Abstract
The Dicistroviridae intergenic region (IGR) internal ribosome entry site (IRES) RNAs drive a cap-independent pathway of translation initiation, recruiting both the small and large ribosomal subunits to the viral RNA without the use of any canonical translation initiation factors. This ability is conferred by the folded, three-dimensional structure of the IRES RNA, which has been solved by X-ray crystallography. Here, we report chemical probing of the Plautia stali intestine virus (PSIV) IGR IRES in the unbound form, in the 40S subunit-bound form, and in the 80S-ribosome bound form. The results, when combined with an analysis of the crystal structures, suggest that parts of the IRES RNA change structure as the preinitiation complex forms. Using mutagenesis coupled with native gel electrophoresis, preinitiation complex assembly assays, and translation initiation assays, we show that these potentially structurally dynamic elements of the IRES are involved in different steps in the pathway of ribosome recruitment and translation initiation. Like tRNAs, it appears that the IGR IRES undergoes local structural changes that are coordinated with structural changes in the ribosome, and these are critical for the IRES’ mechanism of action.
Keywords: IRES RNA, ribosome, structural dynamics, translation initiation, RNA structure, viral RNA
Introduction
Some viral internal ribosome entry site (IRES) RNAs initiate translation of a downstream message using a mechanism that differs dramatically from the canonical protein-based mechanism. In canonical translation initiation, the mRNA’s 5′ 7-methyl guanosine cap first binds eukaryotic initiation factor (eIF) 4E, followed by the action of at least twelve protein factors working in a coordinated, stepwise fashion to recruit the small (40S) ribosomal subunit, scan the mRNA to find the correct start codon, and assemble the 80S ribosome to synthesize the encoded protein.1 Viral IRESs recruit, position, and activate host cell ribosomes to viral RNA in a cap- and end-independent pathway that often requires fewer eIF proteins than are used in the canonical pathway.2,3
IRES RNAs vary in terms of their folded structure and functional requirements for protein factors.4 The most streamlined IRES mechanism yet discovered is found within the intergenic region (IGR) of the Dicistroviridae single-stranded positive-sense RNA viral genomes,5,6 which were first discovered nearly a decade ago.7,8,9 The IGR IRESs do not require any eIFs to bind the 40S and 60S subunits in a GTP hydrolysis-independent fashion, thus they are essentially all-RNA ribosome assembling structures.10,11,12,13 Furthermore, the IGR IRESs initiate translation from the A-site of the ribosome and from a non-AUG codon, they do not require initiator tRNA, and they are able to drive the first translocation step before a peptide bond is formed.7,8,14 These features indicate that the IGR IRESs are active manipulators of the ribosome. Understanding the mechanism by which the IGR IRESs manipulate the eukaryotic translation machinery and initiate translation not only lends insight into IRES function, but also increases our understanding of translation initiation in general and may reveal how the ribosome can be manipulated and regulated by other structured macromolecules.
The IGR IRES RNA’s function is conferred by its structure, which is divided into three secondary structural regions and contains three pseudoknots.15 Although members of the IGR IRES group have different sequences and there are differences in the secondary structure of the type 1 and type 2 IGR IRESs, evidence suggests that their three-dimensional folded structures are very similar.16 Regions 1 and 2 together adopt a compact nested double-pseudoknot structure that folds independently from region 3 and is sufficient to bind the 80S ribosome (Figure 1(a)).10,12,17 The structure of the folded domain 1+2 (“ribosome binding domain; RBD”) from the Plautia stali intestine virus (PSIV) and region 3 (“P-site domain”) from the cricket paralysis virus (CrPV) were recently solved by X–ray crystallography,18,19 yielding an essentially complete structural model for an IGR IRES RNA. These structures help to place the IGR IRES in context with other folded RNAs,20 and when combined with cryo-EM reconstructions can be used to predict the locations of IRES-ribosome contacts.21,22,23 Recent biochemical and genetics experiments are revealing more details of how these structured RNAs interact with the ribosome and the mechanistic features of these streamlined IRESs.24,25,26,27
Figure 1.
Structural and mechanistic features of the PSIV IGR IRES. (a) Secondary structure diagram of the type 1 IGR IRES RNAs. Structural features that are explored in this paper are labeled. Regions 1 and 2, which comprise the ribosome binding domain (RBD), are shaded. Region 3 (P-site domain) is boxed but not shaded. (b) Crystal structure of the RBD of the PSIV IGR IRES. The structure is colored to reflect the crystallographic B-factors; cooler colors (blues, greens) reflect lower B-factors, warmer colors (red, orange, yellow) reflect higher B-factors. Important structural features are labeled.
One interesting feature of the structure of the PSIV IGR IRES RBD is the presence of disordered and thus potentially structurally dynamic regions (Figure 1(b)).18 Specifically, certain parts of the PSIV IRES RNA RBD possess higher crystallographic B-factors. Higher B-factors can be the result of crystal packing but also can indicate parts of the molecule that are flexible in solution. Indeed, some of these regions also appear to be unstructured in cryo-EM reconstructions of the CrPV IRES bound to the ribosome,21,22 suggesting this is an authentic and conserved feature of the RNA structure. If so, the presence and action of these structurally flexible elements may be important for IRES function.
Based on the above observations, we hypothesized that parts of the IGR IRES that are less structured in the crystal are important for interacting with the translation machinery and these elements undergo structural changes during the progression from free IRES to the IRES·40S subunit complex to IRES·80S ribosomes. Furthermore, we hypothesized that mutation of these regions would inhibit the ability of the IRES to form 80S ribosomes. To test these hypotheses, we used the PSIV IGR IRES as a model and used chemical probing to identify IRES elements that change structure upon interacting with the 40S subunit and 80S ribosomes. By using mutagenesis coupled with preinitiation complex assembly assays, we reveal how these potentially conformationally dynamic regions contribute to preinitiation complex formation.
Results
The PSIV IGR IRES RBD crystal structure contains flexible elements
Examination of the refined crystal structure of the PSIV IGR IRES RBD reveals that certain parts of this RNA possess high crystallographic B-factors relative to other parts (Figure 1(b)).18 Specifically, loop L1.1 and adjacent helix P1.1 have higher relative B-factors, and the electron density associated with these portions was of lower quality than in other parts of the RNA. This is also true of pseudoknot (PK) 2 and the adjacent loop L1.2. Higher relative crystallographic B-factors do not prove that these regions are flexible in solution, but they provide the basis for a hypothesis that parts of the IRES are structurally dynamic, and that changes in IRES RNA structure might be functionally important for ribosome assembly on the PSIV IGR IRES.
Local structural changes detected by chemical probing
To explore changes in the structure of the PSIV IGR IRES upon 40S subunit and 80S ribosome binding we performed selective 2′-OH acylation analyzed by primer extension (SHAPE) on RNA in the absence and presence of the ribosome using N-methylisatoic anhydride (NMIA) as the modifying agent.28 We previously reported the use of this experiment to probe the P-site domain of the PSIV IGR IRES;19 here we extend that analysis to the larger RBD. In the unbound state, several areas of the RNA react with NMIA (Figure 2(a)&(b)). Interestingly, these more reactive regions in the RBD include those with higher crystallographic B-factors: L1.1, P1.1, and L1.2. In addition, the apical loops of SL IV and SL V react with the chemical, as do parts of the P-site domain (as previously reported).19 Reactivity of RNA to NMIA is enhanced in areas of local flexibility or at nucleotides that are constrained in a conformation that make the 2′-OH particularly susceptible to modification.28,29 The fact that we observe groups of reactive nucleotides in several regions is consistent with these areas being flexible in the unbound IRES RBD.
Figure 2.
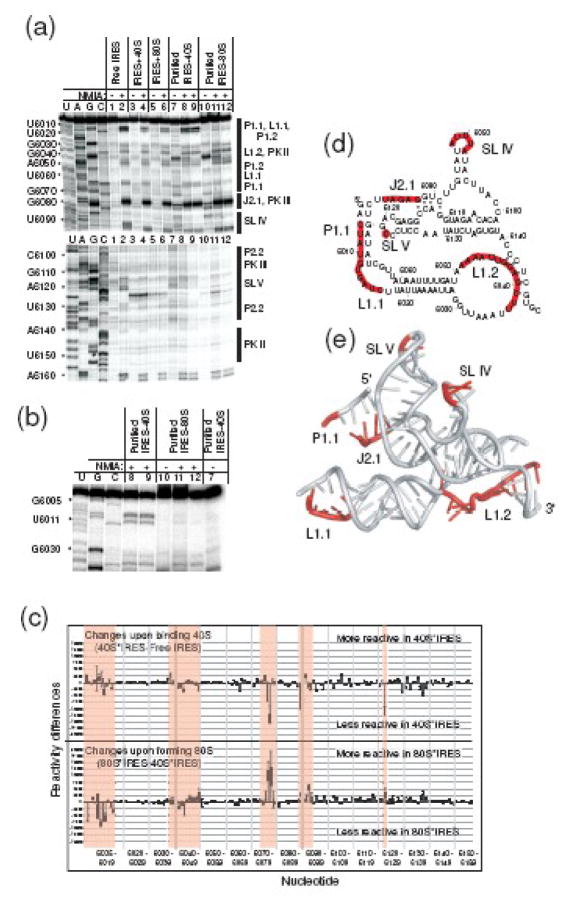
SHAPE analysis of PSIV IGR IRES in the unbound and bound forms. (a) Composite of several SHAPE gels. Nucleotide positions are shown on the left, secondary structural elements are shown on the right. Lanes: U,A,G,C (dideoxy sequencing reactions); 1 (free IRES+DMSO); 2 (free IRES+NMIA); 3 (IRES·40S+DMSO); 4 (IRES·40S+NMIA); 5 (IRES·80S+DMSO); 6 (IRES·80S+NMIA); 7 (purified IRES·40S+DMSO); 8 (purified IRES·40S+NMIA); 9 (identical to lane 8); 10 (purified IRES·80S+DMSO); 11 (purified IRES·80S+NMIA); 12 (identical to lane 11). The IRES+40S and IRES+80S samples consist of the IRES and salt-washed subunits or ribosomes that were probed and the RNA was analyzed directly. The samples “purified IRES·80S” and “purified IRES·40S” were assembled using the salt-washed ribosomes or subunits, probed, then purified on a sucrose gradient prior to analysis of the RNA. (b) Close-up view of a portion of a probing gel, showing the extreme 5′ end of the IRES RNA; lanes are as in panel A. (c) Quantification of data gathered from multiple SHAPE experiments and analyzed using SAFA.40 The top graph is the difference in modification between the 40S subunit-bound and free states, and the bottom is the difference between the 80S ribosome-bound and 40S-subunit bound states. The data used for the calculations were from the purified IRES·80S and purified IRES·40S samples. Error bars denote one standard error from the mean. Parts of the RNA that show substantial changes in modification are shaded; we termed these as hotspots. (d) Secondary structure of the PSIV IGR IRES with hotspots mapped onto the structure. (e) Hotspots mapped onto the crystal structure of the PSIV IGR IRES RBD. Important structural features are labeled.
To examine the RNA conformation in the bound state, purified PSIV IRES RNA was incubated with either isolated salt-washed 40S subunits or salt-washed 80S ribosomes and these complexes were probed by NMIA. Then, the RNA was either directly analyzed by reverse transcription (RT) and gel electrophoresis (IRES+40S, IRES+80S lanes, Figure 2(a)), or the complexes were purified by sucrose gradient electrophoresis and then analyzed by RT and gel electrophoresis (purified IRES+40S, purified IRES+80, Figure 2(a)).19 SHAPE probing on purified IRES·40S subunit shows that when the IRES binds to the 40S subunit the pattern of modification changes when compared to the unbound state. In addition, probing in the context of 80S ribosomes reveals additional changes in the modification pattern (Figure 2(a)&(b)). Although there are nucleotides throughout the RNA that change their modification levels, there are several areas where the changes are grouped, which we termed hotspots (Figure 2(c)&(d)). An example of one of these hotspots is SL IV. Also, SL V is not a hotspot per se, but does contain a nucleotide whose reactivity changes reproducibly when the free, 40S subunit-bound, and 80S ribosome-bound patterns are compared. The fact that SL IV and SL V reactivity generally decreases upon 40S subunit binding is consistent with the fact that they interact directly with this subunit.10,12,17 The observation that their reactivity increases upon joining of the 60S subunit (to form an 80S ribosome) suggests that their interaction with the 40S subunit changes upon 60S binding, perhaps by adopting a more reactive conformation or by partially releasing from the 40S subunit. Other hotspot regions include P1.1, L1.1, and L1.2 (Figure 2(c)&(d)), which are also areas with higher crystallographic B-factors. The fact that the reactive regions correspond to less ordered areas of the crystal provides additional evidence that our probing has identified flexible parts of the RNA. Individual nucleotides within these regions showed variability in their levels of modification (e.g. 6005–6007 versus 6010–6012), but for the purposes of identifying areas of the IRES (other than SL IV and SLV) to be targeted by mutagenesis we looked for groups of modifications and generally did not focus on the behavior of individual nucleotides within these regions.
The hotspot areas were mapped onto a model of the crystal structure of the RBD (Figure 2E). In some cases, the regions of SHAPE change are at or near sites that are proposed to interact directly with the ribosome.19,21 Thus, part of the changes in modification that we observe could be due to a footprinting effect in which the NMIA becomes sterically excluded upon binding of the ribosome to the IRES. However, this type of foot-printing effect can only account for regions that become less reactive upon binding, not those that become more reactive (of which there are several). Regions might become more reactive because they become single-stranded, or they might be locked in a distorted conformation that is particularly reactive,30 while still interacting with the ribosome. Hence, it seems likely that the changes in modification that we observe are due in part to local changes in RNA structure or in the details of their interaction with the ribosome during preinitiation complex assembly. For the purposes of this study, we did not attempt to delineate whether changes in SHAPE were due to changes in flexibility, structure or foot-printing, but rather used these results to guide subsequent mutagenesis coupled with functional studies.
Loop L1.1 is important for 80S ribosome formation
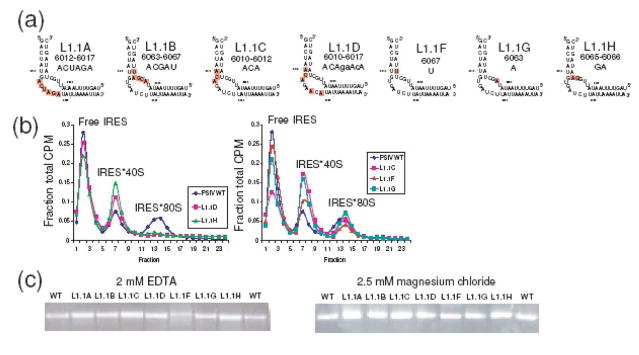
Loop L1.1 is one of the elements identified in our SHAPE probing. Different nucleotides within this region show different reactivity, but when viewed as a whole, the SHAPE probing suggests it is a region whose structure changes during preinitiation complex formation. Cryo-EM, biochemical, and crystallographic studies suggest this element contacts the L1 stalk of the 60S ribosomal subunit and is flexible in the crystal.12,18,21 We previously showed that complete mutation of each side of this loop to its Watson-Crick complement results in IRESs that are not structurally altered in the unbound state and can bind 40S subunits but fail to progress to 80S ribosomes (mutants L1.1A, L1.1B, Figure 3(a)).18 To explore how much of the conserved sequence in L1.1 is necessary for this function, five L1.1 mutant RNAs were subjected to native gel electrophoresis and preinitiation complex assembly assays in rabbit reticulocyte lysate (RRL) (Figure 3(a)–(c)). We chose to use RRL (rather than purified ribosomal components) to observe each mutant’s ability to progress through preinitiation complex formation in a system that contains all translational components and is thus similar to physiological conditions (to include competition for the ribosomal subunits by initiation factors). Although mutations to many other parts of the IRES cause retardation on native gels that are consistent with structural changes,17,18 native gel electrophoresis reveals no distortion of the solution-state structure of the IRES by the L1.1 mutations (Figure 3(c)). However, some mutants (L1.1D and L1.1H) cannot form IRES·80S ribosomes (Figure 3(b)), and thus behaved identically to previously-published mutants L1.1A and L1.1B.18 We note that in these mutants, the amount of IRES·40S subunit is elevated relative to wild-type, which is consistent with the interpretation that these mutants bind to 40S subunit, but then the complex accumulates at the 40S-bound stage as it is unable to progress to 80S ribosomes. In the case of L1.1H, mutation of only two nucleotides, C6065 and U6066, was sufficient to prevent 80S ribosome formation, illustrating the specificity of the IRES interaction with the 60S subunit. Mutants L1.1C, L1.1F, and L1.1G did not have as strong an impact on the IRES’ capability to form IRES·80S ribosomes (Figure 3(b)), thus the mutated bases in these RNAs are not critical for the IRES-60S subunit interaction. Comparison of mutant L1.1D (failed to make IRES·80S ribosomes) with mutant L1.1C (makes IRES·80S ribosomes) shows that they differ only by the mutation of two nucleotides (U6015 and U6017), suggesting both are critical. The observation that mutation of L1.1 does not alter the structure of the unbound IRES RNA, but mutation of as few as two nucleotides in this element is enough to block IRES·80S ribosome formation, is consistent with L1.1 being an unstructured element in the free state that must subsequently form a specific structure to make specific interactions with the 60S subunit.
Figure 3.
Analysis of L1.1 mutants. (a) Diagram of seven mutations to L1.1. L1.1A and L1.1B (at left) were analyzed previously,16 but their sequences are included here for comparison. (b) Ribosome assembly assay of wild type, L1.1D, and L1.1H (middle) and wild type, L1.1C, L1.1F, and L1.1G (right). (c) Nondenaturing (native) gels of wild type PSIV IRES and these seven mutants in the presence of 2 mM EDTA (partially denatured) and 2.5 mM MgCl2 (folded/native).
Helix P1.1 is necessary for forming a stable 80S-IRES complex
In our SHAPE analysis, helix P1.1 is a hotspot and both the crystal structure and cryo-EM reconstructions suggest this helix is disordered in the free state.18,21,22 However, IGR IRES phylogeny suggests it is a conserved element of the IRES secondary structure.15 To determine if helix P1.1 forms and serves a function in the IGR IRES mechanism, we performed preinitiation complex assays using mutants in which each side of the helix was mutated to its Watson-Crick complement (P1.1A and P1.1B), which abrogated formation of the helix (Figure 4(a)). A double mutant in which potential base-pairing was restored (P1.1C) was also analyzed. In addition, the nucleotides mutated in P1.1A are part of one of the strongest hotspots identified in our SHAPE experiments. Native gels in the presence of 2 mM EDTA or 2.5 mM MgCl2 (Figure 4(c)) show that these mutations affect the structure of the unbound RNA. Specifically, in the presence of 2 mM EDTA we expect some secondary structure to form, but not the compact three-dimensional fold, and under this condition both P1.1A and P1.1B run aberrantly compared to wild type. In the presence of MgCl2, P1.1A and P1.1B migrate at a similar rate to wild type, but are more diffuse. The most straightforward interpretation of this result is that when folded, these two RNAs are able to adopt an overall size and shape that is similar to wild-type, but that some conformational exchange has been introduced. If this interpretation is valid, it indicates that P1.1 breathes until interactions with the ribosome occur. The double mutant, P1.1C, runs identically to wild type on both gels thus restoring the base-pairing potential of P1.1 results in wild type folding characteristics.
Figure 4.
Analysis of P1.1 mutants. (a) Diagram of three mutations to P1.1. (b) Ribosome assembly assay of wild type, P1.1A, P1.1B, and P1.1C. Error bars represent one standard error from the mean (c) Nondenaturing (native) gels of wild type PSIV IGR IRES and these three mutants in the presence of 2 mM EDTA (partially denatured) and 2.5 mM MgCl2 (folded/native).
Preinitiation complex assembly assays showed that mutants P1.1A and P1.1B form stable complexes with the 40S subunit, consistent with previously published studies that establish region 2 as the primary determinant of 40S subunit binding (Figure 4(b)). However, both P1.1A and P1.1B were unable to form stable IRES·80S ribosomes, indicating that this helix is essential for stable 60S subunit association. In contrast, P1.1C forms stable IRES·80S ribosomes as efficiently as wild type. Taken together, our results suggest that although helix P1.1 may initially be disordered, its formation is essential to form stable IRES·80S ribosomes. Potentially, this helix constrains the unstructured L1.1 nucleotides so they can become structured and interact with the 60S subunit. Stabilization of P1.1 then would be coupled to the induction of structure in L1.1, and this is consistent with our probing results.
The role of L1.2 in 80S ribosome formation and translation initiation
In the IGR IRES’ mechanism, the P-site domain is placed in the 40S subunit’s decoding groove, and cryo-EM studies show that the position of this domain in relation to the rest of the IRES changes during the progression to 80S ribosomes.22 The P-site domain is connected to the rest of the IRES by PK 2, which is flanked by L1.2. In the crystal structure, L1.2 has higher relative B-factors and parts of this loop are identified in our SHAPE probing (Figure 2(d)), suggesting L1.2 changes shape during preinitiation complex formation on the IRES.
To probe the role of L1.2, we coupled mutagenesis with the preinitiation complex assembly assay and native gel electrophoresis. Because this loop is more conserved in length than sequence, we generated mutants with insertions or deletions (Figure 5(a)). The insertions were made in multiple places in the A-rich regions of the loop in order to increase the overall flexibility of the region and perhaps introduce slack into the connection between the RBD and the P-site domain, and also to reduce this region’s ability to interact with the adjacent helix through A-minor interactions.18 Conversely, the deletion mutants were designed to restrict the flexibility of the loop. In native gels (Figure 5(d)), the insertion mutants (L1.2C, L1.2D, L1.2E) migrated a bit slower compared to wild type, and this retardation increased when the mutants are folded in MgCl2. This suggests that either these mutants are more flexible, which limits them from adopting a shape as compact as wild type, or the difference in size between the mutant and wild type is responsible for the difference in the migration rates. In the case of the deletion mutants (L1.2A and L1.2B), they migrated similarly to wild type when folded, indicating a similar conformation. In fact, L1.1B migrated slightly faster than wild type and thus may have a more compact conformation than wild type. In 2 mM EDTA, the deletion mutants migrated with wild-type, but with subtle differences that can be attributed to the differences in size. All of the mutants were able to form stable 40S·IRES complexes as well as wild type (Figure 5(b)). However, the mutants displayed differing abilities to form stable 80S·IRES ribosomes (Figure 5(b)&(c)). Specifically, the deletion mutant L1.2A was able to form the 80S-IRES complex at least as efficiently as wild type. Another deletion mutant, L1.2B, forms 80S ribosomes similarly to L1.2A, but at slightly lower levels than wild type, based on the statistical analysis (Figure 5(c)). The insertion mutants all show a decrease in their ability to form 80S ribosomes, although none appear to be completely deficient.
Figure 5.
Analysis of L1.2 mutants. (a) Diagram of five mutations to L1.2. (b) Ribosome assembly assay of these five mutants plus wild type. (c) Quantification of multiple preinitiation complex assembly assays of wild type and L1.2A-E, with the ratio of IRES·80S to IRES·40S shown for each RNA. Error bars represent one standard error from the mean. (d) Nondenaturing (native) gels of wild type and these five mutants in the presence of 2 mM EDTA (partially denatured) and 2.5 mM MgCl2 (folded/native).
The fact that the L1.2 mutants are all able to form 80S ribosomes to some degree raises the question of whether or not these ribosomes can synthesize protein. We measured the translation initiation ability of these mutants using a bicistronic (dual luciferase) vector and RRL, and compared the ratio of upstream (non-IRES) translation to downstream (IRES) translation for each mutant. All L1.2 mutants show a decrease in their ability to initiate translation of a downstream reporter (Table 1), with the greatest effect being displayed by L1.2A. Note that L1.2A showed nearly wild-type ability to make 80S ribosomes, therefore the failure of this mutant to make protein must be in a post-80S ribosome formation step. The degree to which this effect is due to a loss of conformational flexibility in L1.2 or an alteration in the ability of the P-site domain to be placed properly through structural changes in PK 2 remains to be explored.
Table 1.
| Mutant | FLUC/RLUC (% of wild type) | 80S/40S ratio (from Fig. 5C) |
|---|---|---|
| Wild-type | 100 % | 1.0 +/− 0.2 |
| L1.2A | 17 +/− 3 % | 0.60 +/− 0.20 |
| L1.2B | 26 +/− 5 % | 0.55 +/− 0.04 |
| L1.2C | 40 +/− 5 % | 0.35 +/− 0.03 |
| L1.2D | 26 +/− 4 % | 0.29 +/− 0.02 |
| L1.2E | N/D | 0.35 +/− 0.05 |
Discussion
The function of structurally complex RNAs often depends not only on their three-dimensional folded structure but also on changes within that structure. Previously published chemical probing, cryo-EM, and X-ray crystallographic studies of the PSIV and CrPV IRESs hinted at conformational flexibility within the IRES RNA.10,12,16,17,18,19,21,22 In this study, we identified elements of the IGR IRES RNAs that may change structure and examined the role of each in preinitiation complex formation.
Our SHAPE probing suggests that parts of the IRES RNA undergo conformational changes during preinitiation complex assembly, but that these changes are likely not global structural rearrangements. This conclusion is based on the observation that the areas of change largely are localized to parts of the structure that are on the exterior of the folded RNA (Figure 2(e)). Were the folded IGR IRES RNA to undergo a dramatic rearrangement in which secondary and tertiary structure change, we would observe reproducible changes in our probing in those regions that are within the tightly folded interior of the structure. The areas identified by SHAPE correlate well with areas with higher B-factors in the crystal structure of the free IRES (Figure 1(b)) and match some predicted sites of IGR IRES-ribosome interactions.21,22 Taken together, these converging results suggest that these specific parts of the IGR IRES are areas of local RNA conformational changes and ribosome interactions that occur at different stages of preinitiation complex formation.
The specific regions we identified in the IRES RBD as undergoing conformational changes include L1.1, P1.1, L1.2, SL IV, and SL V. The mechanistic role of some of these structural features (L1.1, SL IV, SL V) has been partially explored through mutagenesis coupled with functional and biochemical assays.5,10,12,17,18,19,25,27,31 However, for others (P1.1, L1.2), the step at which these elements act, and the effect of altering their sequence or structure, has not been explored previously.
In the case of SL IV and SL V, it is clear that these stem-loops are a critical determinant of initial 40S subunit recruitment to the ribosome.10,12,17 Interestingly, the conformation of the 40S subunit changes when the IGR IRES binds and then changes again when the IRES·80S ribosome is formed.22 We observe that these stem-loops’ SHAPE reactivity changes upon binding to the 40S subunit and then changes again when the 80S ribosome forms, suggesting local changes in the intermolecular interactions that involve the stem-loops. Hence, it is possible that the structural changes we observe in SL IV and SL V during the progression from unbound IRES to 40S subunit-bound IRES to 80S ribosome-bound IRES reflect a changing interaction related to the 40S subunit conformation.
L1.1 has been implicated in making direct contact to the L1 stalk of the 60S subunit,12,18,21 and this element is needed for stable 80S ribosome association with the PSIV and CrPV IGR IRESs and for positioning the P-site domain correctly on the ribosome.18,25 Previously published data show L1.1 is unstructured in the unbound IRES.10,16 Our data suggest that the interaction between the highly conserved L1.1 and the L1 stalk is specific, strongly supporting a mechanism in which an unstructured L1.1 becomes structured when bound to the 80S ribosome.25 This restructuring event then might lead to an allosteric change that propagates through the IGR IRES structure to position the P-site domain.
P1.1 is predicted to form in all known examples of IGR IRESs,5,6,15 but this element showed reactivity in our probing analyses that is consistent with an unstable helix. We found that the formation of P1.1 is not needed for binding of the 40S subunit, consistent with observations that region 2 alone can bind 40S subunits.12 However, our data shows that formation of a P1.1 helical element after 40S subunit binding, and not the sequence of the helix itself, is needed for 80S ribosome formation. Furthermore, the fact that P1.1 is adjacent to L1.1 suggests the action of the two may be coupled in some way.
L1.2 links PK 2 to region 1, and although poorly conserved in terms of sequence, it tends to be A-rich and relatively well-conserved in terms of size. In general, deleting nucleotides from this loop was less disruptive to 80S ribosome formation than was inserting nucleotides, but all of the L1.2 mutants had a decreased ability to initiate protein synthesis. L1.1 and its interaction with the L1 stalk has been shown to be involved in correct placement of the P-site domain on the ribosome,25 and L1.2 links L1.1 to the P-site domain through PK 2. It is therefore tempting to speculate that disruption of the length of the L1.2 strands affects proper transmission of an allosteric signal from L1.1 to the P-site domain upon 60S subunit binding.
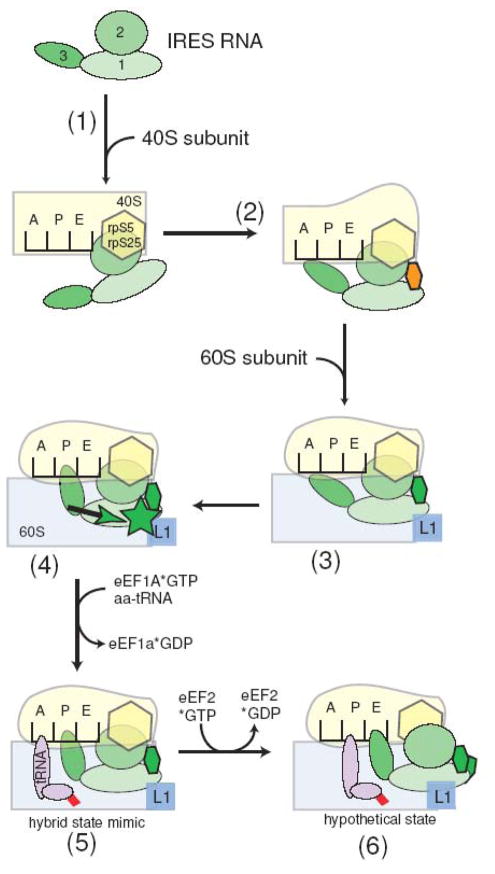
Combining the results reported here with previously reported functional and structural data suggests a model for IGR IRES function that can guide additional experiments (Figure 6). The unbound IGR IRES RNA recruits the 40S subunit using SL IV and SL V that interact (in part) with rpS5 and rpS25 (Figure 6, step (1)).21,22,27,32 This binding event induces local changes in the IRES structure that include partial placement of the P-site domain in the 40S subunit’s decoding groove, and also alteration of the conformation of the 40S subunit (Figure 6, step (2)).21,22 Recruitment of the 60S subunit then leads to an interaction between IRES element L1.1 and the L1 stalk that is made possible by P1.1, and that is critical for stable 80S ribosome formation (Figure 6, step (3)).18,25 The 60S subunit-IRES interaction leads to a structural change that propagates through L1.2 to PK 2 and properly positions the P-site domain (Figure 6, step (4)).25 The idea that L1.2 is a conduit for this structural change is supported by structural features of the IRES. In the crystal structure, L1.2 cradles P2.2 in such a way that region 2 could likely move or slide relative to region 1.18,20 In other words, L1.2 and the adjacent helix could act analogously to a piston that mechanically transmits the structural changes occurring in L1.1 and P1.1 to the P-site domain. This idea provides a hypothetical structural explanation for the functional linkage between L1.1 and the position of the P-site domain reported by Jang et al.25 Once the P-site domain is positioned, several lines of evidence suggest that this domain precisely mimics a codon-anticodon interaction in the P-site,19,31,33 while the entire IRES mimics a P/E hybrid state tRNA.19,26 Subsequent delivery of an aminoacylated tRNA to the A-site by eEF1A leads to a configuration that is recognized by eukaryotic elongation factor 2 (eEF2), which catalyzes the first translocation event and leads to elongation (Figure 6, steps (5)&(6)).26 These last two steps have been explored biochemically, but data regarding the structure of the IRES or its precise position on the ribosome at these stages has not yet been obtained. This mechanistic model is consistent with published functional and structural data, but remains partially speculative and hence must be tested.
Figure 6.
Mechanistic model of IGR IRES function with proposed conformational changes. The IGR IRES is depicted in green with the three major domains labeled, the 40S subunit is pale yellow, the 60S subunit is pale blue, and tRNA is magenta with attached amino acid in red. Ribosomal features (rpS5, rpS25, and the L1 stalk) are shown. The orange oval represents a partially formed helix P1.1 and the green oval represents the state of P1.1 after 60S subunit binding. The green depicts our proposed reordering of L1.1 upon 60S subunit binding, and the green arrow shows the proposed allosteric change that transmits the reorganization of L1.1 to the positioning of the P-site domain.
Coordinated changes in RNA structure are common in complex biological machines. One example are tRNAs, which must maintain a certain three-dimensional structure, but also must change conformation while interacting and moving on the ribosome.34 In addition, the ribosome itself undergoes structural changes that are partially driven by the rRNA.35 In fact, an important feature of IGR IRES function is likely to be coupling of IGR IRES structural changes with ribosome structural changes. Another example is the spliceosome, in which multiple RNAs with many proteins act in a highly coordinated series of conformationally dynamic events that include RNA-RNA rearrangements.36,37 Within viral RNAs, one notable example (among many others) is the 5′ leader of the full-length HIV-1 RNA that can adopt alternate secondary structures that regulate the events occurring on the RNA.38 Overall, evidence from our study and published data suggest that the IGR IRES also uses structural changes in a coordinated series of events that are necessary for proper function. The degree to which this will be true of a greater set of viral IRES RNAs remains an open question.
Materials and Methods
Cloning and plasmid production
A plasmid containing PSIV IRES RNA sequence 6002–6192 (pPSIV7), and a plasmid containing sequence 6002–6146 (pPSIV5) were constructed using standard PCR and ligation cloning methods, as described.17 In both plasmids, the IRES sequence was flanked by a 5′ hammerhead ribozyme and a 3′ hepatitis delta ribozyme that cleaved during transcription to yield homogeneous termini. Mutants to this plasmid were made using the QuikChange kit (Stratagene).
RNA synthesis, purification, and end-labeling
RNA was generated from in vitro transcriptions from template DNA, purified, concentrated, and stored as described.16 RNA generated from a template containing a hammerhead ribozyme at the 5′-end already possesses a 5′-OH and was not dephosphorylated. Labeling with P32 then was done as described.39
40S/80S preinitiation complex assembly assays
To assemble preinitiation complexes in cell-free lysate, ~1,000 CPM of radiolabeled IRES RNA was heat denatured at 85°C for 40 to 60 sec, cooled on the bench, and folding was induced in a buffer containing 30 mM Na-HEPES, pH 7.5 and 2.5 mM MgCl2. The RNA was added to 33 μL of rabbit reticulocyte lysate (RRL) (Promega) and RNase-free water to a final volume of 50 μL. The reactions were incubated at 30°C for 15 min, and then 250 μL of 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT was added and the reaction was placed on ice. Reactions were immediately loaded on 15–30% sucrose gradients in 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, and 1 mM DTT prepared in tubes (Seton) designed for a SW41 rotor using the automated mixing procedure, 15%–30% “long” sucrose, on a BioComp Gradient Master apparatus. Reactions were centrifuged at 4°C for 3 hours at 36,000 RPM, then fractionated using the BioComp Gradient Master. Fractions were analyzed by blotting onto a sandwich of nitrocellulose and nylon membranes and quantified using a phosphorimager and the software ImageQuant. The 80S-IRES and 40S-IRES complex peaks were assigned based on location in the gradient from previous studies.16,25,33
Native gel electrophoresis
An approach similar to the one previously described was used.17 Briefly, approximately 1μg of RNA was denatured at 85°C for 1 min, cooled at room-temperature, and then folded in 66 mM TRIS, 34 mM HEPES, and either 2.5 mM MgCl2 or 2 mM EDTA, and trace amounts of bromophenol blue and xylene cyanol. After incubating at 37°C for 5 min, the RNA was loaded onto the appropriate 10% polyacrylamide gel containing 66 mM TRIS, 34 mM HEPES, and either 2.5 mM MgCl2 or 2 mM EDTA. The samples ran for several hours at 10W at 4°C until the xylene cyanol band migrated 9 cm. The gel was stained with ethidium bromide and imaged on an UV transilluminator.
SHAPE
SHAPE of RNA and RNA-ribosome complexes was conducted using salt-washed ribosomes and 40S subunits as described,19 and analyzed using the program SAFA.40 Quantitated results are the average of six experiments.
Translation initiation assays
Plasmids encoding the wild type and mutant PSIV IGR IRES between two luciferase reporter genes were generated by standard molecular cloning methods. Briefly, DNA containing the IRES sequence was generated by PCR and then ligated into the EcoR1/NcoI sites of the pRL vector (kind gift of Ann Willis),41 resulting in a plasmid with the IRES downstream of the Renilla luciferase gene (RLUC) and upstream of the Photinus luciferase gene (FLUC). Plasmids were amplified in E. coli and sequenced. Plasmids were linearized with BamH1 and used in in vitro transcription reactions. The resultant RNA was treated and used in translation assays with RRL as described.19 Results are based on three independent experiments.
Acknowledgments
The authors would like to thank Dr. Robert Batey, Dr. Thomas Cech, and David Costantino for critical reading of this manuscript. This work was supported by NIH grant R01GM072560 to JSK. JSK is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pestova T, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 2.Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. [Google Scholar]
- 3.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 4.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139:137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 11.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deniz N, Lenarcic EM, Landry DM, Thompson SR. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15:932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieft JS. Comparing the three-dimensional structures of Dicistroviridae IGR IRES RNAs with other viral RNA structures. Virus Res. 2009;130:148–156. doi: 10.1016/j.virusres.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 22.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM Visualization of a Viral Internal Ribosome Entry Site Bound to Human Ribosomes; The IRES Functions as an RNA-Based Translation Factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus IRES. J Biol Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 29.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicens Q, Gooding AR, Laederach A, Cech TR. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galkin O, Bentley AA, Gupta S, Compton BA, Mazumder B, Kinzy TG, Merrick WC, Hatzoglou M, Pestova TV, Hellen CU, Komar AA. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jan E, Thompson SR, Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb Symp Quant Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- 34.Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 35.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 38.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 40.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]