugated polypropylenimine dendrimer as a non-toxic and efficient gene delivery carrier. Biomaterials. 2007;28(11):2061–2067. doi: 10.1016/j.biomaterials.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release. 2006;116(2):123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release. 2006;115(2):216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Chandna P, Saad M, Wang Y, Ber E, Khandare J, Vetcher AA, Soldatenkov VA, Minko T. Targeted proapoptotic anticancer drug delivery system. Mol Pharm. 2007;4(5):668–678. doi: 10.1021/mp070053o. [DOI] [PubMed] [Google Scholar]
- 36.Dharap SS, Minko T. Targeted proapoptotic LHRH-BH3 peptide. Pharm Res. 2003;20(6):889–896. doi: 10.1023/a:1023839319950. [DOI] [PubMed] [Google Scholar]
- 37.Dharap SS, Qiu B, Williams GC, Sinko P, Stein S, Minko T. Molecular targeting of drug delivery systems to ovarian cancer by BH3 and LHRH peptides. J Control Release. 2003;91(1–2):61–73. doi: 10.1016/s0168-3659(03)00209-8. [DOI] [PubMed] [Google Scholar]
- 38.Kam NW, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J Am Chem Soc. 2005;127(36):12492–12493. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 39.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem Biol. 2004;11(8):1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Patil ML, Zhang M, Taratula O, Garbuzenko OB, He H, Minko T. Internally cationic polyamidoamine PAMAM-OH dendrimers for siRNA delivery: effect of the degree of quaternization and cancer targeting. Biomacromolecules. 2009;10(2):258–266. doi: 10.1021/bm8009973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.