Abstract
The Dis3 ribonuclease is a member of the hydrolytic RNR protein family. Although much progress has been made in understanding the structure, function, and enzymatic activities of prokaryotic RNR family members RNase II and RNase R, there are no activity studies of the metazoan ortholog, Dis3. Here, we characterize the activity of the Drosophila melanogaster Dis3 (dDis3) protein. We find that dDis3 is active in the presence of various monovalent and divalent cations, and requires divalent cations for activity. dDis3 hydrolyzes compositionally distinct RNA substrates, yet releases different products depending upon the substrate. Additionally, dDis3 remains active when lacking N-terminal domains, suggesting that an independent active site resides in the C-terminus of the protein. Finally, a study of dDis3 interactions with dRrp6 and core exosome subunits in extracts revealed sensitivity to higher divalent cation concentrations and detergent, suggesting the presence of both ionic and hydrophobic interactions in dDis3-exosome complexes. Our study thus broadens our mechanistic understanding of the general ribonuclease activity of Dis3 and RNR family members.
Keywords: Dis3/Rrp44, RNase R/II, exosome, PIN domain, exoribonuclease
Introduction
Dis3 is an evolutionarily conserved and essential enzyme [1; 2], most known for its association with the RNA surveillance complex, the exosome [3]. Although Dis3 interacts with exosome proteins, the enzyme is functionally distinct from the complex. Dis3 is a homolog of the hydrolytic bacterial RNases, RNase II and RNase R [4]. These enzymes share a conserved set of amino acid motifs that fold into a pocket housing the 3’ to 5’ exoribonuclease activity of the protein, the RNB domain [4]. The budding yeast Saccharomyces cerevisiae Dis3 (ScDis3) enzyme, from which all current biochemical knowledge of Dis3 has emerged, requires both monovalent and divalent ions for RNB-mediated catalysis [5]. In vitro, no other co-factors are necessary for activity [5].
Unlike their prokaryotic counterparts, eukaryotic Dis3 family members contain an N-terminal extension that harbors an additional domain with endoribonuclease activity, the PIN domain [6]. Requirements for PIN-mediated activity are not well established. Work related to the Dis3 PIN and RNB active sites has emerged from six studies of the yeast homolog [5; 7; 8; 9; 10; 11]. Recognition of two enzymatically active domains in Dis3 has made characterizing the ribonuclease activity of the full-length protein a challenge. For example, in vitro, either the RNB alone or the PIN alone is sufficient for activity, and individual RNB and PIN mutations only abrogate their respective activities. This suggests a functional separation of active sites, where one activity does not necessarily rely on the other. However, point mutations in either the ScDis3 RNB or PIN domain result in rRNA processing defects in vivo, suggesting that for some RNA metabolic events, the domains may work together. Presently, the interplay of these domains in the context of full-length Dis3 is poorly understood. In this regard, Dis3 could act on RNA substrates as an exoribonuclease, endoribonuclease, or both.
In addition to the PIN domain, the Dis3 N-terminus (~300 amino acids long) includes several domains that may contribute to the ribonuclease activities of the protein. All Dis3 homologs contain a C3 motif, or a putative iron-sulfur binding domain, and two OB folds, or oligonucleotide-binding folds [8; 10]. These domains have been shown to be important for cell growth in yeast [8] as well as core exosome binding [9; 12], and nuclear targeting [12]. A domain with homology to the mitotic cohesin STAG resides within the Drosophila Dis3 N-terminus, but its function is unclear [12]. A fundamental question is to determine how or whether these domains contribute to Dis3 ribonuclease activities.
Another challenge to understanding Dis3 ribonuclease activities is identification of their substrate specificities and reaction products. The full-length ScDis3 cleaves RNAs regardless of sequence or structure, indicating that the protein is a non-specific ribonuclease [3; 5; 7; 8; 9; 10; 11; 13; 14]. The products of these reactions vary depending on substrate, but typically range in length from 2-5 nucleotides, resulting from either endo- or exoribonuclease activity. ScDis3 activity also releases 5’NMP products, a characteristic of exoribonuclease activity, and RNB point mutations eliminate this ability [5].
Whether metazoan Dis3 homologs have similar activities, ionic requirements, substrate specificity, and reaction products has not been explored. To address this, we characterize Drosophila melanogaster Dis3 (dDis3). Specifically, we assess requirements for in vitro ribonuclease activity, the ability of the enzyme to degrade different RNA substrates, the contributions of distinct domains to activity, and the strength of the dDis3-core exosome interaction. As this study represents a first analysis of a metazoan Dis3, our findings help build a more complete picture of the general features of Dis3 ribonuclease activities.
Materials and Methods
Purification of recombinant proteins
Full-length MBP-dDis3, N-terminally truncated MBP-dDis3, or MBP alone was over-expressed in E. coli strain DH5α overnight at 20°C by the addition of 1 mM IPTG (Denville Scientific) to 500 mL cultures. Cells were lysed by treatment with 1 mg/mL lysozyme (Sigma), and by sonication in buffer (20 mM tris, pH 7.5, 100 mM NaCl) also containing PMSF (Sigma) and EDTA-free protease inhibitor cocktail (Roche). Lysates were loaded onto 1 mL amylose resin (New England BioLabs), and washed with 80 mL of buffer (20 mM tris, pH 7.5, 100 mM NaCl, 0.1 mM PMSF). Proteins were eluted in buffer containing 20 mM tris, pH 7.5, 100 mM NaCl, and 50 mM maltose. Additionally, proteins were dialyzed into buffer containing 20 mM tris, pH 7.5, 100 mM NaCl, and 10% glycerol. Dialyzed proteins were visualized by 12% SDS-PAGE and Coomassie staining. Protein concentrations were determined from Coomassie stained gels by comparison to a BSA standard curve using Quantity One Software.
Preparation of RNA substrates
RNA substrates polyU (32 nucleotides), polyA (30 nucleotides, Dharmacon), polyC (30 nucleotides, Dharmacon), and an RNA of random sequence (31 nucleotides; 3’-AAAACAAAACAAAAUUCGUGGCAUUUCUGCG-5’) were used in ribonuclease activity assays. The substrates were radiolabeled at the 5’ end with γ32P using a T4 polynucleotide kinase (Promega). Radiolabeled nucleotides that were not incorporated were removed using NucAway™ spin columns (Ambion) as per manufacturer's recommendations.
Ribonuclease activity assays
Assays were adapted from those used to analyze ScDis3 in vitro activity [5]. Radiolabeled RNA was incubated alone (lanes labeled “buffer”) or with protein at 37°C in reaction buffer containing 10 mM tris, pH 8.0, 75 mM KCl, and 40 μM MgCl2, unless indicated otherwise. In all reactions, the concentration of RNA was 120 nM, and polyU RNA was used unless indicated otherwise. With the exception of the experiment in Figure 2, protein concentration was 60 nM. In Figure 2, the protein concentrations are as follows: MBP is 60 nM, MBP-dDis31-982 is 60 nM, MBP-dDis3189-982 is 60 nM, MBP-dDis329-982 is 42 nM, and MBP-dDis362-982 is 43 nM. At the time points indicated, 10 μL of the reactions were taken out and stopped with the addition of one reaction volume of formamide loading buffer. Products of the reactions were separated on 12.5% acrylamide, 8M urea denaturing gels or TLC plates that were developed in KH2PO4 buffer, and visualized using autoradiography.
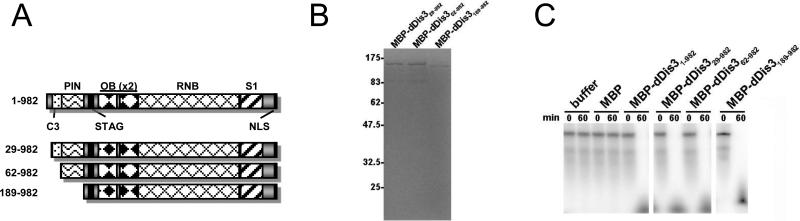
Figure 2.
N-terminally truncated MBP-dDis3 mutants retain ribonuclease activity. (A) Schematic of full-length dDis3 and N-terminal truncation mutants. (B) Purified recombinant MBP-dDis3 mutant proteins. (C) Ribonuclease activity of truncated dDis3 mutants. Enzymatic activity was assessed on a polyU RNA substrate.
Dis3 immunoprecipitation
S2 stable cell lines establishment and maintenance, whole cell extract preparation, and FLAG immunoprecipitation were performed as described previously [12] with the following changes. For experiments in Figure 3A, wash buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 3 mM MgCl2, 0.5 mM EDTA, pH 8.0, 0.5 mM DTT, 1% Triton X (TX)-100, 10% glycerol, and protease inhibitor cocktail (Invitrogen)) was supplemented with NaCl to 1.0 or 1.25 M in the presence or absence of 5% TX-100 or 0.1 M MgCl2. For divalent cation titration experiments, [NaCl] was raised to 0.2 or 1.0 M and MgCl2 was added in at 0.01, 0.2, 0.4, 0.6, 0.8. 1.0. or 1.2 M. Western blotting was performed with appropriate antibodies as described [12; 15].
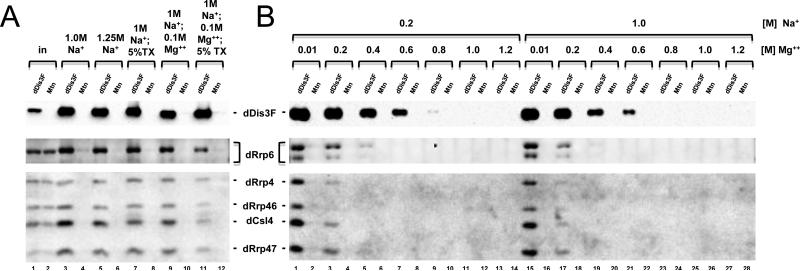
Figure 3.
dDis3-dRrp6 and dDis3-core exosome interactions are stable in vitro. (A) Analysis of dDis3-FLAG interactions with dRrp6 and core exosome subunits in buffer containing high sodium chloride concentrations. (B) dDis3-dRrp6 and dDis3-core exosome interactions in increasing divalent cation concentrations. Note that recovery of dDis3-FLAG itself by the FLAG resin is impeded at MgCl2 concentrations higher than 0.6 M.
Results
Requirements for dDis3 ribonuclease activity in vitro
Although the ribonuclease activity of ScDis3 has been characterized, there is no evidence a metazoan Dis3 homolog degrades RNA in a similar manner. To address this issue, we established a system to assess the activity of Drosophila melanogaster Dis3. Our results show that Drosophila Dis3 (dDis3) is active in vitro on a simple, single-stranded RNA substrate, like ScDis3 [3; 5; 7; 8; 9; 13]. This activity was observed in various denaturing conditions and in a wide pH range (Table 1; Figure S1).
Table 1.
Summary of dDis3 ribonuclease activity analysis
| Construct |
Substrate |
Buffer (ion content) |
Time (min) |
Result |
|---|---|---|---|---|
| 1-982 | U | none | ≤ 60 | activity impaired |
| 1-982 | U | K+ | ≤ 60 | active |
| 1-982 | U | K+, 2Me | ≤ 60 | active |
| 1-982 | U | Na+ | ≤ 60 | active |
| 1-982 | U | NH4+ | ≤ 60 | active |
| 1-982 | U | Cs+ | ≤ 60 | active |
| 1-982 | U | Li+ | ≤ 60 | active |
| 1-982 | U | Mg2+, K+, ±EDTA | ≤ 180 | activity impaired by EDTA |
| 1-982 | U | Mn2+, K+ | ≤ 60 | active |
| 1-982 | U | Mg2+, K+ | 60 | active |
| 1-982 | U | Mn2+ alone | ≤ 60 | activity impaired |
| 1-982 | U | Mg2+ alone | ≤ 60 | activity impaired |
| 1-982 | U | 2Me titration | 60 | active; slight inhibition at higher 2Me concentrations |
| 1-982 | U | pH titration | 60 | active; slight inhibition at more basic pH values |
Optimal ionic conditions for ribonuclease activity can vary from one enzyme to another, even for bacterial orthologs of Dis3 [16]. We sought to establish robust conditions to assess the activity of recombinant maltose binding protein fusion to full-length dDis3 (MBP-dDis3). These results are summarized in Table 1. We first tested the activity of recombinant dDis3 on a polyU sustrate in buffer containing no added monovalent or divalent cations. As expected, little activity was observed under these conditions during the 60 minute reaction period, indicating that dDis3 requires the presence of cations to function properly (Figure S2, tris). Upon addition of any monovalent or divalent cation tested, MBP-dDis3 activity was enhanced (Figures S2 and S3). As a control, MBP alone had negligible activity in any condition over the time course (Figures S2, S3, S4). Moreover, monovalent and divalent cations together in the reaction buffer prompted the most efficient catalysis, as has been shown for other Dis3 homologs (Figure S3).
It has been reported that Dis3 activity specifically requires divalent cations [5]. To confirm this for dDis3, we added EDTA to the reaction buffer to chelate magnesium. The addition of EDTA inhibited degradation of the RNA (Figure S5). We interpret this to mean that, like ScDis3, the ribonuclease activity of MBP-dDis3 requires a divalent cation in vitro.
Analysis of dDis3 substrate specificity and reaction products
To examine the specificity of MBP-dDis3, we tested its activity on several different single-stranded RNA substrates. As expected for sequence non-specific ribonucleases, MBP-dDis3 degraded polyA, polyC, and an RNA containing all four nucleotides within one hour (Figure 1A, top). Within three hours, the enzyme degraded some of the intermediate products that were present at the one hour incubation point (Figure 1A, bottom). The reaction products released by dDis3 activity are observably different between the time points and among the distinct substrates. dDis3 activity on polyA, polyC, and polyU liberates similar ladders of products that are weighted toward the smallest fragment with decreasing amounts of the larger fragments. dDis3 activity on the random RNA yields more discrete major products that are the highest and lowest bands in the product mixture.
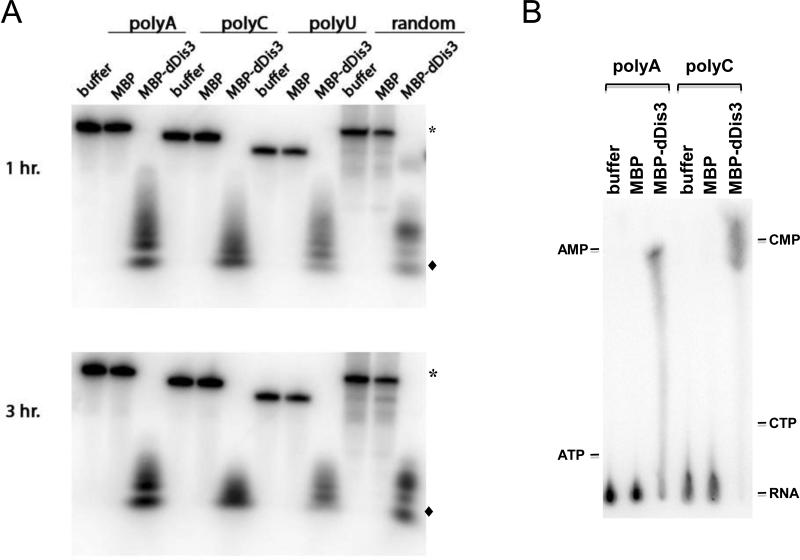
Figure 1.
dDis3 is a sequence non-specific ribonuclease. (A) Ribonuclease activity of dDis3 on multiple RNA substrates. Full-length substrate is indicated by (*) and the smallest degradation product is indicated by (◆); these symbols will be used hereafter. (B) Analysis of products resulting from dDis3 activity. Reaction products that accumulated after 1 hour of incubation were separated on TLC plates. Products were identified by comparison to non-labeled nucleotide standards run on the same TLC plates.
To determine whether the smallest reaction products corresponded to nucleotide monophosphates, we performed thin layer chromatography (TLC) with the reaction mixtures. TLC analysis of the products from the dDis3-polyA or -polyC reactions revealed spots that migrated at the locations of AMP and CMP, respectively (Figure 1B). These data show that dDis3 liberates nucleotide monophosphates during catalysis. This is similar to yeast ScDis3, which has been shown to release NMPs in the presence of the exosome core [5].
Contributions of the dDis3 N-terminus to ribonuclease activity
Our study to this point has analyzed the overall ribonuclease activity of the full-length polypeptide. However, we have not determined where activity is located within Drosophila Dis3, or how domains within the protein contribute to dDis3 activities. To determine the contributions of the N-terminal sequences to ribonuclease activity, we constructed N-terminal truncation mutants that removed the first 28, 61, or 188 amino acids (Figure 2A). These are predicted to remove up to the C3 motif, the C3 motif in its entirety, and the C3 and PIN domains, respectively. Each of these truncated forms was cloned in frame with MBP, and expressed and purified from bacteria to yield a single band at the expected molecular weights (Figure 2B). All three N-terminal truncations behaved similarly to the full-length dDis3, being able to degrade the polyU substrate within 60 minutes (Figure 2C). These data show that the N-terminus, and the PIN domain in particular, is not required to retain functional RNB activity.
Testing the stability of dDis3-dRrp6-core exosome interactions
We have recently shown that Dis3 interacts with Rrp6 and core exosome proteins through its N-terminus [17]. Previous work in yeast showed that the interaction between ScDis3 and the exosome core is also sensitive to divalent cation concentrations, with ScDis3 disassociating at 200 to 800 mM Mg2+ [18]. As the enzymatic activity of dDis3 is sensitive to changes in monovalent and divalent cation concentrations, we envisioned that varying salt conditions could change functionally relevant protein-protein interactions. We first assessed whether monovalent cations affect dDis3-dRrp6-core exosome interactions. To examine this possibility, we performed FLAG immunoaffinity chromatography with dDis3-FLAG isolated from S2 whole cell extracts and measured the recovery of dRrp6, the nuclear cofactor dRrp47, as well as three core exosome subunits: dRrp46, dRrp4, and dCsl4. The control for these experiments was extracts from cells expressing the vector alone (Mtn). We saw no effect on the ability of dDis3F to recover these proteins at concentrations up to 1.25 M Na+, an ~8-fold increase over normal wash buffer conditions (Figure 3A, lanes 3-10). This recovery was also stable in spite of the presence of 100 mM Mg2+, an ~33-fold increase over normal buffer conditions. Interestingly, we only observed a difference at these high salt concentrations in the presence of 5% TritonX-100, with approximately a 2-fold reduction in co-immunoprecipitation efficiency (Figure 3A, lane 11). This suggests sensitivity of the interactions to detergent, a reagent commonly used to disrupt hydrophobic interactions.
To determine whether dDis3-dRrp6-core exosome interactions were sensitive to divalent cations, we titrated in the divalent cation and held the monovalent cation concentration constant (and at two different concentrations). At either low (200 mM Na+) or high (1.0 M Na+) monovalent and divalent (10 mM Mg2+) salt concentrations, we observed quantitative recovery of the assessed proteins (Figure 3B, lanes 1 and 15). A 20-fold increase in Mg2+ concentration elicited a modest effect on dDis3F binding to the FLAG antibody yet caused a 2-5-fold reduction in dDis3F-mediated recovery of dRrp4, dRrp46, dCsl4, and dRrp47, with a lesser effect, approximately 2-fold, on dRrp6 recovery. Increasing the Mg2+ concentration to 400 mM led to a significant reduction of dDis3F recovery by the resin and a consequential loss of binding of the other proteins as well. Again, the dDis3-dRrp6 interaction appeared to be less sensitive to salt concentration than dRrp4, dRrp46, dCsl4, and dRrp47 (Figure 3B, lanes 5 and 19). Additional findings are complicated as higher Mg2+ concentration (600 mM – 1.2 M) reduced or eliminated dDis3F interaction with the FLAG resin. Regardless, these data show that dDis3-dRrp6-core exosome interactions are sensitive to divalent cation concentration.
Discussion
This study provides insight into both the requirements for and specificity of Drosophila Dis3 enzymatic function. First, our study has uncovered that full-length, recombinant dDis3 is active in vitro. Like other RNR family members, dDis3 in vitro activity requires a divalent cation [5; 16]. Analyses of ScDis3 and RNase II crystal structures have suggested that magnesium is associated with the RNB active site [10; 19; 20]. Other functional assays have suggested that magnesium coordinates 3’→5’ exoribonuclease activity mediated by RNB domain amino acids in dDis3 homologs [5; 16]. Our data shows that full-length dDis3 is able to release NMP products from cleavage of 5’ end-labeled polynucleotide RNAs in the presence of magnesium, which is consistent with 3’→5’ exoribonuclease activity. We conclude that magnesium-activated exoribonuclease activity is conserved among Dis3 homologs.
dDis3 substrate and product diversity points to overlap of ribonuclease activities
Dis3 activities may be dependent upon RNA substrate identity. In our study, dDis3 cleavage of polynucleotide RNAs produces a ladder of products that is weighted differently depending upon the sequence of the substrate. Consistent with this, previous studies show that ScDis3 degrades a variety of RNAs, but the reaction products generated from in vitro ribonucleolytic cleavage depends on sequence and/or structure [13]. These data suggest that RNA sequence could influence whether exo- and/or endoribonuclease activity is used for degradation. On one hand, dDis3 degradation of polyA produces AMP, consistent with an exoribonuclease activity. On the other, dDis3-mediated cleavage of the random RNA substrate produces approximately equivalent amounts of the largest and smallest products, eliciting several interpretations. First, the reaction products are created by exoribonucleolytic degradation alone, in which NMPs are cleaved off up to the last few nucleotides. At this point dDis3 no longer binds the substrate with the same efficiency, and thus releases the last few nucleotides as smaller polynucleotide fragments. A similar mechanism of action has been suggested for RNase II/R [16; 19]. Second, exoribonuclease activity is impeded by sequence composition or secondary structure. This stalling would create product fragments of various sizes. Third, dDis3 may use both exo- and endo-ribonuclease activity on this substrate. Exoribonuclease activity would produce the smallest NMP product, whereas endoribonucleolytic cleavage would produce the larger fragments. Regardless of the explanation, our results suggest that the type of activities dDis3 utilizes to cleave a particular RNA may depend upon its sequence.
The N-terminus of dDis3 is not necessary for activity
Our analysis shows that the C-terminus of dDis3 by itself contains ribonuclease activity, presumably within the RNB domain, as has been reported for ScDis3 [5; 10; 11]. However, we expected that removal of N-terminal domains via truncation would result in some alteration of activity, as the N-terminus contains the PIN domain, a putative site for endoribonuclease activity. Because our truncated mutants retain activity, this suggests that the C-terminal RNB domain functions independently of N-terminal domains to cleave single-stranded RNA substrates in vitro. Further analysis is required to determine if the RNB active site always acts independently of the N-terminal PIN domain to degrade substrates, or if this phenomena may be substrate or condition specific.
dDis3 protein-protein interactions are stable in vitro
Interactions between dDis3 and core exosome subunits remain stable at varying monovalent cation (Na+) concentrations. Only increasing concentrations of divalent cations (Mg2+) above physiologic concentrations result in their dissolution. This confirms similar observations for ScDis3 [5; 18]. Interestingly, our data also shows that core exosome subunits dRrp4, dRrp46, dCsl4, and dRrp47 lose interactions with dDis3 at lower magnesium concentrations than dRrp6, suggesting that the dDis3-dRrp6 interaction is more stable. These data are consistent with our finding that dDis3 and dRrp6 form a complex independent of the core exosome [12]. We also show that dDis3 interactions with core exosome subunits are sensitive to detergent. This suggests that not only ionic interactions, but hydrophobic interactions within the dDis3-core exosome complex lend to the stability of those protein-protein interactions.
Conclusions
As this represents the first study of a Dis3 ribonuclease from a multicellular organism, our observations highlight the conservation of activity among Dis3 homologs. Given the relationship of Dis3 homologs to cell cycle checkpoints ([1; 2; 14; 21], our unpublished work) and cancer progression [22; 23], understanding the mechanisms of dDis3 RNase activities is an important and topical endeavor. With the essential function of Dis3 still awaiting identification, biochemical studies to define the interface of its domains, substrates, and enzyme requirements may reveal general and conserved properties of Dis3 family members required for viability.
Supplementary Material
Figure S1. MBP-dDis3 is active in a range of reducing and pH conditions. (A) dDis3 activity in reducing conditions. Ribonuclease activity was assessed in buffer containing 75 mM KCl, 40 μM MgCl2, 10 mM tris, pH 8.0, and the designated 2-mercaptoethanol concentrations. Reactions were stopped and products were analyzed after 60 minutes. (B) dDis3 activity in various pH conditions. Reactions here also contained 75 mM KCl, 40 μM MgCl2, and 10 mM tris at the designated pH values. Again, reactions were incubated for 60 minutes.
Figure S2. dDis3 is active in the presence of various monovalent cations. (A) Ribonuclease activity of dDis3 in monovalent cation-containing buffer. Assays were performed as described in Materials and Methods. Here, the reaction buffer either contained 10 mM tris, pH 8.0 alone or tris and 75 mM of the monovalent cation indicated. Additionally, 1 mM 2-mercaptoethanol was present in the buffer of one experiment as indicated (labeled K+ + 2Me). (B) Quantification of dDis3 exoribonuclease activity. The amounts of RNA remaining at each time point were quantified by densiometry using ImageQuant software. To obtain % polyU remaining, each lane was normalized to the zero time point lane for that particular reaction; this method of quantification was used for the remaining experiments. The MBP control line (•) represents data averaged for all of the reaction conditions (14 experiments). The MBP control data has also been separated for each reaction condition, which can be seen in Figure S4. The remaining lines on the graph are as follows: Tris ■; KCl ▲; KCl + 2-Me ▼; NaCl ◆; NH4Cl ○; CsCl △; LiCl □. These represent data averaged for two independent experiments.
Figure S3. dDis3 is activated by divalent cations. (A) dDis3 activity in divalent ion-containing buffer. The reaction buffer for the first panel contained 10 mM tris, pH 8.0, 75 mM KCl, 40 μM MnCl2, and 1 mM 2-mercaptonethanol (labeled “Mn2+/K+”). The reaction buffer for the two remaining experiments contained 10 mM tris, pH 8.0, and 40 μM of either divalent cation, as indicated. (B) Quantification of dDis3 activity. % polyU remaining for the MBP control reactions again represents data averaged for all of the reaction conditions shown (6 experiments total). See Supplemental Figure S4 for MBP control data from each individual reaction condition. For MBP-dDis3 reactions, averages of two independent experiments for each reaction condition are shown.
Figure S4. MBP is not active on polyU under any condition. MBP was incubated with 5’-end labeled RNA for 60 minutes in reaction buffer containing 10 mM tris, pH 8.0 with or without monovalent cation or divalent cations added to the reaction buffer, as specified. % polyU was calculated as described in Figure S2. Data shown represents averages of two independent experiments for each condition. (A) MBP activity in monovalent cation-containing buffer. Tris ■; KCl ▲; KCl + 2-Me ▼; NaCl ◆; NH4Cl ○; CsCl △; LiCl □. (B) MBP activity in divalent ion-containing buffer. MnCl2 + KCl •; MnCl2 ■; MgCl2 ▲.
Figure S5. dDis3 is inactive in the presence of chelating agent. Assays were carried out essentially as described before. Here, however, proteins were incubated with polyU RNA for 10 minutes total. Reactions on the left were incubated in buffer containing 10 mM tris, pH 8.0, 75 mM KCl, and 40 μM MgCl2. Bracket indicates reaction products. Reactions on the right were incubated in buffer containing the same components plus 5 mM EDTA. Images are representative of two independent experiments.
Acknowledgements
The authors thank Peter Harte and Alan Tartakoff and members of the Andrulis lab for reviewing the manuscript, and Dr. Eckhard Jankowsky for the polyU stubstrate. This work is supported by grant GM072820 from the NIH (E.D.A). E.D.A. is a Mount Sinai Health Care Foundation Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kinoshita N, Goebl M, Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol. 1991;11:5839–47. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. Embo J. 1988;7:1465–73. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'-->5' exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 4.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–26. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 6.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–90. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 7.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 8.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–40. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the RNase II Nuclease Family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–31. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome, and importin-alpha3. Traffic. 2009;10:499–513. doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 14.Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease Activity of Dis3 Is Required for Mitotic Progression and Provides a Possible Link between Heterochromatin and Kinetochore Function. PLoS ONE. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 17.Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome, and importin-alpha3. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 20.Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–56. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, Nishimoto T. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. Embo J. 1996;15:5595–605. [PMC free article] [PubMed] [Google Scholar]
- 22.Rozenblum E, Vahteristo P, Sandberg T, Bergthorsson JT, Syrjakoski K, Weaver D, Haraldsson K, Johannsdottir HK, Vehmanen P, Nigam S, Golberger N, Robbins C, Pak E, Dutra A, Gillander E, Stephan DA, Bailey-Wilson J, Juo SH, Kainu T, Arason A, Barkardottir RB, Nevanlinna H, Borg A, Kallioniemi OP. A genomic map of a 6-Mb region at 13q21-q22 implicated in cancer development: identification and characterization of candidate genes. Hum Genet. 2002;110:111–21. doi: 10.1007/s00439-001-0646-6. [DOI] [PubMed] [Google Scholar]
- 23.Lim J, Kuroki T, Ozaki K, Kohsaki H, Yamori T, Tsuruo T, Nakamori S, Imaoka S, Endo M, Nakamura Y. Isolation of murine and human homologues of the fission-yeast dis3+ gene encoding a mitotic-control protein and its overexpression in cancer cells with progressive phenotype. Cancer Res. 1997;57:921–5. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. MBP-dDis3 is active in a range of reducing and pH conditions. (A) dDis3 activity in reducing conditions. Ribonuclease activity was assessed in buffer containing 75 mM KCl, 40 μM MgCl2, 10 mM tris, pH 8.0, and the designated 2-mercaptoethanol concentrations. Reactions were stopped and products were analyzed after 60 minutes. (B) dDis3 activity in various pH conditions. Reactions here also contained 75 mM KCl, 40 μM MgCl2, and 10 mM tris at the designated pH values. Again, reactions were incubated for 60 minutes.
Figure S2. dDis3 is active in the presence of various monovalent cations. (A) Ribonuclease activity of dDis3 in monovalent cation-containing buffer. Assays were performed as described in Materials and Methods. Here, the reaction buffer either contained 10 mM tris, pH 8.0 alone or tris and 75 mM of the monovalent cation indicated. Additionally, 1 mM 2-mercaptoethanol was present in the buffer of one experiment as indicated (labeled K+ + 2Me). (B) Quantification of dDis3 exoribonuclease activity. The amounts of RNA remaining at each time point were quantified by densiometry using ImageQuant software. To obtain % polyU remaining, each lane was normalized to the zero time point lane for that particular reaction; this method of quantification was used for the remaining experiments. The MBP control line (•) represents data averaged for all of the reaction conditions (14 experiments). The MBP control data has also been separated for each reaction condition, which can be seen in Figure S4. The remaining lines on the graph are as follows: Tris ■; KCl ▲; KCl + 2-Me ▼; NaCl ◆; NH4Cl ○; CsCl △; LiCl □. These represent data averaged for two independent experiments.
Figure S3. dDis3 is activated by divalent cations. (A) dDis3 activity in divalent ion-containing buffer. The reaction buffer for the first panel contained 10 mM tris, pH 8.0, 75 mM KCl, 40 μM MnCl2, and 1 mM 2-mercaptonethanol (labeled “Mn2+/K+”). The reaction buffer for the two remaining experiments contained 10 mM tris, pH 8.0, and 40 μM of either divalent cation, as indicated. (B) Quantification of dDis3 activity. % polyU remaining for the MBP control reactions again represents data averaged for all of the reaction conditions shown (6 experiments total). See Supplemental Figure S4 for MBP control data from each individual reaction condition. For MBP-dDis3 reactions, averages of two independent experiments for each reaction condition are shown.
Figure S4. MBP is not active on polyU under any condition. MBP was incubated with 5’-end labeled RNA for 60 minutes in reaction buffer containing 10 mM tris, pH 8.0 with or without monovalent cation or divalent cations added to the reaction buffer, as specified. % polyU was calculated as described in Figure S2. Data shown represents averages of two independent experiments for each condition. (A) MBP activity in monovalent cation-containing buffer. Tris ■; KCl ▲; KCl + 2-Me ▼; NaCl ◆; NH4Cl ○; CsCl △; LiCl □. (B) MBP activity in divalent ion-containing buffer. MnCl2 + KCl •; MnCl2 ■; MgCl2 ▲.
Figure S5. dDis3 is inactive in the presence of chelating agent. Assays were carried out essentially as described before. Here, however, proteins were incubated with polyU RNA for 10 minutes total. Reactions on the left were incubated in buffer containing 10 mM tris, pH 8.0, 75 mM KCl, and 40 μM MgCl2. Bracket indicates reaction products. Reactions on the right were incubated in buffer containing the same components plus 5 mM EDTA. Images are representative of two independent experiments.