Summary
Many disease genes encode proteins that are difficult to target directly using small molecule drugs. Improvements in libraries based on synthetic compounds, natural product and other types of molecules may ultimately allow some challenging proteins to be successfully targeted; however, these developments alone are unlikely to be sufficient. A complementary strategy exploits the functional interconnectivity of intracellular networks to find druggable targets, lying upstream, downstream or in parallel to a disease-causing gene, where modulation can influence the disease process indirectly. These targets can be selected using prior knowledge of disease-associated pathways or identified using phenotypic chemical and genetic screens in model organisms and cells. These approaches should facilitate the development of effective drugs for many genetic disorders.
Keywords: druggable genome, chemical genetics, small molecule, RNAi, synthetic lethal
A. Disease and the Druggable Genome
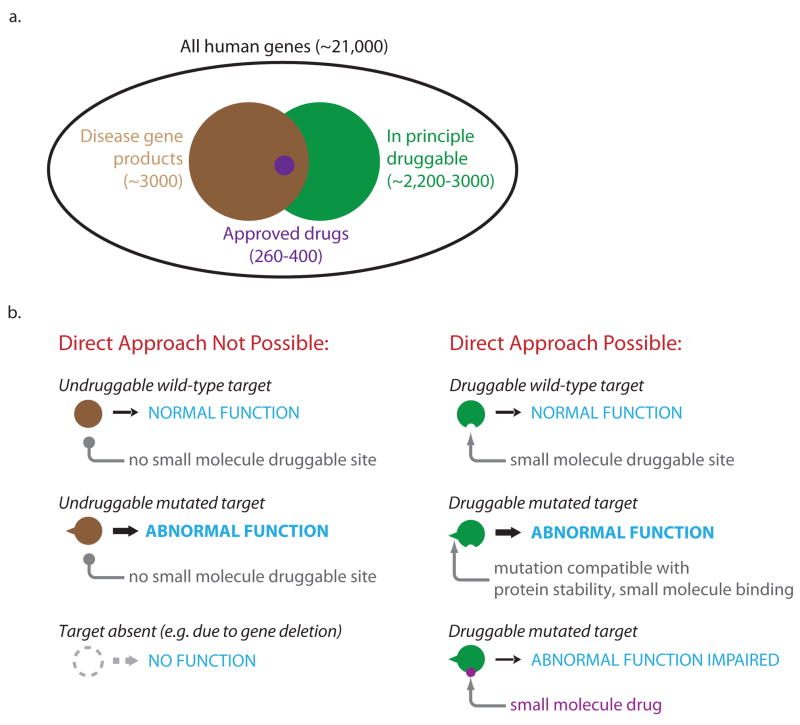
The development of drugs to combat human genetic disorders, including cancer and neurodegenerative disease, is a high priority. In recent years, new DNA sequencing and genotyping technologies have enabled the discovery of a slew of novel disease-causing mutations and disease-associated DNA sequence variants [1–5]. Transforming this knowledge into a set of validated drug targets poses a significant challenge. It is sobering to consider that, to date, only ~2% of all predicted human gene products (260–400) have been successfully targeted with small molecule drugs [6,7] (Figure 1a). In part, this may reflect the fact that only 10–15% of all human genes (2,200–3,000) are thought to be in principle ‘druggable’ (e.g. encode proteins similar in sequence to those that have already been targeted with small molecules) [8,9], and that the overlap between druggable genes and known disease genes is only on the order of 25% [10,11] (Figure 1a). Adding to the challenge, mutated human disease genes can give rise to protein targets that differ only subtly in structure or abundance compared to their wild-type counterparts, or eliminate the production of a specific gene product altogether (e.g. due to mRNA destabilization or gene deletion) (Figure 1b). These considerations suggest that few gene products, disease-associated or not, are likely to represent direct small molecule drug targets. How then can we find targets that are disease-specific, druggable, and that can be modulated with small-molecule-based drugs or other reagents to bring about a desired therapeutic effect. Here we review several strategies used to discover useful molecular targets for human genetic disorders.
Figure 1.
The druggable genome in relation to disease. a. Venn diagram illustrating the relationship between all potential human proteins, those proteins that are in principle druggable (green), those proteins encoded by disease genes (brown), and those proteins targeted by approved therapeutics (purple). The size of the ovals approximates the number of gene products in each category. While not considered here, it should be noted that one gene may give rise to multiple gene products through alternate splicing. b. Cartoon depicting disease gene products that are in principle undruggable, either because a suitable drug-binding fold is not present or because the disease-causing mutation eliminates protein production, and gene products that are druggable (e.g. accessible to a small molecule modulator). Small molecule modulators of druggable targets could in principle act to either impair the abnormal function of a target resulting from a gain of function mutation, or restore the impaired function of a target resulting from a partial loss of function mutation (not shown). Part (a) is in part adapted from Reference 11.
B. Direct Targeting of Disease Gene Products
Conceptually, the simplest approach to treating a genetic disorder is to modulate the function of a disease-causing gene product directly (Figure 1b), as illustrated by the use of the small molecule imatinib (Gleevec) to inhibit the constitutively active kinase produced by the BCR-ABL1 fusion gene found in patients with chronic myeloid leukemia [12]. The number of disease-associated gene products considered druggable is small (see above) but continues to slowly expand. For example, in early surveys, the disease-associated E3 ubiquitin ligase Mdm2, which is amplified in many cancers, was thought to be undruggable [8]. It has since been shown that the crucial Mdm2-p53 binding interface can be disrupted by the nutlin family of small molecule inhibitors, leading to stabilization of p53 and cancer cell death [13] (Figure 2a). These results suggest that extensive searching of existing chemotypes may yield direct modulators of additional disease gene products.
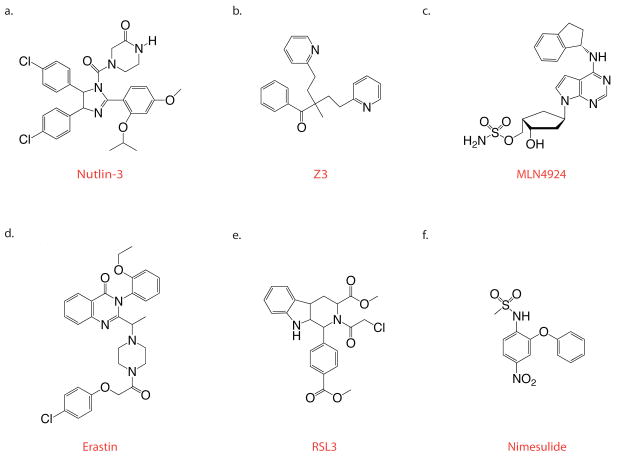
Figure 2.
Chemical structures of small molecule compounds discussed in the text. a. Nutlin-3 directly inhibits the binding of p53 to Mdm-2. b. Z3 is a Janus kinase (Jak) inhibitor discovered through in silico screening. c. MLN4924 is a novel anti-cancer agent that selectively inhibits the NEDD8 activating enzyme (NAE). d. Erastin is an oncogenic Ras-selective lethal compound discovered through unbiased phenotypic screening. Erastin kills tumor cells through binding to the mitochondrial voltage-dependent anion channels 2 and 3. e. RSL3 is functionally similar to erastin but structurally distinct. The RSL3 target is unknown. f. Nimesulide is active in a zebrafish model of AML, counteracting the effects of the AML1-ETO oncogene on granulocytic blast cell differentiation.
Both re-screening of existing chemotypes, and de novo computationally-assisted drug design [14], will be facilitated by new models of protein structures and protein-interaction interfaces. For example, starting with an existing model of the Jak3 tyrosine kinase domain, Sayyah et al first used orthology modeling to develop a model of the Jak2 kinase domain, which is mutated in several cancers [15]. This model was then used to screen 20,000 known compounds in silico for those likely to bind adjacent to the ATP binding site and inhibit kinase activity [15]. This screen resulted in the identification of six candidate compounds, one of which, Z3, was subsequently shown to specifically inhibit Jak2 function in several cell culture and disease models [15] (Figure 2b). The application of similar approaches to other targets may greatly decrease the amount of actual screening that needs to be performed in the future, and help characterize novel direct modulators of disease gene product function.
C. Exploiting the Functional Interconnectivity of Biological Systems to Find Alternate Druggable Targets
It is not always possible to target a disease gene product itself directly. However, normal and disease genes do not function in isolation: genes, gene products and metabolites interact with one another to form functionally interconnected genetic, protein and metabolic interaction networks of exquisite complexity [16–19]. Genetic diseases perturbing one or more genes alter the connectivity of these networks, as reflected in disease-specific patterns of gene expression, protein-protein interactions and metabolite production [20–22]. Changes in network connectivity induced by disease gene activity (or lack thereof) may expose unique genetic or chemical sensitivities due to a loss of biological redundancy, feedback regulation and/or the up- or down-regulation of alternate, druggable target genes [23–26]. If suitable drugs are available to modulate these indirect targets, it becomes possible to exploit acquired chemical sensitivities to achieve a desired phenotypic outcome, such as cancer-cell selective cell death [27–29]. Indirect targets can be identified using a number of approaches that embrace the functional connectivity of cellular networks (see below).
Target selection guided by existing knowledge of disease-associated pathways
Tractable indirect targets can be selected using knowledge of disease-associated biological processes. A recent example involves the link between cancer, protein degradation and the cell cycle. Cullin-RING type E3 ubiquitin ligases regulate the degradation of numerous cell cycle proteins. To function properly, these enzymes require post-translational modifications by the ubiquitin-like protein NEDD8. It was hypothesized that inhibiting the NEDD8 modification of Cullin-RING E3 ubiquitin ligases would specifically disrupt the proteolytic turnover of cell cycle proteins, thereby inhibiting the growth of tumor cells [30]. High-throughput small molecule screening was used to identify compounds specifically inhibiting the NEDD8-activating enzyme (NAE), which is required to conjugate NEDD8 to target proteins [30]. One small molecule, MLN4924, was identified as a potent inhibitor of NAE and shown to be effective in preventing the growth of tumor cell lines and human tumor xenografts in mice [30] (Figure 2c). This study illustrates the power of using existing knowledge to guide the selection of novel druggable targets.
Target selection guided by systematic genetic and biochemical screens
The selection of candidate indirect targets need not be guided solely by educated guesswork; systematic high-throughput genetic and biochemical methods can be used to pinpoint targets functionally connected with disease genes. Follow-up studies can then assess the druggability of candidate ‘hits’.
For example, genome-wide RNAi libraries, based on endoribonuclease-prepared or chemically synthesized small interfering RNAs (esiRNAs or siRNAs), or virally-encoded short hairpin RNAs (shRNAs) [31–34], can be used in screens to identify synthetic lethal genetic interactions with cancer-specific mutations. This involves searching for gene products that, when depleted, cause lethality only in the presence of a second genetic alteration; this demonstrates a degree of functional connectivity between the depleted mRNA and the genetic change. One screen examined multiple shRNAs targeting 1,006 human genes, including 571 kinases, to identify genes that were synthetic lethal with oncogenic Ras alleles in two different cell lines [35]. This screen identified a single gene, CSNK1E, that when disrupted resulted in apoptosis in cancer cells, but not in matched non-tumorigenic cells [35]. Subsequently, it was demonstrated that inhibition of the protein product of this gene, casein kinase 1 epsilon, by the small molecule inhibitor IC261, recapitulated the effects of shRNA-mediated knockdown, suggesting that inhibition of this kinase may be an effective method to kill some tumor cells [35]. More recently, larger screens examining more cell lines [36] or a larger number of shRNAs [37] have been undertaken to find KRAS-specific synthetic lethal interactions. This work identified STK33, encoding a serine/threonine kinase, and PLK1, encoding a Polo-like kinase, as oncogenic KRAS synthetic lethal interactors in several different cancers. Notably, both genes are good candidate drug targets. One interesting application of this general approach is to tailor drug treatment to individual cancer patients based on cellular profiles of RNAi sensitivity [38].
In addition to genetic approaches, target selection can be guided by high-throughput biochemical profiling. Recently, the activation status of 46 tyrosine kinases was profiled in a large panel of cancer cell lines using a high-throughput, bead-based immunosandwich assay [39]. This screen determined that the non-receptor tyrosine kinase Src is activated in a large proportion of human tumor cell lines [39]. This was unexpected, because there is little evidence for mutation or amplification of the SRC gene in human cancers [39]. It was subsequently shown that the previously developed small molecule tyrosine kinase inhibitor Dasatanib could be used to inhibit Src activity and that this treatment killed glioblastoma tumors, where Src was active [39]. These results demonstrate the power of systematic functional studies to identify druggable, indirect targets. Crucially, functional screens allow for the identification of targets that would not be predicted from DNA sequencing alone.
Target Identification Using Unbiased Phenotypic Screens
Complementary to the above approaches are those involving phenotype-based chemical screening. In this approach, a screen is conducted to identify a small molecule or other chemical perturbagen that yields a desired phenotypic outcome, such as cell death or an alternative cell fate. Importantly, small molecules can induce both loss- and gain-of-function phenotypes in target proteins, potentially broadening the number of observable phenotypes compared to perturbagens such as RNAi, which can only induce loss-of-function effects in targets. Follow-up studies can then determine the relevant protein target and mechanism of action. Overall, the most compelling advantage of this approach is that it can potentially identify useful drug leads directly.
Cell death
It is possible to identify small molecules that are synthetically lethal in cancer cells in the context of specific gene mutations. For example, screening of ~ 70,000 compounds using lung, bone and engineered cancer cells harboring oncogenic gain-of- function KRAS, NRAS and HRAS alleles, respectively, identified three compounds exhibiting tumor-cell selective synthetic lethality: erastin, RSL3 and RSL5 [40–42] (Figure 2d,e). The mode of cell death induced by these compounds is a novel form of oxidative death distinct from necrosis or apoptosis [40,42]. The cellular targets of two of these compounds (erastin and RSL5) were identified as mitochondrial Voltage Dependent Anion Channels 2 and 3 (VDAC2/3), which are upregulated by oncogenic Ras [40]. How erastin modulation of VDAC2 and VDAC3 brings about tumor-cell-specific cell death is under investigation, but appears to involve iron and mitochondrial respiratory activity, as well as VDACs [42]. VDAC2 and 3 represent examples of targets that would not have been selected as relevant to cancer a priori, because they themselves are not mutated in disease. Similar phenotype-driven approaches have recently been used to identify compounds synthetically lethal in VHL null and KRAS mutant cells [43,44], suggesting that this approach will be useful for a broad range of mutations.
Chemical Suppression
Cell death is only one potentially useful phenotype. In other cases, it is sufficient or preferable to channel the cell to an alternate cell fate. One recent example involves synthetic interactions in a model of acute myelogenous leukemia (AML). In one form of AML, caused by the AML1-ETO oncogene, the differentiation of granulocytic blast cells is slowed, resulting in their overproduction, which in turn may serve as a source of stem cells contributing to cancer [45]. Recently, a screen of 2,000 bioactive small molecules conducted using a transgenic zebrafish model of AML1-ETO-induced AML determined that nimesulide, an inhibitor of the prostaglandin-endoperoxide synthase (PTGS) family of enzymes, suppressed AML1-ETO-oncogene-mediated cell fate transformation by preventing prostaglandin E2 synthesis [46] (Figure 2f). It was subsequently shown that a related compound could inhibit cell fate transformation in human cells expressing this oncogene [46]. In both zebrafish and human cells, PTGS enzymes are upregulated at the transcriptional level, possibly accounting for why they become useful targets [46]. This screen is also notable for the use of a live animal model, which allows for the effects of drugs on whole-animal development and physiology to be assessed. Chemical suppressor screens have also been used with cultured cells to identify small molecules that suppress cell death in models of neurodegenerative disorders, such as Huntington’s disease and Parkinson’s Disease [47–50], suggesting that phenotypic suppression may be a useful general approach to discovering small molecules with therapeutic potential.
D. Conclusions & Future Directions
Finding new drugs to treat genetic disorders presents a grand challenge for 21st century medicine. No one existing approach is likely to be suitable in all cases and each has distinct disadvantages. It is conceivable that many disease genes will continue to elude efforts at direct targeting with existing chemical libraries. Functional-connectivity-based approaches to target identification, even though guided by high-throughput genetic and biochemical screening technologies, may simply suggest undruggable proteins; thus improved chemical libraries will always be useful. The effectiveness of small molecules discovered through phenotype-based approaches will only be as good as the cellular models used to discover them, and drug-target identification remains a technical hurdle.
These concerns are being addressed in a number of ways. For example, new compound collections can provide access to previously uncharted regions of chemical space, potentially opening up the possibility of finding new direct or indirect modifiers for a given gene product or disease model. These libraries may be based on proven ligand scaffolds, or structures found in natural products or generated through novel small molecule synthetic routes [12,51–54] (Figure 3). Fragment-based screening [55] and de novo computational design [56] may also be helpful in broadening the number of targets that can be modulated (Figure 3). Non-small molecule-based reagents, such as stabilized, stapled peptide helices [57,58], cyclic peptides, peptide-like molecules, DNA and RNA aptamers [59] or even siRNA molecules, may also prove useful in some circumstances to inactivate the function of specific gene products (Figure 3). From a screening perspective, the development of new functional RNAi screening approaches, such as genome-wide pooled screens [36,60] and in vivo screens [61], is opening the door to comprehensive annotation of the role of most gene products and broadening the potential for finding useful candidate druggable genes. New proteomic methods to identify the targets of small molecule ligands should speed the process of characterizing the function of small molecules discovered in unbiased phenotypic screens [62]. Together, these improvements in small molecule library design, high-throughput target identification and functional connectivity screening should improve our ability to take full advantage of the druggable genome to treat disease.
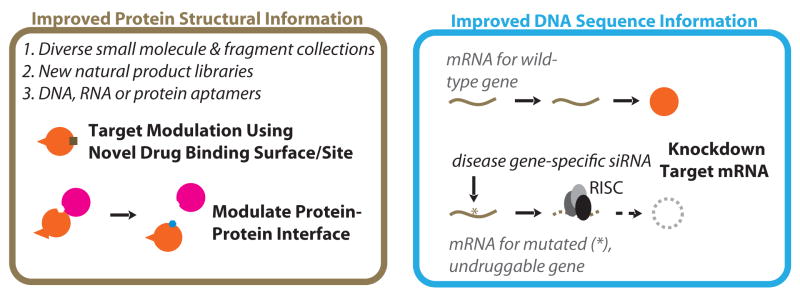
Figure 3.
Approaches to increase the size of the druggable genome, thereby facilitating both direct and indirect disease gene targeting. New chemical libraries, based on small molecule, fragment, or aptamer approaches may allow new protein folds or interaction interfaces to be targeted. In cases where these approaches fail, purely genetic approaches, involving the delivery of siRNAs directly to cells to silence mRNA expression may prove availing. RISC = RNAi-induced silencing complex, which binds and cleaves siRNA-mRNA double-stranded RNA duplexes.
Acknowledgments
The authors thank Andras Bauer and David Clarke for comments on the manuscript. S.J.D. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research and by a grant to B.R.S. (R01CA097061). B.R.S. is funded by NIH (R01GM085081, R01CA097061), NYSTAR and the Arnold and Mabel Beckman Foundation.
Footnotes
Conflict of Interest Statement
The authors are not aware of any conflicts of interest that would materially impact the content of this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. The first whole genome re-sequencing project to examine both cancer cells and control fibroblasts from the same patient, allowing for the unambiguous identification of several novel, somatically-acquired, cancer-specific mutations. Cancer genome sequencing is poised to come into widespread use and is likely to flood biologists with information on thousands of new, cancer-associated mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 6.Landry Y, Gies JP. Drugs and their molecular targets: an updated overview. Fundam Clin Pharmacol. 2008;22:1–18. doi: 10.1111/j.1472-8206.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- 7.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 9.Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 10.Billingsley ML. Druggable targets and targeted drugs: enhancing the development of new therapeutics. Pharmacology. 2008;82:239–244. doi: 10.1159/000157624. [DOI] [PubMed] [Google Scholar]
- 11.Sakharkar MK, Sakharkar KR, Pervaiz S. Druggability of human disease genes. Int J Biochem Cell Biol. 2007;39:1156–1164. doi: 10.1016/j.biocel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 14.van Montfort RL, Workman P. Structure-based design of molecular cancer therapeutics. Trends Biotechnol. 2009;27:315–328. doi: 10.1016/j.tibtech.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Sayyah J, Magis A, Ostrov DA, Allan RW, Braylan RC, Sayeski PP. Z3, a novel Jak2 tyrosine kinase small-molecule inhibitor that suppresses Jak2-mediated pathologic cell growth. Mol Cancer Ther. 2008;7:2308–2318. doi: 10.1158/1535-7163.MCT-08-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 17.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 18.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 19.Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 20.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 21.McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Lehar J, Krueger A, Zimmermann G, Borisy A. High-order combination effects and biological robustness. Mol Syst Biol. 2008;4:215. doi: 10.1038/msb.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. Resolves the apparent paradox of gene redundancy in the yeast S. cerevisiae by showing that the majority of yeast gene (97%) are essential for viability under at least one of 400 different tested environmental or small molecule growth conditions. These results help illuminate the limits of biological robustness in eukaryotic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 27.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 28.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 29.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. An important new indirect drug target in cancer is discovered using insightful biological sleuthing and in-depth small molecule screening. [DOI] [PubMed] [Google Scholar]
- 31.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 32.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 33.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 34.Zheng L, Liu J, Batalov S, Zhou D, Orth A, Ding S, Schultz PG. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9:R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyner JW, Deininger MW, Loriaux MM, Chang BH, Gotlib JR, Willis SG, Erickson H, Kovacsovics T, O’Hare T, Heinrich MC, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc Natl Acad Sci U S A. 2009;106:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 42.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji Z, Mei FC, Lory PL, Gilbertson SR, Chen Y, Cheng X. Chemical genetic screening of KRAS-based synthetic lethal inhibitors for pancreatic cancer. Front Biosci. 2009;14:2904–2910. doi: 10.2741/3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nimer SD, Moore MA. Effects of the leukemia-associated AML1-ETO protein on hematopoietic stem and progenitor cells. Oncogene. 2004;23:4249–4254. doi: 10.1038/sj.onc.1207673. [DOI] [PubMed] [Google Scholar]
- 46.Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5:236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varma H, Voisine C, DeMarco CT, Cattaneo E, Lo DC, Hart AC, Stockwell BR. Selective inhibitors of death in mutant huntingtin cells. Nat Chem Biol. 2007;3:99–100. doi: 10.1038/nchembio852. [DOI] [PubMed] [Google Scholar]
- 48.Varma H, Cheng R, Voisine C, Hart AC, Stockwell BR. Inhibitors of metabolism rescue cell death in Huntington’s disease models. Proc Natl Acad Sci U S A. 2007;104:14525–14530. doi: 10.1073/pnas.0704482104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci U S A. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 51.Shang S, Tan DS. Advancing chemistry and biology through diversity-oriented synthesis of natural product-like libraries. Curr Opin Chem Biol. 2005;9:248–258. doi: 10.1016/j.cbpa.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen TE, Schreiber SL. Towards the optimal screening collection: a synthesis strategy. Angew Chem Int Ed Engl. 2008;47:48–56. doi: 10.1002/anie.200703073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tse BN, Snyder TM, Shen Y, Liu DR. Translation of DNA into a library of 13,000 synthetic small-molecule macrocycles suitable for in vitro selection. J Am Chem Soc. 2008;130:15611–15626. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129:13137–13143. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hesterkamp T, Whittaker M. Fragment-based activity space: smaller is better. Curr Opin Chem Biol. 2008;12:260–268. doi: 10.1016/j.cbpa.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Evensen E, Joseph-McCarthy D, Weiss GA, Schreiber SL, Karplus M. Ligand design by a combinatorial approach based on modeling and experiment: application to HLA-DR4. J Comput Aided Mol Des. 2007;21:395–418. doi: 10.1007/s10822-007-9119-x. [DOI] [PubMed] [Google Scholar]
- 57.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 60.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]