Abstract
The induction of key pro-inflammatory genes is regulated by the SWI/SNF class of ATP-dependent remodeling complexes. In particular, the catalytic ATPase subunit, Brg1, is distinctly involved in the chromatin remodeling required for activating pro-inflammatory genes in a temporally, ordered fashion. Despite advances in our understanding of the role for Brg1 in the kinetics of inflammatory responses, little is known about the precise mechanisms which down-regulate Brg1 activity. Biochemical studies implicate a role for the proteasome in the regulation of SWI/SNF assembly and function; however, it is unclear if proteasome-dependent mechanisms modulate its remodeling activity or recruitment to chromatin in order to regulate inflammatory gene transcription. We now demonstrate for the first time that proteasome function represents an important mechanism for limiting inducible association of Brg1 with promoters of SWI/SNF-regulated, inflammatory genes. As a result, catalytic activity of the proteasome fine-tunes the kinetics of inflammatory gene transcription by inhibiting both premature and persistent chromatin remodeling at SWI/SNF-regulated genes. These results provide mechanistic insight into the interplay between nucleosome remodeling, inflammation and proteasome, and underscore the critical role of the proteasome in controlling both extent and duration of inflammatory responses.
Keywords: Proteasome, SWI/SNF, Brg1, inflammation, gene regulation, chromatin remodeling
1. Introduction
Alteration of chromatin architecture by ATP-dependent remodeling complexes is considered a pivotal step in transcriptional regulation of many eukaryotic genes. Based on the makeup of its catalytic component, the ATP-dependent remodeling complexes are divided into four major subfamilies, SWI/SNF, ISWI, NuRD, and INO80 (Wu et al., 2009). Among these multiprotein complexes, SWI/SNF has been implicated in a wide range of cellular events including cell cycle regulation and differentiation. As a central catalytic ATPase of the SWI/SNF complex, Brg1 not only plays an essential role in embryogenesis and tumor suppression but is also fundamentally involved in immune phenomena such as early thymocyte development and antiviral and inflammatory responses (Trotter and Archer, 2008;Agalioti et al., 2000;Chi et al., 2003;Ramirez-Carrozzi et al., 2006). For instance, through its specific and precisely-timed nucleosome remodeling activity, the Brg1-containing SWI/SNF complex regulates the kinetics of inflammatory responses (Agalioti et al., 2000;Ramirez-Carrozzi et al., 2006).
Based on the contribution of SWI/SNF-mediated chromatin remodeling, inflammatory genes induced in LPS-stimulated macrophages have been broadly categorized as primary and secondary response genes (Ramirez-Carrozzi et al., 2006). Since early primary response genes are characterized by promoters which are constitutively permissive for transcription, these genes are induced with faster kinetics than secondary response genes. In contrast, late primary response genes and secondary response genes, like IL-6, are induced with delayed kinetics because they require stimulus-dependent recruitment of SWI/SNF. Despite advances in our understanding of SWI/SNF-dependent regulation of the induction of inflammatory responses, little is known about the mechanisms which antagonize its function. Consequently, it is unclear if dysregulation in these inhibitory mechanisms underlies the excessive inflammation accompanying pathological conditions, such as septic shock, autoimmunity, and aging (Foster and Medzhitov, 2009;Licastro et al., 2005). Given the physiological significance of these inhibitory mechanisms, we sought to understand how SWI/SNF activity is terminated.
Recent biochemical studies demonstrate a role for the proteasome in the destabilization of the SWI/SNF complex due to degradation of key SWI/SNF complex components, such as Brg1 (Chen and Archer, 2005;Sohn et al., 2007). Besides regulation of complex formation, the degradation of SWI/SNF catalytic subunits inhibits its chromatin remodeling activity. For example, proteasome suppresses Brg1 recruitment at an intergenic, regulatory region associated with B cell development in order to maintain pluripotency in embryonic stem cells (Szutorisz et al., 2006). Similarly, proteasome is implicated in the protein turnover of the Brg1 paralog, Brm, as a means of inhibiting its activity during cellular events such as mitosis and apoptosis (Sif et al., 1998;Muchardt et al., 1996;Biggs et al., 2001). Given its ability to modulate SWI/SNF assembly and function, proteasome-dependent mechanisms seem a plausible means of down-regulating Brg1 activity at inflammatory, SWI/SNF-regulated genes.
In lieu of recent studies, the 26S proteasome is emerging as an essential regulatory mechanism for controlling the extent and duration of inflammatory responses (Natoli and Chiocca, 2008). By targeting nuclear NF-κB for proteolytic degradation, the proteasome contributes to the termination of NF-κB activity and prevents excessive induction of genes involved in inflammation. As proteasome inhibition results in sustained transcription of particular inflammatory genes (Saccani et al., 2004), this implies that such exaggerated transcription stems from both prolonged nuclear NF-κB activity and persistently remodeled chromatin. Accordingly, we hypothesize that proteasomal regulation of SWI/SNF subunits antagonizes its nucleosome remodeling activity at secondary response genes, thereby limiting the duration of inflammatory responses.
One challenge in defining the in vivo role of proteasome catalytic function at secondary response genes is that, in the presence of proteasome inhibitor, LPS stimulation does not sufficiently induce these genes. Since proteasome inhibition sequesters NF-κB in the cytoplasm, the addition of proteasome inhibitor results in inadequate NF-κB signaling during the primary response, which in turn hampers induction of secondary response genes (Kayama et al., 2008). For this reason, we employed an approach which bypasses the requirement for proteasome in NF-κB induction (Imbert et al., 1996), thereby permitting us to better define the regulatory role of proteasome at the SWI/SNF-regulated gene, IL-6. This alternative strategy relies on the atypical, NF-κB stimulus, pervanadate (PV), which activates NF-κB signaling by potently inhibiting tyrosine phosphatases. Based on our previous work with a murine stromal epithelial cell line (ILU-18), proteasome inhibition is accompanied by persistent recruitment of PV-induced NF-κB p65/RelA at the IL-6 promoter; correspondingly, persistently recruited NF-κB led to sustained transcription of IL-6 (manuscript submitted). Given this prolonged promoter association of NF-κB p65/RelA, we reasoned that, due to sustained SWI/SNF activity, prolonged chromatin accessibility must also occur when proteasomal activity is inhibited.
We now present evidence that proteasomal degradation of the SWI/SNF ATPase subunit, Brg1, actively promotes its removal from chromatin. As a result, the catalytic activity of the 26S proteasome fine-tunes the kinetics of inflammatory gene transcription by inhibiting both premature and persistent chromatin remodeling at SWI/SNF-regulated genes. Accordingly, these results provide a molecular basis for the increased inflammation in physiological conditions marked by lowered proteasomal function, such as during aging and in geriatric diseases (Das et al., 2007;Licastro et al., 2005;Ponnappan et al., 1999).
2. Materials and Methods
2.1. Antibodies & Reagents
Brg1 (H-88) and SNF5 (H-300) antibodies were from Santa Cruz Biotechnology (Carlsbad, CA). Sug1 and 20S Proteasome (PW 8155) antibodies were from Biomol (Plymouth Meeting, PA). Aclacinomycin was from Calbiochem (La Jolla, CA) and electrophoresis supplies and molecular weight standards were from Biorad (Richmond, CA). Unless otherwise mentioned, all fine chemicals were from Sigma (St. Louis, MO).
2.2. Cell Culture
Murine bone marrow-derived stromal cell line, ILU-18, was kindly provided by Dr. Charles O’Brien (UAMS, Little Rock, AR) (Lin et al., 1997). ILU-18 cells were maintained in DMEM medium supplemented with 2mM glutamine, 100units/mL penicillin, 100μg/mL streptomycin, and 10% fetal bovine serum.
2.3. Pervanadate & Aclacinomycin Treatment
Aliquots of 200mM Sodium orthovanadate, pH 10.0, were prepared according to a protocol from Millipore (Billerica, MA) and stored at −20°C. To ensure that Sodium orthovanadate is in its depolymerized form, each aliquot was boiled for 5 minutes, prior to use. Upon cooling, 10μL of 200mM Sodium Orthovanadate and 8μL of phosphate-buffered saline were combined with 2μL of 3% (w/w) H2O2. Following appropriate dilution with DMEM media, the pervanadate solution (10mM) was incubated on a rocker for 15 minutes to remove excess H2O2. Cells were treated with a final concentration of pervanadate of 100μM. PV treatment was carried out for 2, 4, or 8 hours, as indicated in specific experiments. For cells undergoing proteasome inhibition, ILU-18 cells were pretreated with Aclacinomycin (0.25μM) for 2 hours to ensure inhibition of proteasome prior to activation with PV.
2.4 Microarray Analysis
Microarray analysis employing Illumina cDNA arrays was performed by the University of Tennessee Health Sciences Center, Microarray Facility. Identification of differentially expressed genes was carried out using Bead Studio Gene Expression module. Thresholds for selecting significant genes were set at a relative difference >1.35-fold and absolute intensity differences between experimental groups, p<0.01. Genes that met these criteria simultaneously were considered as significant changes. We selected four of the most robustly expressed (>2.0-fold difference between 2h Acla + 4h PV vs. 2h Acla + 8h PV) SWI/SNF-regulated genes; it should be noted that, in addition to these four genes, IL-6 also met this criteria. Both IL-6 and Ifit3 were validated by qPCR.
2.5 Reverse Transcription and Quantitative PCR (RT-qPCR)
Total RNA was isolated using TRIzoL Reagent (Invitrogen Corporation, Carlsbad, CA). Total RNA was reverse transcribed with an oligo dT primer using M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA). Complementary DNA was analyzed by qPCR in duplicate using SYBR Green Master Mix (SuperArray Biosciences Corporation) on the BioRad iCycler PCR system. Primer sets were specifically designed to amplify the mRNA target of interest by spanning exon-exon boundaries and unique products were tested by melt curve analysis. Data was analyzed by the relative standard curve method with GAPDH as the endogenous control and untreated sample used as the calibrator sample. Primer sequences are available upon request.
2.6. Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed with the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer’s protocol. Briefly, untreated or treated ILU-18 cells were subjected to cross-linking with 1% formaldehyde at 37°C for 10 minutes, followed by cell lysis using SDS Lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl - pH 8.1, and protease inhibitors). Sonication was used to shear the genomic DNA into fragments with an average size of ~500bp which were then diluted 10-fold in ChIP dilution buffer. After removing an aliquot for evaluation as input DNA, the lysates were pre-cleared with Salmon Sperm DNA/Protein G Agarose. The pre-cleared lysates were immunoprecipitated with the appropriate antibody by overnight incubation and immune complexes were precipitated with Salmon Sperm DNA/Protein G Agarose. The precipitates were washed sequentially with buffers, including low salt wash buffer, high salt wash buffer, LiCl wash buffer, and TE buffer. Antibody-DNA complexes were eluted twice with extraction buffer (100mM NaHCO3, 1% SDS, 20μg yeast tRNA). The eluates were heated overnight at 65°C to dissociate the cross-linking. Genomic DNA fragments were extracted and purified by phenol:chloroform extraction and Ethanol precipitation before proceeding to detection of specific chromatin fragments by PCR. The amplification conditions were 94°C for 45 seconds, 60°C for 1 minute, and 72°C for 1 minute for 30–36 Cycles. PCR primer sequences are as follows: IL-6 Promoter, 5′-GACATGCTCAAGTGCTGAGTCAC-3′ (Forward) and 5′-AGATTGCACAATGTGACGTCG-3′ (Reverse); IFIT3 Promoter, 5′-ACTCTTTTCCTCCCAGAGGG-3′ (Forward) and 5′-GGGCTGAGCAGTTCAGAAA-3′ (Reverse). PCR products were analyzed by 3% agarose gel electrophoresis and stained with ethidium bromide. Analysis of immunoprecipitated DNA by quantitative PCR was performed with SYBR Green Master Mix (SuperArray Biosciences Corporation) using the BioRad iCycler PCR system. Each sample was normalized to input DNA, with the final result expressed as fold induction relative to untreated control.
2.7. Chromatin Accessibility using PCR
Experiments were performed as described previously (Rao et al., 2001;Weinmann et al., 1999), with slight modifications. ILU-18 cells (6×106) underwent one wash with ice-cold 1X PBS before lysing the cells on ice for 5 minutes with ice-cold NP-40 lysis buffer (10mM Tris [pH 7.4], 10mM NaCl, 3mM MgCl2, 0.5% NP-40, 0.15mM spermine, and 0.5mM spermidine) to obtain nuclear fraction. Nuclei were isolated by pelleting at 1000 RPM and subsequently washed with RE buffer (10mM Tris [pH 7.4], 50mM NaCl, 10mM MgCl2, 0.2mM EDTA, 0.2mM EGTA, 1mM β-Mercaptoethanol, 0.15mM spermine, and 0.5mM spermidine). Upon resuspension with the appropriate New England Biolabs (NEB) buffer, nuclei were incubated with 20U HincII for 60 min at 37°C. Following overnight digestion with proteinase K, DNA was purified using QIAquick PCR purification kit (Qiagen). Purified DNA (2μL) was used in PCR reactions, employing either primers encompassing the HincII site in the IL-6 promoter or primers which spanned a GAPDH promoter region lacking HincII/HindII sites. PCR amplification was employed with the following primers: IL-6 promoter, 5′-TCGATGCTAAACGACGTCAC-3′ (Forward) and 5′-TGAGCTACAGACATCCCCAGT-3′ (Reverse); GAPDH promoter 5′-AGTGCCAGCCTCGTCCCGTAGACAAAATG-3′ (Forward) and 5′-AAGTGGGCCCCGGCCTTCTCCAT-3′ (Reverse).
3. Results
3.1. Proteasomal activity limits Brg1 association with the IL-6 promoter in a timely manner
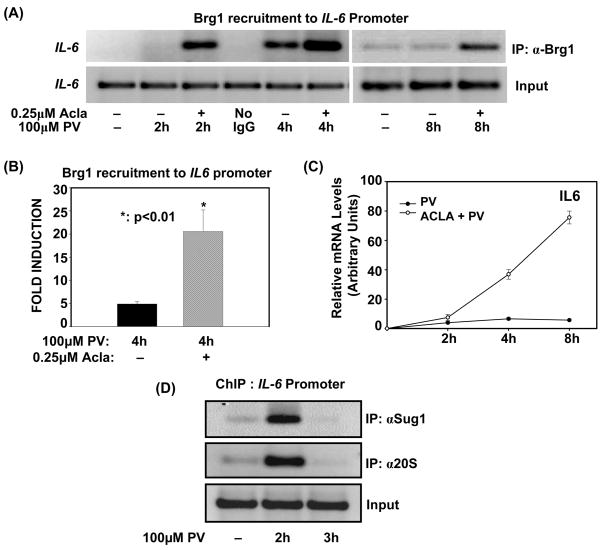
In the absence of a scaffold protein, key subunits of the SWI/SNF complex, including Brg1, are susceptible to proteolytic degradation (Chen and Archer, 2005;Sohn et al., 2007). Apart from destabilizing the complex, proteasomal regulation of Brg1 also modulates its recruitment in order to suppress its remodeling activity (Szutorisz et al., 2006). To demonstrate that proteasome limits the induction of SWI/SNF-regulated inflammatory genes by targeting Brg1 for degradation, we determined if proteasomal degradation of Brg1 is required for its removal from the IL-6 promoter. Hence, we induced IL-6 expression with PV, in cells deficient (Acla-treated) or sufficient in proteasome activity and then performed ChIP assay with an antibody to Brg1. As depicted in Fig. 1A, inhibition of the chymotryptic activity of the proteasome by Aclacinomycin results in enhanced retention of Brg1 at the IL-6 promoter, both at 2h and 4h post-activation. Analysis of Brg1 recruitment at 4h post-activation by qPCR validated that proteasome inhibition significantly enhances the association of Brg1 with the IL-6 promoter (Fig. 1B). Furthermore, Brg1 is found at the IL-6 promoter as late as 8h post-activation when proteasome is inhibited (Fig. 1A). Since Brg1 is persistently recruited when proteasome is inhibited, these results indicate that proteasome activity regulates the timely removal of Brg1 from the promoter. Consistent with the prolonged and sustained recruitment of Brg1 accompanying proteasome inhibition, IL-6 expression was significantly and more persistently up-regulated by PV in cells pretreated with proteasome inhibitor compared to cells treated with PV alone (Fig. 1C). Thus, proteasome inhibition is accompanied by both persistent recruitment of Brg1 at the IL-6 promoter and sustained transcription of IL-6.
Figure 1. Proteasome activity limits Brg1-promoter association with kinetics consistent with transcriptional induction and shut-off.
(A). ILU-18 cells were either untreated or treated with PV (100μM) for 2, 4 and 8 hours, with (+) or without (-) pretreatment with Acla (0.25μM) for 2 hours. ChIP assay employing anti-Brg1 was performed using immunoprecipitated DNA amplified with promoter-specific primers for IL-6
(B). ChIP assays were performed on ILU-18 cells treated with PV (100μM) for 4 hours, with (+) or without (−) pretreatment with Acla (0.25μM) for 2 hours using Brg1 antibody for the immunoprecipitation. The immunoprecipitated chromatin was submitted to qPCR analysis using primers amplifying the promoter region of IL-6. Fold induction represents anti-Brg1 ChIP relative to control ChIP (immunoprecipitated DNA from untreated cells). Statistically significant differences are denoted by * (p value < 0.05)
(C). ILU-18 cells were stimulated with PV (100μM), with (○) or without (●) pretreatment with Acla (0.25μM) for 2 hours. Total RNA was extracted at indicated times and analyzed by RT-qPCR for mRNA expression levels of IL-6. The relative mRNA levels were determined from two independent experiments and are presented as mean ± standard error.
(D). ChIP assays employing antibodies recognizing Sug1 and 20S were performed using immunoprecipitated DNA amplified with promoter-specific primers for IL-6. Chromatin was obtained from ILU-18 cells which were stimulated with PV (100μM) for 0, 2, and 3 hours.
Our earlier studies have demonstrated that PV-stimulation induces NF-κB and RNA pol II recruitment at 4h post-activation (manuscript submitted). Since previous studies have shown that Brg1 recruitment precedes that of the transcriptome (Kayama et al., 2008;Ramirez-Carrozzi et al., 2006), we analyzed recruitment of Brg1 to the IL-6 promoter at 2h post-activation. In agreement with previous observations, Brg1 recruitment preceded that of NF-κB and RNA pol II; unexpectedly, however, its association with chromatin at 2h post-activation was only detected in the presence of proteasome inhibitor (Fig. 1A). To further establish that proteasome regulates Brg1 at this timepoint, we performed ChIP assays with antibodies recognizing components comprising the 19S regulatory particle and 20S catalytic core of the 26S proteasome. As depicted in Fig. 1D, components of both the 19S and 20S proteasome complexes are indeed recruited following 2h PV-stimulation. This suggests that proteasome regulates steps associated with priming the promoter for transcriptome recruitment. Collectively, these results show that proteasome activity is responsible for removing Brg1 from the IL-6 promoter, with kinetics consistent with both transcriptional induction and shutoff (Fig. 1C).
3.2. Proteasome function removes Brg1 from the promoter of an additional SWI/SNF-regulated gene
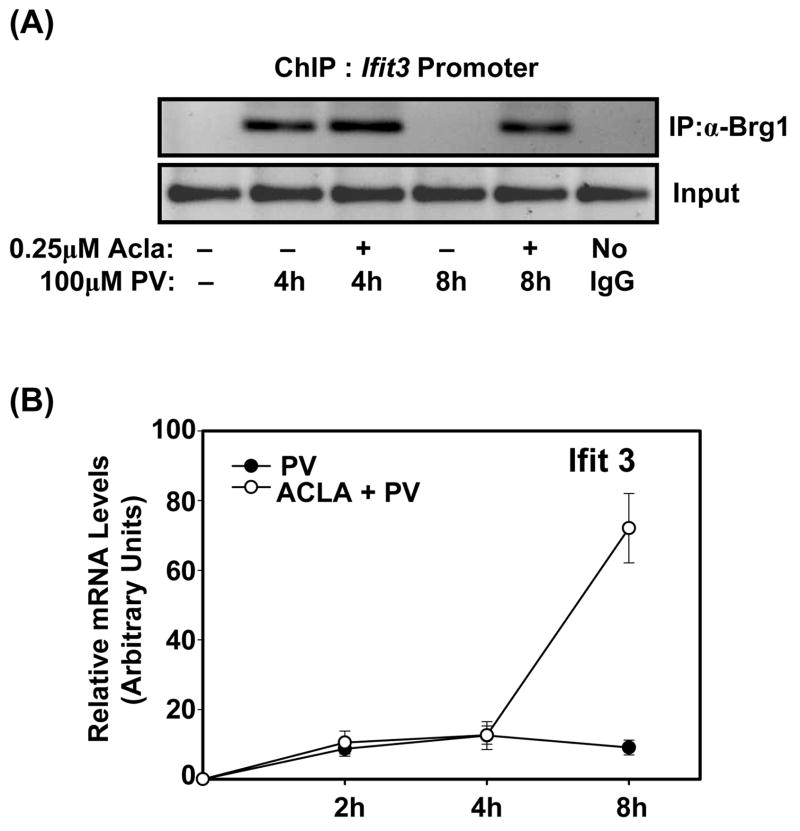
Next, we investigated if proteasome function tightly controls Brg1 recruitment at additional SWI/SNF-regulated genes. To identify genes, we conducted microarray analysis and then focused on genes for which pretreatment with proteasome inhibitor led to a persistent increase in expression. To do so, we compared genes from cells pretreated with Aclacinomycin followed by treatment with PV for either 4h or 8h (2h Acla + 4h PV vs. 2h Acla vs. 8h PV). From this list of genes, we selected genes which are known to be regulated by the SWI/SNF complex (Cui et al., 2004;Ramirez-Carrozzi et al., 2006). This resulted in a list of genes, whose expression is dependent on the function of the SWI/SNF complex and is persistently up-regulated by proteasome inhibition by at least 2-fold: CXCL10, CCL2, Ifit2, and Ifit3. Among these genes, we selected Ifit3 as its expression is induced with delayed kinetics akin to those of IL-6. Hence, we performed ChIP assays to delineate if proteasomal degradation of Brg1 removes it from the Ifit3 promoter. Similar to the IL-6 promoter, inducible association of Brg1 with the Ifit3 promoter is not promptly terminated when proteasome activity is inhibited (Fig. 2A). Moreover, persistent Brg1 recruitment at the Ifit3 promoter coincides with sustained induction of Ifit3, as determined by qPCR analysis (Fig. 2B). Thus, proteasome represents an important mechanism for not only limiting inducible association of Brg1 with the promoters of SWI/SNF-regulated genes but also for terminating the transcription of these SWI/SNF-regulated genes.
Figure 2. Treatment with proteasome inhibitor results in persistent association of Brg1 with the Ifit3 promoter.
(A). ChIP assay employing anti-Brg1 was performed using immunoprecipitated DNA amplified with promoter-specific primers for Ifit3. Chromatin was obtained from ILU-18 cells either left untreated or treated with PV (100μM) for 4 and 8 hours, with (+) or without (−) pretreatment with Acla (0.25μM) for 2 hours
(B). ILU-18 cells were stimulated with PV (100μM), with (○) or without (●) pretreatment with Acla (0.25μM) for 2 hours. Total RNA was extracted at indicated times and analyzed by RT-qPCR for mRNA expression levels of Ifit3. The relative mRNA levels were determined from two independent experiments and are presented as mean ± standard error.
3.3. Activity of the proteasome limits sustained nucleosome remodeling at the IL-6 promoter
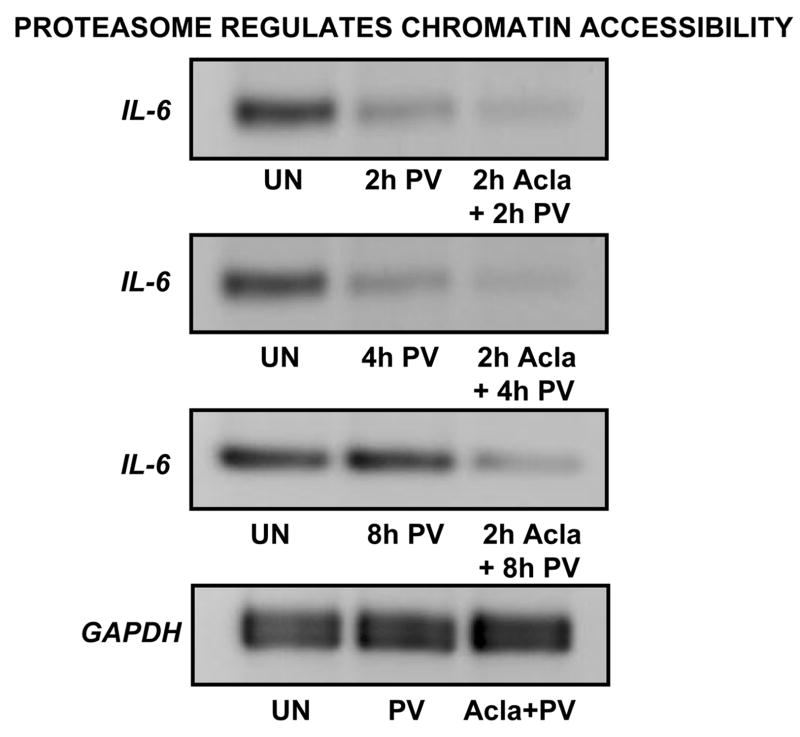
If proteasome activity is blocked, Brg1 is clearly not removed from promoters of genes regulated by Brg1-containing SWI-SNF complexes. Therefore, we sought to determine how chromatin accessibility is affected by this sustained recruitment of Brg1. To do this, we employed an accessibility assay in which DNA purified from nuclei treated with restriction enzyme, Hinc II, is subjected to PCR using promoter-specific primers encompassing the cleavage site. As described previously (Rao et al., 2001), stimulation is accompanied by increased cleavage and as a consequence of this cleavage, PCR amplification across this region is impaired. Thus, due to digestion, DNA from stimulated cells yields less PCR product than DNA from unstimulated cells.
Using this assay, we observed a relative increase in Hinc II accessibility upon PV-stimulation, both in absence and presence of proteasome inhibitor (Fig. 3). Consistent with results obtained by ChIP, PV-dependent cleavage was detected at both 2h- and 4h-post activation. Additionally, we know from data obtained with GAPDH primers that the PV-dependent cleavage observed at 2h- and 4h-post activation is indeed derived from Hinc II-specific cleavage. Most notably, following long-term PV exposure, Hinc II accessibility at the IL-6 promoter was only detected in the presence of proteasome inhibitor. This result supports our original hypothesis that loss in proteasome function results in persistently remodeled chromatin at the inflammatory gene, IL-6. Coupled with results from ChIP analyses, these data indicate that the timely removal of Brg1 from the promoter represents a mechanism by which proteasome antagonizes sustained SWI/SNF remodeling.
Figure 3. Proteasome inhibition leads to prolonged chromatin accessibility at IL-6 promoter.
Intact nuclei were isolated from ILU-18 cells which were either unstimulated or stimulated with PV (100μM) for 2, 4, or 8 hours, with or without pretreatment with Acla (0.25μM) for 2 hours. Nuclei were incubated with 20U of Hinc II for 60 min at 37°C. The purified DNA was subjected to semiquantitative PCR with IL-6 promoter primers spanning the Hinc II site. Representative control data, obtained by amplifying DNA with GAPDH primers, is presented.
4. Discussion
To influence the kinetics of inflammatory responses, Brg1-containing SWI/SNF complexes must be recruited in a timely manner and its nucleosome remodeling activity must be promptly shut off. By demonstrating that proteasome antagonizes SWI/SNF function, we have shown how proteasome intrinsically regulates the kinetics of inflammatory responses. As proteasome has already been shown to regulate nuclear NF-κB activity (Natoli and Chiocca, 2008), it remains to be determined if there is a relationship between proteasome, NF-κB, and chromatin remodeling. A model proposed recently hypothesizes that, similar to the glucocorticoid receptor, the rapid rate with which NF-κB dissociates from DNA is due to the transient reversal of chromatin accessibility (Natoli and Chiocca, 2008). Intriguingly, oscillatory binding of SWI/SNF underlies the rapid exchange between the GR and the chromatin template (Nagaich et al., 2004). Moreover, the observation that chromatin remodeling mechanisms promote rapid exchange of the GR at the promoter is juxtaposed with the finding that proteasome activity is also required for rapid GR exchange (Nagaich et al., 2004;Stavreva et al., 2004). In lieu of our observations, we propose that proteasomal degradation of Brg1 is ultimately responsible for the transient reversal of chromatin remodeling that displaces transcription factors, such as GR and NF-κB. To test this hypothesis, future studies will need to employ proteolysis-resistant, Brg1 mutants to confirm that proteolysis of Brg1 transiently limits chromatin accessibility.
In agreement with previous studies, we observed that restricted chromatin remodeling appears to be an essential facet of priming the IL-6 promoter. As previously reported (Ramirez-Carrozzi et al., 2006), this particular promoter undergoes a time-dependent increase in chromatin accessibility. Due to issues of assay sensitivity, it is difficult to discern from our data if a similar time-dependent increase in accessibility occurs following PV-stimulation. Nonetheless, results from our ChIP analyses clearly indicate that during the priming period of 2h post-activation, Brg1 association with the IL-6 promoter is severely limited by the proteasome. This observation strengthens the likelihood that restricted chromatin accessibility is achieved by limiting the association of Brg1-containing SWI/SNF complexes with the IL-6 promoter. Furthermore, it implicates proteasome as a contributing factor in the restricted chromatin accessibility accompanying IL-6 promoter priming.
Promoters of inflammatory genes are rendered permissive for transcriptional induction through the combined activities of ATP-dependent chromatin remodeling complexes and histone-modifying enzymes (Foster and Medzhitov, 2009). While not addressed in this study, it is becoming clear that histone modifications are significantly impacted by loss in proteasome function. For instance, proteasome inhibition causes global alterations in the levels of trimethylated H3K4, ubiquitinated H2A, and acetylated H3K27 (Dantuma et al., 2006;Kinyamu and Archer, 2007;Oliva et al., 2009). Thus, it is not surprising that dysregulation in epigenetic mechanisms accompanies physiological conditions under which proteasomal function is compromised, i.e. advancing age and geriatric diseases (Chambers et al., 2007;Kawahara et al., 2009). Such perturbations in histone modifications, particularly H3K4 methylation, are to be expected because proteasome-mediated degradation of histone demethylases equilibrates histone demethylase and histone methyltransferase activities in the cell (Mersman et al., 2009). Likewise, coactivators and histone acetyltransferases, CBP and p300, are subject to proteasomal regulation in certain cellular contexts. For example, exposure to inhibitors of histone deacetylases, such as valproic acid and sodium butyrate, results in the proteolytic degradation of CBP and p300 (St-Germain et al., 2008;Li et al., 2002). Clearly, these observations signal that, either by itself or through a synergism with other signaling pathways, proteasome inhibition can have a striking effect on the stability, and in turn, the function of histone modifying enzymes. Given the proteasomal regulation of histone modifying enzymes, studies are currently underway to delineate if persistently modified histones also contribute to the sustained PV-induced IL-6 and Ifit3 expression observed during proteasome inhibition.
In summary, these studies expand our understanding of proteasome-dependent mechanisms underlying the down-regulation of inflammatory responses. By demonstrating that proteasomal activity aids in the timely removal of Brg1 from promoters of inflammatory genes, we have defined a precise mechanism by which proteasome modulates SWI/SNF activity to fine-tune the kinetics of SWI/SNF-regulated inflammatory genes.
Acknowledgments
This work was supported by NIH grants AG13081, AG030599 and AG025220 to UP and in part by the UAMS Graduate Student Research Fund to SJC. We gratefully acknowledge Dr. Charles O’Brien for providing us with ILU-18 cell line.
Abbreviations used in the manuscript
- SWI/SNF
mating-type switching and sucrose non-fermenting
- Brg1
Brahma-related gene-1
- PV
Pervanadate
- Acla
aclacinomycin
- CCL2
CC Chemokine ligand 2
- CXCL10
CXC Chemokine ligand 10
- Ifit2
Interferon-induced protein with tetratricopeptide repeats 2
- Ifit3
Interferon-induced protein with tetratricopeptide repeats 3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Biggs JR, Yang J, Gullberg U, Muchardt C, Yaniv M, Kraft AS. The human brm protein is cleaved during apoptosis: the role of cathepsin G. Proc Natl Acad Sci U S A. 2001;98:3814–3819. doi: 10.1073/pnas.071057398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol. 2004;24:4476–4486. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Ponnappan S, Ponnappan U. Redox regulation of the proteasome in T lymphocytes during aging. Free Radic Biol Med. 2007;42:541–551. doi: 10.1016/j.freeradbiomed.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130:7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert V, Rupec RA, Livolsi A, Pahl HL, Traenckner EB, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle PA, Peyron JF. Tyrosine phosphorylation of I kappa B-alpha activates NF-kappa B without proteolytic degradation of I kappa B-alpha. Cell. 1996;86:787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayama H, Ramirez-Carrozzi VR, Yamamoto M, Mizutani T, Kuwata H, Iba H, Matsumoto M, Honda K, Smale ST, Takeda K. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IkappaBzeta. J Biol Chem. 2008;283:12468–12477. doi: 10.1074/jbc.M709965200. [DOI] [PubMed] [Google Scholar]
- Kinyamu HK, Archer TK. Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol Cell Biol. 2007;27:4891–4904. doi: 10.1128/MCB.02162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Su A, Chen J, Lefebvre YA, Hache RJ. Attenuation of glucocorticoid signaling through targeted degradation of p300 via the 26S proteasome pathway. Mol Endocrinol. 2002;16:2819–2827. doi: 10.1210/me.2002-0154. [DOI] [PubMed] [Google Scholar]
- Licastro F, Candore G, Lio D, Porcellini E, Colonna-Romano G, Franceschi C, Caruso C. Innate immunity and inflammation in ageing: a key for understanding age-related diseases. Immun Ageing. 2005;2:8. doi: 10.1186/1742-4933-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Yamate T, Taguchi Y, Borba VZ, Girasole G, O’Brien CA, Bellido T, Abe E, Manolagas SC. Regulation of the gp80 and gp130 subunits of the IL-6 receptor by sex steroids in the murine bone marrow. J Clin Invest. 1997;100:1980–1990. doi: 10.1172/JCI119729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Sci Signal. 2008;1:e1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- Oliva J, Dedes J, Li J, French SW, Bardag-Gorce F. Epigenetics of proteasome inhibition in the liver of rats fed ethanol chronically. World J Gastroenterol. 2009;15:705–712. doi: 10.3748/wjg.15.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnappan U, Zhong M, Trebilcock GU. Decreased proteasome-mediated degradation in T cells from the elderly: A role in immune senescence. Cell Immunol. 1999;192:167–174. doi: 10.1006/cimm.1998.1418. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, Seong RH. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- St-Germain JR, Chen J, Li Q. Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics. 2008;3:342–349. doi: 10.4161/epi.3.6.7203. [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]