Abstract
The rapid evolution of telomere proteins has hindered identification of orthologs from diverse species and created the impression that certain groups of eukaryotes have largely non-overlapping sets of telomere proteins. However, the recent identification of additional telomere proteins from various model organisms has dispelled this notion by expanding our understanding of the composition, architecture and range of telomere protein complexes present in individual species. It is now apparent that versions of the budding yeast CST complex and mammalian shelterin are present in multiple phyla. While the precise subunit composition and architecture of these complexes vary between species, the general function is often conserved. Despite the overall conservation of telomere protein complexes, there is still considerable species specific variation, with some organisms having lost a particular subunit or even an entire complex. In some cases, complex components appear to have migrated between the telomere and the telomerase RNP. Finally, gene duplication has created telomere protein paralogs with novel functions. While one paralog may be part of a conserved telomere protein complex and have the expected function, the other paralog may serve in a completely different aspect of telomere biology.
Key terms: Shelterin, CST, OB-fold, Myb repeat, gene duplication, POT1, TRF1
Telomere proteins and telomere function
Telomeres have two main functions: to protect the chromosome terminus from unwanted nuclease and DNA repair activities, and to provide a mechanism to compensate for the inability of DNA polymerase to replicate the 5′ end of a linear chromosome (Verdun and Karlseder, 2007). Telomeric DNA is packaged by a core group of proteins that bind the DNA duplex and the 3′ G-overhang on the chromosome terminus (Rhodes et al., 2002). These telomere proteins form a protective cap that prevents the chromosome end from being sensed as DNA damage and eliciting a DNA damage response (Palm and de Lange, 2008). If the cap structure is defective, the resulting damage response leads to a cell cycle checkpoint and/or attempts to repair the chromosome end by non-homologous end joining (NHEJ) (Riha et al., 2006). The NHEJ leads to end-to-end fusion of chromosomes followed by chromosome breakage.
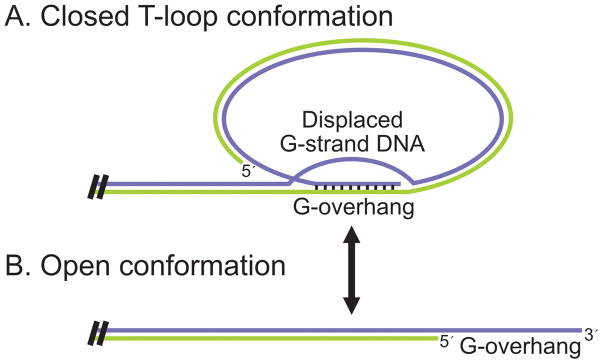
Although it is essential to have the DNA terminus sequestered from nuclease and DNA repair activities for much of the cell cycle, during S-phase the telomere must be made accessible to telomerase and the DNA replication machinery. To achieve this change in access, the overall chromatin structure of the telomere is thought to cycle from a closed protective configuration into a more open, S-phase conformation (Figure 1) (Gilson and Geli, 2007; Teixeira et al., 2004). In the protective configuration, access to the DNA terminus appears to be reduced by the telomeric tract folding back on its self to form a loop or hairpin type structure. In some organisms, the fold-back structure is stabilized by protein-protein interactions. However, in plants, vertebrates and some other organisms, the DNA appears to form a T-loop where the G-overhang invades and base-pairs with segments of the telomere duplex (Cesare et al., 2008; Cesare et al., 2003; de Lange, 2004; Griffith et al., 1999). During S-phase, the T-loop is probably removed by passage of the replication fork.
Figure 1.
Telomere cycling between (A) closed and (B) open conformations.
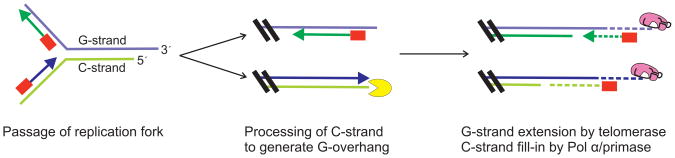
During DNA replication, most of the telomere is replicated by a standard replication fork. However, a number of extra steps are needed at the DNA terminus to ensure that telomere length is maintained and the protective cap structure is reassembled (Figure 2) (Gilson and Geli, 2007; Verdun and Karlseder, 2007). These steps include DNA processing to generate or extend the 3′ G-overhang on the leading or lagging strand telomere, extension of the overhang by telomerase, and fill in of the complementary C-strand by Pol α-primase. The process of telomere replication is achieved through the combined action of replication factors, telomere proteins and components of the DNA repair machinery (Verdun and Karlseder, 2007). The telomere proteins help recruit telomerase and Pol α-primase, and some may also facilitate passage of the replication fork through the telomeric duplex DNA (Bianchi and Shore, 2008; Miller et al., 2006; Sfeir et al., 2009). The repair factors recognize the DNA terminus and appear to initiate a transient DNA damage response that is an integral part of the replication process (Bianchi and Shore, 2008; Verdun and Karlseder, 2007). The initial damage response seems to result in phosphorylation of telomere proteins and modification of their activity (Tseng et al., 2006; Wu et al., 2007). However, activation of a full DNA damage response is prevented, most likely by binding of the telomere proteins at the DNA terminus (Palm and de Lange, 2008). Thus, telomere proteins function at multiple levels in telomere replication and chromosome end-protection.
Figure 2.
Steps during telomere replication. Adapted from (Price, 2009). Color figure available online.
In addition to telomere proteins and replication or repair factors, a large number of other proteins can be present at a telomere. For example, approximately 200 different proteins were identified when stretches of telomeric chromatin were isolated from human cells and analyzed by mass spectrometry (Dejardin and Kingston, 2009). In general, these 200 proteins differ from dedicated telomere proteins in that they tend to have well established, non-telomeric functions and to exhibit evolutionary conservation in their primary sequence. The telomeric role of most of these proteins is unclear but some may be needed to transcribe and package TERRA RNA (Telomere Repeat containing RNA) (Azzalin et al., 2007), while others may be involved in organizing higher order telomeric chromatin structure (Blasco, 2007).
Rapid evolution of telomere proteins
Given that telomeres from diverse organisms play the same essential role in chromosome maintenance (Bianchi and Shore, 2008), one might expect telomere proteins to be highly conserved. However, a simple amino acid sequence comparison quickly reveals that this is not the case (Table I). Even within a single taxonomic class such as mammals, telomere proteins demonstrate less conservation than other chromosomal proteins or DNA replication and repair factors. Some telomere proteins have clear orthologs in several phyla, but even these proteins have obviously undergone a rapid rate of change during evolution. Other telomere proteins that are essential for a particular function in one species, appear to be absent from a related species and that function is served by a completely different set of proteins. This striking variation in telomere protein composition initially suggested the telomere protein complexes from different organisms might have evolved independently.
Table I.
Percent identity between telomere proteins (TRF1, TRF2, RAP1, POT1), histones (H3), DNA repair factors (MRE11) and DNA replication factors (POLα and MCM2) from various species. Human (Hs), mouse (Mm), chicken (Gg), frog (Xl), or fission yeast (Sp).
| Protein | Hs vs Mm | Hs vs Gg | Hs vs Xl | Hs vs Sp |
|---|---|---|---|---|
| TRF1 | 65 | 42 | 34 | 11** |
| TRF2 | 82 | 33 | 36 | 9** |
| RAP1 | 84 | 39 | 32 | 10 |
| POT1 | 75(72)* | 61 | 50 | 10 |
| H3 | 98 | 97 | 97 | 91 |
| MRE11 | 88 | 74 | 71 | 34*** |
| POLα | 88 | 72 | 71 | 34 |
| MCM2 | 95 | 88 | 88 | 51 |
Mouse Pot1a (Pot1b)
SpTaz1
SpRad32
Subsequent structural analysis of telomere proteins from yeast, humans and ciliates revealed the presence of conserved protein motifs in the DNA-binding domain. This finding indicated a conservation of structure and function despite the lack of sequence identity. Recently, an even more striking level of conservation has become apparent. The identification of many additional telomere proteins has made it feasible to compare the overall protein composition of telomeres from multiple organisms. These comparisons indicate that, despite the rapid rate of telomere protein evolution, the core telomere protein complexes are in fact quite conserved. However, they show organism-specific variations in composition and function. In the following sections, we will explore this apparent “variation on a theme” by first examining the similarities and differences between the well characterized telomere protein complexes from budding yeast and humans. We will then discuss a series of newly identified proteins that have revealed the basal conservation but functional and compositional fluidity of telomere protein complexes.
Budding yeast versus mammalian telomere proteins: a different means to the same end?
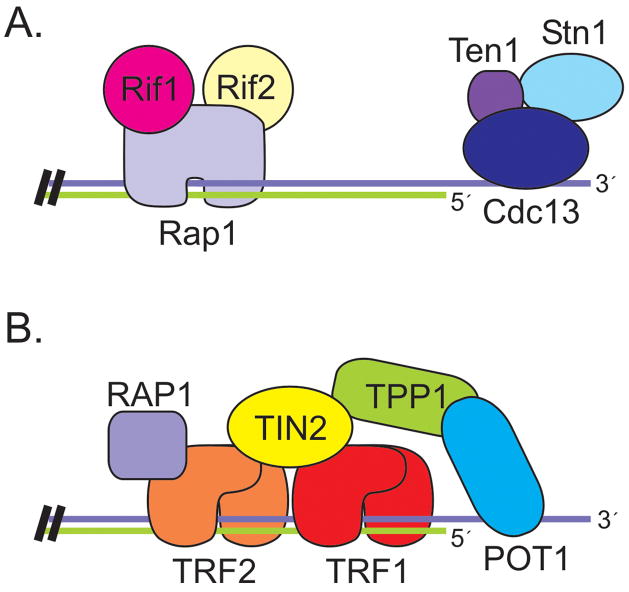
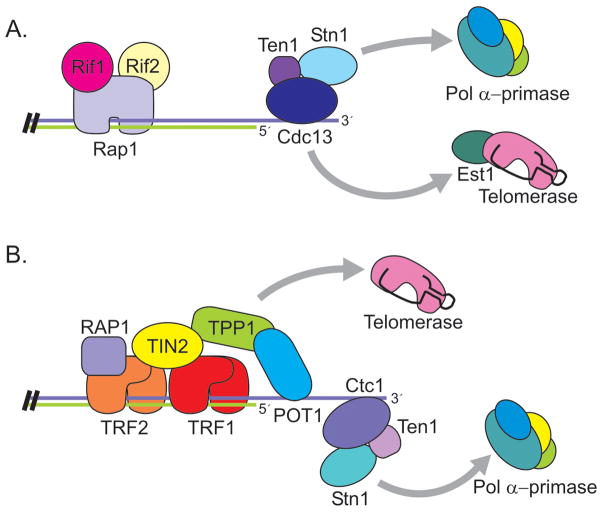
Telomeres from the budding yeast, S. cerevisiae, are bound by two separate protein complexes (Figure 3A): one that binds the G-strand overhang and one that binds the DNA duplex (Lundblad, 2006). The complex that interacts with the G-overhang does so through sequence-specific binding of the Cdc13 protein. Cdc13 recruits two binding partners, Stn1 and Ten1, to form a trimeric complex (Gao et al., 2007). The Cdc13/Stn1/Ten1 (CST) complex is essential for chromosome end-protection and defects in any one CST component lead to degradation of the C-strand, accumulation of ss G-rich telomeric DNA, a DNA-damage response and cell cycle arrest (Garvik et al., 1995; Grandin et al., 2001; Grandin et al., 1997). Stn1 and Ten1 seem to be primarily responsible for telomere protection as overexpression of Stn1 plus Ten1 rescues the lethality of Cdc13 depletion (Grandin et al., 1997; Petreaca et al., 2006).
Figure 3.
Organization of telomere protein complexes in (A) S. cerevisiae and (B) Human cells. Color figure available online.
The CST complex also plays a key role in telomere replication. Cdc13 recruits telomerase to the telomere through a direct interaction with the telomerase subunit Est1 (Bianchi et al., 2004; Pennock et al., 2001). In contrast, Stn1 is thought to inhibit telomerase action by competing with Est1 for Cdc13 binding (Chandra et al., 2001; Li et al., 2009; Puglisi et al., 2008). The interaction between Cdc13 and Est1 is mediated by a Cdk1-dependent phosphorylation of Cdc13 which promotes preferential binding to Est1 over Stn1 (Li et al., 2009). In addition to recruiting telomerase, CST recruits Pol α-primase, the enzyme needed for telomeric C-strand synthesis (Grossi et al., 2004; Qi and Zakian, 2000). Cdc13 interacts with Pol1, the catalytic subunit of Pol α-primase, while Stn1 interacts with the Pol12 accessory subunit. Thus, CST coordinates synthesis of both strands of the telomere. Cdc13 first mediates G-strand synthesis by direct recruitment of telomerase. Additional elongation is then blocked by Stn1, with Cdc13 and Stn1 subsequently recruiting the C-strand synthesis machinery (Chandra et al., 2001; Li et al., 2009; Puglisi et al., 2008).
The protein complex associated with the DNA duplex is bound to DNA by the Rap1 protein (Lundblad, 2006). The C-terminal domain of Rap1 then recruits two additional proteins, Rif1 and Rif2, which are responsible for telomere length regulation (Marcand et al., 1997). Length regulation occurs via a negative feedback loop where the increased binding of Rif1 and Rif2 at longer telomeres inhibits Tel1 (ATM) association (Hirano et al., 2009). Since Tel1 is required for efficient telomerase recruitment (Goudsouzian et al., 2006), this leads to less frequent extension of long telomeres (Bianchi and Shore, 2007; Chang et al., 2007; Hector et al., 2007; Sabourin et al., 2007; Viscardi et al., 2007). In addition to recruiting Rif1 and Rif2, Rap1 is also capable of recruiting a second group of proteins, Sir3 and Sir4, that promote heterochromatin formation and telomere silencing (Lundblad, 2006).
Human telomeres are bound by a protein complex called shelterin that contains six different proteins: TRF1, TRF2, TIN2, RAP1, TPP1, and POT1 (Liu et al., 2004a; Palm and de Lange, 2008). With the exception of RAP1, none of these proteins share obvious sequence identity with the telomere proteins from S. cerevisiae and even the homology between the Rap1 proteins is quite limited (Li et al., 2000). Unlike the yeast CST and Rap1 complexes, shelterin contains both dsDNA and G-overhang binding proteins. As a result, the telomere duplex and the G-overhang are physically linked (Figure 3B). The duplex DNA is bound by TRF1 and TRF2 while POT1 binds to the G-overhang. TIN2 and TPP1 then provide the bridge between TRF1/2 and POT1 (Houghtaling et al., 2004; Liu et al., 2004b; Ye et al., 2004a; Ye et al., 2004b). TIN2 is a central component of the complex as it binds simultaneously to TRF1, TRF2 and TPP1 while TPP1 binds POT1 and links it to TIN2. RAP1 interacts with TRF2. All six shelterin components can be isolated together from nuclear extracts, suggesting that they form a stable complex in the absence of telomeric DNA (Liu et al., 2004b; Ye et al., 2004b). A number of sub-complexes have also been identified, however their functions remain largely unknown (Kim et al., 2008; Liu et al., 2004b).
Although all six shelterin components help protect the chromosome terminus and prevent it from being recognized as DNA damage, TRF2 and POT1 play a key role in this process. POT1 prevents the G-overhang from activating an ATR-mediated DNA damage response by competing with RPA for binding to the G-strand DNA (Churikov and Price, 2008; Denchi and de Lange, 2007; Guo et al., 2007). TRF2 prevents removal of the G-overhang, activation of an ATM-mediated DNA damage response, and end-to-end fusion of chromosomes (Celli and de Lange, 2005; Denchi and de Lange, 2007; Konishi and de Lange, 2008). It is not clear exactly how TRF2 functions in this capacity, but its ability to assemble T-loops is probably important. Formation of a T-loop would protect the G-overhang from being removed by a nuclease, thus preventing the DNA terminus from becoming a substrate for NHEJ. TRF2 can promote the formation of higher-order DNA structures by binding DNA as a multimer and inducing positive supercoiling (Amiard et al., 2007; Poulet et al., 2009). The resulting DNA unwinding leads to strand invasion, a key step in T-loop formation (Stansel et al., 2001). It is notable that both human and yeast telomere proteins play an essential role in protecting the DNA terminus, but in humans the process is mediated in part by alteration of the DNA structure not merely through envelopment of the DNA by protein molecules.
As in yeast, human telomere proteins are important for telomere length regulation; however, their mode of action is not fully understood. Studies with TRF1, TRF2 and POT1/TPP1 suggest that shelterin components may function at several different levels (Smogorzewska and de Lange, 2004). First, loading of shelterin on the DNA duplex by TRF1 and TRF2 probably enhances formation of T-loops or another protective chromatin structure that restricts telomerase access (Amiard et al., 2007; Baker et al., 2009; Stansel et al., 2001). Second, shelterin binding may impact telomere length by increasing the amount of POT1 at the DNA terminus (Hockemeyer et al., 2007; Loayza and De Lange, 2003). POT1 appears to play a complex role in telomere length regulation. In vitro assays indicate that it can inhibit telomerase action by sequestering the DNA terminus (Lei et al., 2005). In vivo studies support this inhibitory activity as removal of POT1 from the telomere leads to telomerase-dependent telomere growth (Churikov and Price, 2008; Loayza and De Lange, 2003). Formation of a POT1/TPP1 dimer increases the in vitro affinity of POT1 for G-strand DNA by ~10 fold (Xin et al., 2007). This suggests that the in vivo association of POT1 with TPP1 would enhance binding of POT1 to the overhang and hence might increase sequestration of the DNA terminus from telomerase. However, in vitro studies indicate that this may not be the case. Rather, TPP1 can interact directly with telomerase and the POT1/TPP1 heterodimer actually enhances telomerase activity by increasing the processivity of repeat addition (Wang et al., 2007; Xin et al., 2007). Thus, POT1/TPP1 seems to act both as a positive and a negative regulator of telomerase. Currently, it is unclear how the protein complex switches between these two modes.
While the differences in composition and architecture of human and S. cerevisiae telomeres are striking, the general principals of telomere protein function are the same. In both cases, they protect against unwanted nuclease and repair activities by sequestering the DNA terminus, they actively promote telomerase activity through direct interactions with telomerase subunits, and they regulate telomere length by formation of a chromatin structure which limits telomerase access. These mechanistic similarities, together with the conserved nature of telomere maintenance by telomerase, have led to debate about the apparent lack of conservation in telomere protein composition and telomere architecture and whether this merely reflects an incomplete understanding of the composition and structure of the underlying protein complexes.
Conservation of Structural Domains
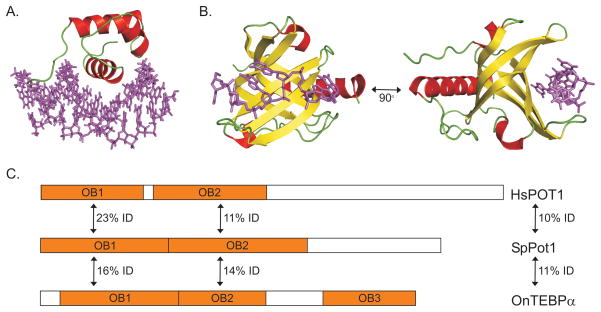
Although telomeric duplex and G-overhang binding proteins from different species frequently show minimal sequence identity, they do share common structural motifs. This first became apparent when S. cerevisiae Rap1 and human TRF1 were both found to contain Myb motifs in their DNA binding domain (DBD) (Bilaud et al., 1996; Chong et al., 1995; Konig and Rhodes, 1997). Myb motifs were subsequently found in mammalian TRF2, and in telomere duplex binding proteins from a variety of other organisms including S. pombe Taz1, Arabidopsis TRFLs and Trypanosome TRF (Bilaud et al., 1997; Broccoli et al., 1997; Cooper et al., 1997; Karamysheva et al., 2004; Li et al., 2005). The Myb motif has a well defined consensus sequence and it is composed of a distinct tertiary structure containing three helices with the second and third helices forming a helix-turn-helix variant motif (Figure 4A) (Ogata et al., 1994). The motif was originally found in the c-Myb transcriptional activator and it is frequently found in transcription factors (Klempnauer and Sippel, 1987). Interestingly, the physical arrangement of the Myb motifs used for DNA binding can vary. At least two Myb motifs are required for high affinity binding (Bianchi et al., 1997; Ogata et al., 1994), however, these may or may not be within the same protein subunit. S. cerevisiae Rap1 contains two motifs and can bind DNA as a monomer (Konig and Rhodes, 1997). In contrast, TRF1 and TRF2 contain only one Myb motif and they must form homodimers to generate a functional DNA-binding domain (Court et al., 2005). Human RAP1 resembles TRF1 and TRF2 in that it also contains only one Myb motif (Hanaoka et al., 2001). However, RAP1 does not dimerize which explains why it cannot bind DNA directly but must instead be tethered to shelterin via TRF2.
Figure 4.
Structural domains of telomeric DNA binding proteins. (A) Ribbon diagram of the Myb domain from human TRF1 bound to ds telomeric DNA. (B) OB-fold 1 (OB1) from human POT1 bound to telomeric G-strand DNA. Purple, telomeric DNA; red, α-helicies; yellow, β-strands; green, loops. (C) Domain structure of POT1 homologs from H. Sapiens, S. pombe and O. nova with % identity shown between OB1, OB2, or the full-length protein. Color figure available online.
Structural conservation between G-overhang binding proteins was more difficult to discern because the underlying protein motif, the OB- (Oligonucleotide/Oligosaccharide-Binding) fold, lacks a well defined consensus sequence (Theobald et al., 2003). Structurally, the OB-fold is composed of a five-stranded β-sheet that forms a closed β-barrel structure, usually capped by an α-helix (Figure 4B) (Murzin, 1993). It is a common protein domain that is frequently used to recognize single-stranded nucleic acids (Croy and Wuttke, 2006). The OB-fold was first identified as a telomeric G-strand DNA-binding motif when the crystal structure of Oxytricha nova TEBP (telomere end-binding protein) was solved (Horvath et al., 1998). TEBP functions as a heterodimer with the α-subunit binding the DNA via tandem OB-folds. The β-subunit alone does not bind DNA, but it also has an OB-fold through which it contacts the DNA, thus increasing the binding affinity of the heterodimer. The Pot1 protein family was eventually identified because the human and S. pombe proteins showed weak sequence homology (~15%) to the first OB-fold in the TEBPα DBD (Figure 4C) (Baumann and Cech, 2001). However, it only became apparent that the OB-fold motif is characteristic of telomeric G-strand binding proteins when the structure of the Cdc13 DBD was solved and found to contain an OB-fold closely related to those from TEBPα (Mitton-Fry et al., 2002). The DBDs of human and S. pombe Pot1 were subsequently crystallized and also found to contain related OB-folds (Lei et al., 2003; Lei et al., 2004).
Until recently, the absence of a conserved consensus sequence made it difficult to identify OB-fold-containing proteins from their primary sequence. However, an increase in the number of crystal structures, together with advances in protein threading programs, now allows OB-fold-containing regions to be recognized with reasonable certainty (Croy and Wuttke, 2006; Theobald and Wuttke, 2004). This has led to the identification of new telomeric ssDNA-binding proteins and to the detection of OB-folds in previously known telomere or telomerase-associated proteins. Two such newly identified telomere proteins include the C. elegans proteins CeOB1 and CeOB2 (Raices et al., 2008). CeOB1 appears to contain an OB-fold that is related to OB2 from human POT1 (Figure 4C) while CeOB2 is predicted to contain an OB-fold that is related to OB1. Interestingly, CeOB1 binds telomeric G-strand DNA while CeOB2 binds telomeric C-strand DNA.
OB-folds have now been identified in a surprisingly large number of previously characterized telomere or telomerase-associated proteins. Discovery of an OB-fold in a protein has in some cases lead to the realization that this protein is the structural and functional homolog of a telomere protein from another species. Human TPP1 is a case in point. TPP1 had been recognized as a POT1 interacting protein for some years; however, it only became apparent that TPP1 might be the mammalian homolog of O. nova TEBPβ when structure prediction suggested that TPP1 might contain an OB-fold. X-ray crystallography then revealed that the OB-folds from TPP1 and TEBPβ are closely related, while biochemical analysis indicated that TPP1 resembles TEBPβ in that it increases the affinity of the TPP1/POT1 (TEBPα/β) heterodimer for telomeric DNA (Wang et al., 2007; Xin et al., 2007).
Interestingly, the Est3 telomerase subunits from S. cerevisiae and C. albicans each appear to contain a single OB-fold that is structurally related to the OB-folds from TPP1 and TEBPβ (Lee et al., 2008; Yu et al., 2008). This structural similarity, together with the observation that TPP1 and TEBPβ both physically interact with telomerase and enhance telomerase catalytic activity (Paeschke et al., 2008; Xin et al., 2007), has lead to the suggestion that Est3, TPP1 and TEBPβ have evolved from a common ancestor (Lee et al., 2008; Yu et al., 2008). Presumably such a scenario would have involved the switching of the ancestoral protein from one multiprotein complex to become part of a different protein complex. Other examples of such subunit shuffling are discussed in a later section.
The new view of telomere protein complexes: Variations on a theme
The past couple of years have seen important advances in our overall understanding of the general form and function of several key telomere protein complexes. The critical step was identification of additional telomere proteins and telomere protein complexes from a range of organisms including Schizosaccharomyces pombe and Arabidposis thaliana. Analysis of these proteins or complexes has revealed that versions of shelterin and the CST complex exist in many organisms where they play similar, but not necessarily identical, roles.
Until recently, the organization of S. pombe telomeres remained enigmatic because the known telomere proteins resembled components of vertebrate shelterin rather than subunits of S. cerevisiae CST (Kanoh and Ishikawa, 2003). Specifically, S. pombe Pot1 shows no apparent sequence identity to Cdc13 but instead is orthologous to mammalian POT1 (Baumann and Cech, 2001). Likewise, Taz1 resembles TRF1 and TRF2 rather than budding yeast Rap1 in that it contains a single Myb motif and must form a homodimer in order to bind telomeric DNA (Cooper et al., 1997; Spink et al., 2000). S. pombe Rap1 also resembles mammalian Rap1 as it contains a single Myb motif but cannot dimerize and bind DNA directly (Kanoh and Ishikawa, 2001). It instead associates with the telomere via Taz1.
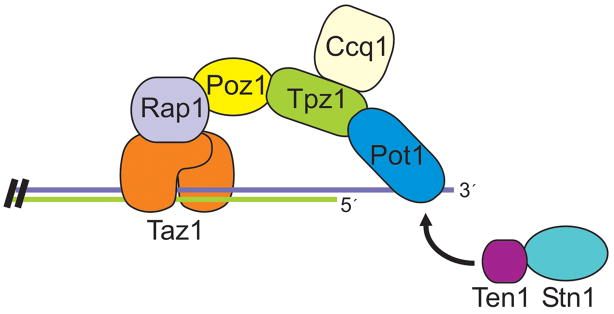
Despite the conservation between S. pombe and mammalian Pot1, Taz1/TRF1/2 and Rap1, other components of shelterin cannot be discerned in the S. pombe genome. Thus, it appeared that the overall organization of the telomere protein complexes must be different. However, the recent identification of a series of Pot1-interacting proteins has dispelled this idea. Purification of tagged Pot1 from S. pombe cells revealed a Pot1 complex that contained three additional proteins: Tpz1, Poz1 and Ccq1 (Miyoshi et al., 2008). Tpz1 and Poz1 are novel proteins with no apparent sequence identity to known telomere proteins. Ccq1 had previously been identified in a histone deacetylase complex and shown to function in bouquet formation during meiosis (Flory et al., 2004; Sugiyama et al., 2007). Analysis of the interactions between components of the Pot1 complex revealed a striking organizational similarity to shelterin (Figure. 5). Tpz1 binds to Pot1 and to Poz1 and seems to be the S. pombe homolog of TPP1. Poz1 appears to be functionally equivalent to mammalian TIN2 as it links the Pot1 complex to Taz1 and the DNA duplex via an interaction with Rap1. Thus, as in shelterin, the S. pombe telomere protein complex connects the G-strand overhang to the double-stranded region of the telomeric DNA. Ccq1 turns out to be a telomerase recruitment factor (Tomita and Cooper, 2008). It interacts directly with telomerase and serves as a bridge between telomerase and the G-overhang.
Figure 5.
Organization of the shelterin complex from S. pombe. Color figure available online.
It is striking that shelterin and the S. pombe protein complex not only have a similar architecture, but they also play much the same role in chromosome end-protection and telomerase recruitment. Although the fine details differ in terms of which duplex associated protein contacts the Pot1 complex or which protein recruits telomerase, the general principals are the same, suggesting that the S. pombe complex should also be called shelterin. It is notable that certain components of mammalian and S. pombe shelterin are obviously conserved, while other proteins have either evolved extremely rapidly or arisen from different origins. It will be interesting to learn the extent to which Tpz1 and Poz1 resemble TPP1 and TIN2 from a functional standpoint, as this will indicate the degree of variation tolerated in shelterin.
Given that fission yeast are more closely related to budding yeast than to mammals, it has been perplexing that fission yeast appeared to lack a CST complex. However, this conundrum was solved when analysis of the S. pombe genome revealed a gene related to S. cerevisiae STN1 (Martin et al., 2007). Subsequent analysis revealed that S. pombe has a Stn1/Ten1 complex that is present at telomeres and is essential for chromosome end-protection. Deletion of either STN1 or TEN1 leads to rapid telomere loss and cell death, a phenotype similar to that observed after Pot1 depletion. However, although the Stn1/Ten1 and Pot1 (shelterin) complexes both bind to the G-overhang there is no evidence that they interact. Moreover, ChIP analysis indicates that during telomere replication, the two complexes bind the telomere with different kinetics (Moser et al., 2009). Thus, S. pombe contains two separate G-overhang binding complexes that are likely to have distinct functions. To date, a Cdc13 homolog has not been identified in S. pombe. However, recent findings with Arabidopsis and mammalian cells suggests that S. pombe Stn1 and Ten1 are likely to be part of a larger CST complex with the remaining Cdc13-like subunit too diverged to detect by primary sequence analysis.
Evidence that Arabidopsis contains a CST complex has come from two sets of studies. First, a potential STN1 ortholog was identified in the Arabidopsis genome and the encoded protein was shown to co-localize with telomeres (Song et al., 2008). Disruption of Arabidopsis STN1 causes severe morphological defects and an acute telomere deprotection phenotype. The telomere phenotype includes rapid telomere shortening, a large increase in G-overhang and frequent telomere fusions. Additional information about Arabidopsis CST was obtained when a genetic screen for mutants with dysfunctional telomeres uncovered a novel gene, CTC1 (Conserved Telomere maintenance Component 1) (Surovtseva et al., 2009). Disruption of CTC1 results in essentially the same morphological defects and telomere deprotection phenotype observed after STN1 disruption. Moreover, the ctc1/stn1 double mutant has the same phenotype as the single mutants, indicating that CTC1 and STN1 function in the same telomere protection pathway. The two proteins also interact in pull-down assays. These findings suggest that CTC1 and Stn1 are part of a CST complex. In support of this idea, a TEN1 homolog has been found in the Arabidopsis genome and the protein shown to interact with Stn1 (D. Shippen, personal communication). CTC1 has no apparent sequence homology to Cdc13. However, like Cdc13 it is a large protein that appears to contain multiple OB-folds.
Database searches revealed CTC1 homologs in a wide range of plant species and in many vertebrates. ChIP and immunolocalization studies indicate that mammalian CTC1 is present at telomeres, while knockdown of human CTC1 causes chromatin bridges, an increase in G-overhang length and sporadic telomere loss (Miyake et al., 2009; Surovtseva et al., 2009). Like the Arabidopsis protein, mammalian CTC1 interacts with orthologs of Stn1 and Ten1. The three proteins co-purify from cell extracts and they co-localize at telomeres. Thus, a CST complex is present in mammalian cells and it is needed for telomere maintenance. However, mammalian CST may have both telomeric and non-telomeric functions because not all CST localizes with telomeres and CTC1 knockdown causes DNA damage at non-telomeric locations.
Interestingly, the CTC1 and Stn1 subunits of mammalian CST turn out to be identical to the two subunits of Alpha Accessory Factor (AAF) (Casteel et al., 2009). AAF was identified in 1989 as a factor that stimulates Pol α-primase activity, but the sequence of its subunits (AAF132/CTC1 and AAF44/Stn1) was not published until 2009. AAF functions by binding single-stranded DNA and enhancing Pol α-primase association with a DNA template. The finding that human CTC1-STN1 modulates Pol α-primase activity is striking because this indicates that, as for budding yeast CST, mammalian CTC1-STN1 provides a link to the lagging strand replication machinery. Given the role played by S. cerevisiae CST in telomeric C-strand synthesis, one attractive model for mammalian CST function is that it recruits Pol α-primase to the telomeric G-strand after telomerase action and/or C-strand processing (Figure 6).
Figure 6.
Model for telomere replication in budding yeast and mammals. (A) Role of CST in S. cerevisiae. Cdc13 recruits telomerase via direct interaction with Est1. Cdc13 and Stn1 interact with subunits of Pol α-primase, recruiting it for C-strand synthesis. (B) Role of shelterin and CST in mammalian cells. TPP1 promotes telomerase action while CST recruits Pol α-primase for C-strand synthesis. Color figure available online.
In Arabidopsis, the phenotype of CTC1 or STN1 depletion is severe telomere deprotection, whereas knockdown of human CTC1 has a more modest effect. While this difference may simply reflect the partial depletion of the human protein, it is also possible that in plants the CST complex is needed for both chromosome end-protection and telomere replication, but in mammals it only functions in telomere replication. As discussed in the next section, Arabidopsis POT1 has evolved to become a component of telomerase rather than a G-overhang binding protein. Hence, as in S. cerevisiae, Arabidopsis CST may be the only telomere protein complex available to protect the chromosome terminus.
New roles for telomere proteins: gene duplication and shifts in function
The first evidence for evolution of telomere proteins via gene duplication was obtained in the 1990s with discovery of the Pot1 orthologs TEBPα and RTP (Replication Telomere Protein) in the ciliate Euplotes crassus (Wang et al., 1992), and TRF1 and TRF2 in mammals (Broccoli et al., 1997). Characterization of these proteins quickly revealed that telomere proteins can exist as gene families with family members showing considerable divergence in both sequence (Table I) and function. However, it has only recently become apparent that gene duplication and functional divergence of the encoded telomere proteins is quite widespread and a stochastic process with closely related species exhibiting quite different levels of duplication. For example, the human genome contains only one POT1 gene while the mouse genome contains two (Hockemeyer et al., 2006). Likewise, the Arabidopsis thaliana genome contains three POT1 genes however Carica papaya, a member of a closely related family in the Brassicales order, has only one (Shakirov et al., 2009). The Arabidopsis genome also harbors gene duplications of TRF1/2-Like (TRFL) genes (Karamysheva et al., 2004). At least six of the TRFL genes encode proteins with a single C-terminal Myb repeat that dimerize and bind telomeric DNA. It is interesting that not all telomere protein genes are subject to duplication, instead this phenomenon seems to be limited to genes encoding TRF1/2 and POT1-related proteins.
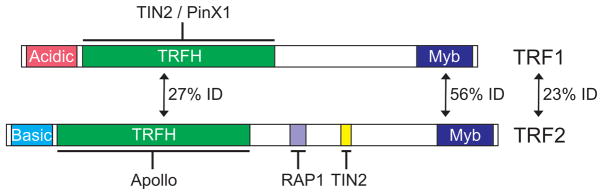
Proteins in the TRF1/2 family tend to have related functions in that they bind telomeric duplex DNA and participate in telomere length regulation and chromosome end protection (Karamysheva et al., 2004; Palm and de Lange, 2008). However, as illustrated by human TRF1 and TRF2, changes within individual domains alter the specific details of protein function. Overall, TRF1 and TRF2 have a similar domain structure as they both have a C-terminal Myb DNA binding motif and a TRFH (TRF homology) dimerization domain (Figure 7). However, TRF2 has a basic N-terminal domain whereas TRF1 has an acidic domain (Broccoli et al., 1997). The acidic domain of TRF1 has evolved quite rapidly and is missing in Xenopus and chickens (Crumet et al., 2006; De Rycker et al., 2003). The different roles played by TRF1 and TRF2 within the shelterin complex reflect their dissimilar N-termini and the significant sequence divergence in the TRFH domain. The basic domain of TRF2 is required for the protein to modulate DNA topology and stimulate strand invasion, thus this domain is thought to be critical for T-loop assembly and stabilization (Amiard et al., 2007; Fouche et al., 2006). The TRFH domains of TRF1 and TRF2 exhibit some primary sequence conservation and are structurally quite similar (Fairall et al., 2001). However, sequence changes at key amino acids result in the two proteins using this domain to interact with a very different set of binding partners (Chen et al., 2008; Kim et al., 2009). For example, TRF2 uses the TRFH domain to interact with Apollo but not TIN2, while TRF1 uses this domain to interact with TIN2 and PinX1 but not Apollo.
Figure 7.
Functional domains within human TRF1 and TRF2. Arrows indicate percent identity between domains or the full-length proteins. Interactions partners are indicted and regions mediating the interaction are underlined. Color figure available online.
Duplication of the POT1 gene is found in multiple phyla, and while some POT1 paralogs exhibit considerable similarity in both sequence and function, other POT1 paralogs are highly diverged and have evolved to fulfill novel telomeric functions. The mouse POT1 genes, POT1a and POT1b, represent more closely related family members (72% identical) that both participate in chromosome end protection (Hockemeyer et al., 2006). They have partially redundant functions but Pot1a is most important for preventing a telomeric DNA-damage response while Pot1b regulates resection of the C-strand. In contrast, the POT1 paralogs from ciliates and plants are more diverged and some family members function in telomere replication or new telomere synthesis rather than chromosome end-protection (Jacob et al., 2007; Surovtseva et al., 2007; Wang et al., 1992).
Tetrahymena thermophila resembles mouse in that it has two Pot1 proteins, however Tetrahymena Pot1a and Pot1b are only 42% identical and they have very different functions. Pot1a is present at telomeres in vegetative cells where it plays the role of a canonical Pot1 protein in that it regulates telomere length and prevents a telomeric DNA-damage response (Jacob et al., 2007). Pot1a can be isolated from cells in a complex with two other proteins: p61 and p45. p45 is required for telomerase to access the telomere while p61 is probably the Tetrahymena homolog of TPP1 (B. Linger and C. Price, unpublished results). Tetrahymena Pot1b differs from Pot1a in that it is present only in cells that have mated and are undergoing the process of new telomere synthesis during development of the macronucleus. Thus, this protein is developmentally regulated and appears to function in de novo telomere synthesis rather than end-protection (S. Heyse and C. Price, unpublished results). TEBPα from Euplotes crassus seems to be the Euplotes ortholog of Tetrahymena Pot1a because it binds to telomeres in vegetative cells and is present throughout the cell cycle (Skopp et al., 1996). Surprisingly, Euplotes RTP is not the functional ortholog of Tetrahymena Pot1b. RTP is expressed only in S-phase cells where it localizes to replication bands (the sites of DNA replication) (Skopp et al., 1996). Thus, RTP appears to play a role in telomere replication rather than end-protection or new telomere synthesis.
The three POT1 proteins from Arabidopsis, POT1a, POT1b, and POT1c, are also highly diverged. POT1a and POT1b both have two N-terminal OB-folds but their sequence is only 31% identical (Schrumpfova et al., 2008; Shakirov et al., 2005). POT1c has only a single OB-fold and is a truncated version of POT1a. Although POT1a and POT1b have the OB-fold architecture common to the DNA-binding domains of other Pot1 proteins, neither Arabidopsis protein binds telomeric DNA. For POT1a, this lack of DNA binding appears to reflect evolution of the protein to become part of the telomerase RNP rather than a stable component of the telomere (Rossignol et al., 2007; Surovtseva et al., 2007). POT1a has been shown to physically associate with the active telomerase RNP and plants null for POT1a show a dramatic decrease in telomerase activity and a progressive telomere shortening phenotype similar to that observed after TERT disruption (Surovtseva et al., 2007). Most recent studies indicate that POT1b and POT1c are also telomerase components (D. Shippen, personal communication). Thus, in Arabidopsis, CST may be the only protein complex responsible for chromosome end-protection.
While much remains to be learned about the function of Arabidopsis POT1 proteins, the available data lead one to speculate that POT1a is a telomerase recruitment factors that has migrated from the telomere to become a subunit of the telomerase RNP. It may function in much the same way as the S. cerevisiae Est1 telomerase subunit, which binds simultaneously to telomerase RNA and Cdc13, thus serving to bridge telomerase and the CST complex on the G-overhang (Bianchi and Shore, 2008). Given that the POT1 proteins from Arabidopsis and other plants seem unable to bind telomeric DNA directly (Shakirov et al., 2009), it is possible that, as in S. cerevisiae, plant CST is solely responsible for chromosome-end protection. However, in contrast to S. cerevisiae which appears to have lost the POT1 gene, plants have retained POT1 but have shuffled it into the telomerase complex. Thus, plants illustrate an extreme form of telomere protein evolution.
Conclusions
The rapid evolution of telomere proteins means that it is hard to identify orthologous proteins via bioinformatics approaches. The resulting difficulty in identifying telomere proteins from different organisms has lead to key telomere components remaining undiscovered for many years and has made it appear as if distinct telomere protein complexes arose independently during evolution. However, recent identification of new telomere proteins from various organisms has revealed that this is unlikely to be the case. Instead, it appears that a common eukaryotic ancestor probably had several distinct telomere protein complexes with separate functions. During the course of evolution, some organisms appear to have lost or gained subunits from these complexes while other organisms have lost an entire complex, requiring that the remaining telomere proteins take on new functions.
The telomerase RNP also appears to have entered into this evolutionary mix as in some organisms, telomere proteins have evolved into telomerase components. The opposite may also be true. For yet other proteins, it is difficult to know whether they should be referred to as a telomere protein or a telomerase subunit as they form a bridge between telomerase and the telomere and association with both entities is quite transient. Given, the conserved nature of telomere function, the extreme fluidity in the composition and function of telomere protein complexes was quite unexpected. However, it now seems likely that additional variations on a theme remain to be discovered and analysis of telomere proteins from model organisms will reveal further insight into the intriguing complexity of nature.
Acknowledgments
We thank Mary Chaiken, Rachid Drissi, Chunying Du and Dorothy Shippen for helpful comments and the NIH for support (grant GM041803 to CMP).
Footnotes
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Baker AM, Fu Q, Hayward W, Lindsay SM, Fletcher TM. The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. How telomerase reaches its end: mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. Embo J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD. Telomere looping in P. sativum (common garden pea) Plant J. 2003;36:271–279. doi: 10.1046/j.1365-313x.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Crumet N, Carlson RL, Drutman SB, Shampay J. A truncated acidic domain in Xenopus TRF1. Gene. 2006;369:20–26. doi: 10.1016/j.gene.2005.10.006. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- De Rycker M, Venkatesan RN, Wei C, Price CM. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Flory MR, Carson AR, Muller EG, Aebersold R. An SMC-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol Cell. 2004;16:619–630. doi: 10.1016/j.molcel.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint [published erratum appears in Mol Cell Biol 1996 Jan;16(1):457] Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Goudsouzian LK, Tuzon CT, Zakian VA. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. Embo J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004;18:992–1006. doi: 10.1101/gad.300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J Mol Biol. 2001;312:167–175. doi: 10.1006/jmbi.2001.4924. [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p Preferentially Associates with Short Telomeres to Stimulate Their Elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Else T, Daniels JP, Takai KK, Ye JZ, Keegan CE, de Lange T, Hammer GD. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. Composition and conservation of the telomeric complex. Cell Mol Life Sci. 2003;60:2295–2302. doi: 10.1007/s00018-003-3245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE. A C-terminal Myb Extension Domain Defines a Novel Family of Double-strand Telomeric DNA-binding Proteins in Arabidopsis. J Biol Chem. 2004;279:47799–47807. doi: 10.1074/jbc.M407938200. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- Kim SH, Davalos AR, Heo SJ, Rodier F, Zou Y, Beausejour C, Kaminker P, Yannone SM, Campisi J. Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes. J Cell Biol. 2008;181:447–460. doi: 10.1083/jcb.200710028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer KH, Sippel AE. The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 1987;6:2719–2725. doi: 10.1002/j.1460-2075.1987.tb02565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–47. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- Li B, Espinal A, Cross GA. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol Cell Biol. 2005;25:5011–5021. doi: 10.1128/MCB.25.12.5011-5021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O’Connor MS, Qin J, Songyang Z. Telosome -- A mammalian telomere associated complex formed by multiple telomeric proteins. J Biol Chem. 2004a doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004b;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;243:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Lundblad V. In: Budding yeast telomeres. Telomeres T, de Lange, Lundblad V, Blackburn E, editors. Cold Spring Harbor Laboratory Press; 2006. pp. 345–386. [Google Scholar]
- Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast [see comments] Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci U S A. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian CTC1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the POT1 pathway. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.08.009. In Press. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol. 2006;8:748–755. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM. Antics at the telomere: uncoupled polymerases solve the end replication problem. Embo J. 2009;28:795–796. doi: 10.1038/emboj.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. Embo J. 2008 doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Fairall L, Simonsson T, Court R, Chapman L. Telomere architecture. EMBO Rep. 2002;3:1139–1145. doi: 10.1093/embo-reports/kvf246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrumpfova PP, Kuchar M, Palecek J, Fajkus J. Mapping of interaction domains of putative telomere-binding proteins AtTRB1 and AtPOT1b from Arabidopsis thaliana. FEBS Lett. 2008;582:1400–1406. doi: 10.1016/j.febslet.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, McKnight TD, Shippen DE. POT1-independent single-strand telomeric DNA-binding activities in Brassicaceae. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopp R, Wang W, Price C. rTP: a candidate telomere protein that is associated with DNA replication. Chromosoma. 1996;105:82–91. doi: 10.1007/BF02509517. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink KG, Evans RJ, Chambers A. Sequence-specific binding of Taz1p dimmers to fission yeast telomeric DNA. Nucleic Acids Res. 2000;28:527–533. doi: 10.1093/nar/28.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. Embo J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Surovtseva Y, Churikov D, Bolts K, Song X, Lamb J, Warrington R, Leehy K, Heacock M, Price C, Shippen D. Conserved Telomere maintenance Component 1 interacts with STN! and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.09.017. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. Embo J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic Acid Recognition by OB-Fold Proteins. Annu Rev Biophys Biomol Struct. 2003 doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure (Camb) 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. MRX-dependent DNA damage response to short telomeres. Mol Biol Cell. 2007;18:3047–3058. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wang W, Skopp R, Scofield M, Price C. Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res. 1992;20:6621–6629. doi: 10.1093/nar/20.24.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as comediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007;14:832–840. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O’Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 Binds TRF1 and TRF2 Simultaneously and Stabilizes the TRF2 Complex on Telomeres. J Biol Chem. 2004a;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004b;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]