Abstract
Since Neisseria gonorrhoeae and Neisseria meningitidis are obligate human pathogens, a comparison with commensal species of the same genus could reveal differences important in pathogenesis. The recent completion of commensal Neisseria genome draft assemblies allowed us to perform a comparison of the genes involved in the catalysis, assembly and regulation of the denitrification pathway, which has been implicated in the virulence of several bacteria. All species contained a highly conserved nitric oxide reductase (NorB) and a nitrite reductase (AniA or NirK) that was highly conserved in the catalytic but divergent in the N-terminal lipid modification and C-terminal glycosylation domains. Only Neisseria mucosa contained a nitrate reductase (Nar), and only Neisseria lactamica, Neisseria cinerea, Neisseria subflava, Neisseria flavescens and Neisseria sicca contained a nitrous oxide reductase (Nos) complex. The regulators of the denitrification genes, FNR, NarQP and NsrR, were highly conserved, except for the GAF domain of NarQ. Biochemical examination of laboratory strains revealed that all of the neisserial species tested except N. mucosa had a two- to fourfold lower nitrite reductase activity than N. gonorrhoeae, while N. meningitidis and most of the commensal Neisseria species had a two- to fourfold higher nitric oxide (NO) reductase activity. For N. meningitidis and most of the commensal Neisseria, there was a greater than fourfold reduction in the NO steady-state level in the presence of nitrite as compared with N. gonorrhoeae. All of the species tested generated an NO steady-state level in the presence of an NO donor that was similar to that of N. gonorrhoeae. The greatest difference between the Neisseria species was the lack of a functional Nos system in the pathogenic species N. gonorrhoeae and N. meningitidis.
INTRODUCTION
There are 12 species and biovars in the genus Neisseria isolated from humans, while only two species, Neisseria gonorrhoeae and Neisseria meningitidis, are frequently pathogenic (Vedros, 1984). Neisserial species can be differentiated by characteristics such as patterns of acid production from carbohydrates, the capacity to reduce nitrate, and the ability to produce polysaccharide from sucrose (Knapp, 1988). Human Neisseria species can be divided into two major groups. The first group includes N. gonorrhoeae, N. meningitidis, Neisseria lactamica, Neisseria cinerea, Neisseria flavescens and Neisseria polysaccharea. Members of this group generally grow as non-pigmented, translucent colonies, with the exception of N. flavescens, which is yellow-pigmented. The second group of neisserial species includes the saccharolytic commensals Neisseria subflava (including the N. subflava biovars perflava and flava), Neisseria sicca and Neisseria mucosa. Colonies of these species are generally opaque and yellow-pigmented (Knapp, 1988).
Of the Neisseria species, only N. gonorrhoeae is considered pathogenic at all times. Gonococcal infection can occur within a variety of mucosal surfaces including the cervix, urethra, rectum and nasopharynx, causing symptomatic or asymptomatic infections. N. meningitidis is also capable of causing disease in healthy humans, and pathogenicity is associated with specific serogroups or serotypes. From a total of 13 serogroups of meningococci, encapsulated strains belonging to the serogroups A, B, C and W-135 have been most frequently associated with epidemics (Knapp, 1988). Meningococcal strains may also be carried asymptomatically in the oro- or nasopharynx (Gold et al., 1978). Many commensal Neisseria spp. have been sporadically isolated from disseminated sites, blood and cerebrospinal fluid, but no correlation has been established between any species and syndrome (Morse & Knapp 1987). Therefore, these Neisseria spp. are not true pathogens, but instead appear to be opportunistic pathogens in immunocompromised individuals (Herbert & Ruskin, 1981; Lauer & Fisher, 1976). These commensals are usually found within the nasopharynx and/or oropharynx where they may exist as part of the normal flora. The carriage rate of N. lactamica is generally higher than that of N. meningitidis in children (Blakebrough et al., 1982), with 59 % of children colonized by N. lactamica by the age of 4 years (Gold et al., 1978). When evaluating the overall carriage rates of commensal Neisseria species, 45 % of adults harboured N. sicca, 40 % carried N. perflava, and N. subflava–flava appeared in 11 % of individuals tested. It must be noted that this study did not recognize N. mucosa (Knapp, 1988).
As one of the most extensively studied Neisseria species, N. gonorrhoeae was originally believed to be an obligate aerobe until it was found that cultures obtained from urethral exudates could remain viable when incubated anaerobically (Short et al., 1982). Nitrite was subsequently identified as the terminal electron acceptor supporting anaerobic growth (Knapp & Clark, 1984). Several gonococcal outer-membrane proteins are either induced or repressed in the absence of oxygen, suggesting that growth under oxygen-limited conditions is a relevant physiological state (Clark et al., 1987). The major anaerobically induced protein has been identified as a copper-containing nitrite reductase, AniA (formerly Pan1), responsible for reducing nitrite to nitric oxide (NO), representing one of the enzymic reactions of the denitrification pathway (Cardinale, 1999; Mellies et al., 1997).
Denitrification is a microbial process in which oxidized nitrogen compounds are used as alternative electron acceptors for energy production. A bacterium capable of reducing nitrate to dinitrogen gas contains four independent metalloenzymes that are usually induced sequentially under anaerobic conditions (for in-depth reviews of denitrification, see Zumft, 1997; Tavares et al., 2006). In recent years, the denitrification pathway has been implicated in the virulence of several bacterial species, including Brucella and Pseudomonas (Baek et al., 2004; Van Alst et al., 2007). Denitrification has also been implicated in the pathogenicity of N. meningitidis, where active denitrification has been linked to modulation of host cytokine responses, enhanced intracellular survival, and inhibition of apoptosis in a macrophage model (Stevanin et al., 2005, 2007; Tunbridge et al., 2006). In the gonococcus, inactivation of the denitrification pathway hinders biofilm formation and prevents establishment of anti-inflammatory NO steady-state levels (Barth & Clark, 2008; Cardinale & Clark, 2005; Falsetta et al., 2008).
Comparative genomics can be a useful method to evaluate the role of specific proteins in neisserial pathogenesis, as only two neisserial species are capable of causing disease in healthy individuals. Since N. gonorrhoeae is an obligate human pathogen, it has been a challenge to establish an animal model that would allow direct testing of the involvement of denitrification in virulence. Recent completion of genome sequencing projects for the commensal Neisseriaceae has allowed for a genomic and biochemical survey of the denitrification pathway, with a focus on comparison of pathogenic (N. gonorrhoeae and N. meningitidis) with commensal Neisseria species (N. lactamica, N. cinerea, N. subflava, N. mucosa and N. flavescens).
In this paper we explore the genetic organization and amino acid sequence of each of the four respective reductases (Nar, Nir, Nor and Nos) involved in denitrification within Neisseria species. The ability of commensal Neisseria species to grow under anaerobiosis, produce and degrade NO, and to establish an NO steady-state concentration during denitrification was examined. We show that there is high sequence conservation within the catalytic domains of the reductases involved in denitrification, while more variation exists in other domains of the nitrite reductase. There are also major differences in the nitrous oxide reductase regulon between the pathogenic and commensal Neisseria species.
METHODS
Bacterial strains and growth conditions.
The strains used for biochemical studies were N. gonorrhoeae F62, a collection of clinical gonococcal isolates, and commensal Neisseria strains, acquired from the Neisseria Reference Laboratory, Centers for Disease Control and Prevention (CDC), Atlanta, GA. N. meningitidis strains used were acquired from the CDC (Table 1). Strains were grown on Difco gonococcal (GC) medium base (Becton Dickinson) plates with 1 % Kellogg's supplement (GCK) (Kellogg et al., 1963), either aerobically in a 5 % CO2 incubator or anaerobically in a Coy anaerobic chamber (Coy Laboratory Products) with an atmosphere of 85 % N2, 10 % H2 and 5 % CO2, as previously described (Clark et al., 1987). Anaerobic plates were subcultured three times to ensure full induction of Nir and Nor. Nitrite was provided for anaerobic growth by placing 40 μl 20 % (w/v) NaNO2 solution on a sterile cellulose disk in the centre of the plate. For examination of anaerobic growth of commensal Neisseria strains, nitrite was added either on a sterile cellulose disk or directly to plates at concentrations of 10, 25 and 50 mM. All incubations were at 37 °C.
Table 1.
Bacterial strains used in this study for biochemical analysis of the denitrification pathway
| Species | Strain | Source* |
|---|---|---|
| Pathogenic species | ||
| N. gonorrhoeae | F62 | Laboratory collection |
| N. meningitidis | RUN 5645 | CDC |
| RUN 5647 | CDC | |
| Commensal species | ||
| N. lactamica | RUN 5071 | NRL |
| RUN 5106 | NRL | |
| N. cinerea | RUN 5066 | NRL |
| RUN 5084 | NRL | |
| N. mucosa | RUN 5100 | NRL |
| RUN 5101 | NRL | |
| N. subflava | RUN 5075 | NRL |
| N. flava | RUN 5068 | NRL |
| N. perflava | RUN 5073 | NRL |
| N. sicca | RUN 5074 | NRL |
*CDC, Centers for Disease Control and Prevention, Atlanta, GA; NRL, Neisseria Reference Laboratory, University of Washington, Seattle, WA.
blast analysis of denitrification proteins and regulatory components.
Basic local alignment search tool (blast) was used to compare amino acid sequences of the denitrification reductases (Nar, Nir, Nor and Nos) and the transcriptional regulators involved in controlling their expression (FNR, NarP, NarQ and NsrR) in various Neisseria species. Sequences were obtained from the STDgen database (http://stdgen.northwestern.edu/), those deposited within the National Center for Biotechnology Information (NCBI), and an additional 10 unpublished gonococcal and meningococcal aniA sequences completed in our laboratory. Annotated genomes were surveyed using the blastp algorithm using a protein query against the protein database. The filter was set to default and the expected value was set to 10. For those bacterial genomes that are currently at the draft assembly stage, N. gonorrhoeae FA1090 amino acid sequences were used to perform analysis using the tblastn algorithm with the query set as protein against the genomic database. The filter and expected value were set to default. The genomic sequence of the appropriate contig from the sequencing project was analysed in the Clone Manager Professional software program. The translate tool was utilized to provide amino acid sequence, and alignments were performed in MultAlin (Corpet, 1988).
Biochemical assays.
Examination of anaerobic growth, nitrite and nitric oxide reductase activities, and the establishment of an NO steady-state concentration during denitrification were done with neisserial strains identified as either clinical isolates or laboratory strains (Table 1). These do not represent the specific isolates that have been, or are in the process of, having their complete genomes sequenced.
Measurement of NO concentrations.
An Apollo 4000 free radical analyser with ISO-NOP probes (World Precision Instruments) allowed for real-time measurement of NO concentrations, as previously described (Cardinale & Clark, 2005). All of the standard curves and reaction assays were performed in a temperature-controlled reaction vessel (World Precision Instruments) at a constant 37 °C.
Nitrite reductase (Nir) and nitric oxide reductase (Nor) assays.
Nir activity was measured by the reduction in nitrite concentration and Nor activity was measured using the short half-life NO donor, 2-(N,N-diethylamino)-diazenolate-2-oxide (DEA/NO) (Sigma Chemical Company), as previously described (Cardinale & Clark, 2005).
Determination of NO steady-state values.
Cells were resuspended in GCK broth and the OD600 was adjusted to 0.02. The cells were allowed to incubate for 10 min followed by addition of nitrite to a final concentration of 2 mM or DETA/NO [2,2′-(hydroxynitrosohydrazono) bis-ethanimine; Sigma Chemical Company] as a source of exogenous NO, at a final concentration of 0.3 mM. The range of NO concentrations during an established steady state in three independent trials was used to calculate the mean NO concentration and standard error for each condition.
RESULTS AND DISCUSSION
Extent of the denitrification pathway within Neisseria species
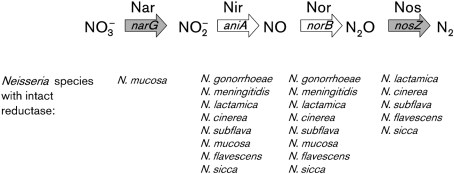
A bioinformatics approach was used in order to identify the genetic organization and conservation of each enzyme responsible for catalysing the reactions of denitrification. Fig. 1 depicts the complete denitrification pathway, where the reduction of nitrogen oxides to dinitrogen gas is catalysed by four independent enzymes. Previous studies have identified that both pathogenic species, N. gonorrhoeae and N. meningitidis, contain the internal enzymic steps of the pathway. In these internal steps, nitrite is reduced to NO by the nitrite reductase AniA (Cardinale, 1999; Mellies et al., 1997), and NO is reduced to nitrous oxide by the nitric oxide reductase, NorB (Lissenden et al., 2000). We performed a genetic survey using the 11 N. gonorrhoeae strains, four N. meningitidis strains and six commensal species whose genomes have been either sequenced or had draft assemblies completed and deposited in NCBI. blast analysis used the tblastn algorithm of amino acid sequences from Ralstonia eutropha H16, a soil bacterium that has been shown to encode a nitric oxide reductase with a high level of identity to that of N. gonorrhoeae FA1090 NorB (61 %) and to contain the full complement of denitrification enzymes, which was used to examine reductase orthologues within each sequenced Neisseria species (Householder et al., 2000). Genomic analysis of all sequenced Neisseria species demonstrates that each species contains a complete Nir (AniA) and Nor (NorB). Only N. mucosa contains the three subunits of a nitrate reductase (NarGHI), and has been shown to reduce nitrate (Berger, 1961). The commensal species N. lactamica, N. cinerea, N. subflava, N. flavescens and N. sicca contain the Nos regulon (NosRZDFYLX) in its entirety, having no premature stop codons, partial deletions or frameshift mutations (Table 2).
Fig. 1.
Denitrification pathway within Neisseria species. The enzymic steps of the denitrification pathway are shown in addition to the intermediate NO substrates. Above each arrow the specific enzymes that catalyse each reaction are given (Nar, nitrate reductase; Nir, nitrite reductase; Nor, nitric oxide reductase; Nos, nitrous oxide reductase), while the genes encoding each catalytic subunit are written within the arrows. All Neisseria species that have been investigated in this study encode enzymes responsible for the internal reactions (Nir and Nor, indicated by white arrows), with the exception of one meningococcal strain (053442) that lacks aniA in its entirety. Only some of the commensal Neisseria species encode complete Nar and Nos (grey arrows) without premature stop codons or deletions.
Table 2.
Denitrification reductases within Neisseria species compared with R. eutropha H16
| Species and strain | Nitrate reductase | Nitrite reductase | Nitric oxide reductase | Nitrous oxide reductasea |
|---|---|---|---|---|
| N. gonorrhoeae FA 1090 | † | NirK: aniA | NorZ: norB | NosR*Z*D*FYLX* |
| N. meningitidis Z2491 | † | NirK: aniA | NorZ: norB | NosRΔZ†DΔF*YΔLX* |
| N. meningitidis MC58 | † | NirK: aniA | NorZ: norB | NosRΔZ†DΔF*YΔLX* |
| N. meningitidis FAM18 | † | NirK: aniA | NorZ: norB | NosRΔZ†DΔF*YΔLX* |
| N. meningitidis 053442 | † | † | NorZ: norB | NosRΔZ†DΔF*YΔLX* |
| N. lactamica ATCC 23970 | † | NirK: aniA | NorZ: norB | NosRZDFYLX |
| N. cinerea ATCC 14685 | † | NirK: aniA | NorZ: norB | NosRZDFYLX |
| N. subflava NJ9703 | † | NirK: aniA | NorZ: norB | NosRZDFYLX |
| N. mucosa ATCC 25996 | NarG | NirK: aniA | NorZ: norB | NosRZ*DFΔYLX |
| N. flavescens NRL 30031 | † | NirK: aniA | NorZ: norB | NosRZDFYLX |
| N. sicca ATCC 29256 | † | NirK: aniA | NorZ: norB | NosRZDFYLX |
| R. eutrophab H16 | NarG NapA | NirS | NorZ: norB NorZ: norB2 | NosRZDFYLX |
aThe full Nos regulon is shown in order to indicate which accessory, assembly or catalytic subunits are present in their entirety versus those strains having coding regions with mutations.
†Gene is absent from the genome in its entirety.
*Gene contains a premature stop codon.
ΔGene is partially deleted.
bThe specific amino acid sequences used to search for denitrification reductases within Neisseria species. R. eutropha H16 contains both a membrane-bound nitrate reductase (NarG: NP_942905, PHG270) and a periplasmic nitrate reductase (NapA: NP_942849, PHG211), a cytochrome cd1-type nitrite reductase (NirS: YP_841789, H16_B2277), two qNor-type (NorZ) nitric oxide reductases (NorB: NP_942879, PHG244; NorB2: YP_841835, H16_B2323), and a nitrous oxide reductase (NosZ: NP_942887, PHG252). All of the reductases are found on megaplasmid pHG1 (AY305378), with the exception of NirS and one copy of NorB encoded on chromosome 2 (AM260480). These contain the genes encoding the catalytic portions of each respective reductase used as the amino acid query sequence within the tblastn algorithm of blast analysis. The R. eutropha H16 genome sequence accession numbers and locus numbers are also listed above when applicable.
Genomic analysis of the nitrate reductase within N. mucosa
N. mucosa contains an orthologue to a NarG-type membrane-bound nitrate reductase (NEIMUCOT_00642). Such reductases are made up of three subunits. narG encodes the catalytic α subunit, narH encodes the soluble β subunit, and narI encodes the γ subunit; NarH and NarI contain electron transfer centres (Zumft, 1997). Each of the subunits, NarG, NarH (NEIMUCOT_00688) and NarI (NEIMUCOT_00686), shows high amino acid conservation compared with orthologues in R. eutropha, with 70, 77 and 68 % identity, respectively. Orthologues of the major facilitator superfamily responsible for nitrate/nitrite transport (NarK1, NarK2) are also encoded by N. mucosa. None of the Neisseria species encodes an orthologue to the periplasmic nitrate reductase NapA (Potter et al., 2001).
Genomic analysis of the neisserial nitrite reductase
The gonococcal nitrite reductase, AniA, is classified as a dissimilatory, copper-containing nitrite reductase (CuNir), primarily found in Gram-negative bacteria (Fenderson et al., 1991; Kakutani et al., 1981). Crystallographic data for the soluble domain of AniA reveal that it is analogous to similar proteins from soil bacteria in that a type I copper centre (site of electron transfer from a proteinaceous electron donor) and a type II copper centre (site of nitrite reduction) are highly conserved (Boulanger & Murphy, 2002). The AniA protein contains three distinct domains, including an N-terminal lipid modification domain (amino acids 1–100) containing a signal peptidase II cleavage/modification sequence, ALAAC; a catalytic domain (amino acids 101–350); and a proline-rich domain (amino acids 351–394) recently identified as being a target of the O-glycosylation pathway in N. gonorrhoeae (Ku et al., 2009).
blast analysis using a tblastn algorithm indicated that AniA is highly conserved within the catalytic domain across all Neisseria species (92–99 % identity), with significantly lower sequence conservation within the other two domains (Table 3 and Supplementary Table S1). Each of the sequenced N. gonorrhoeae strains displayed similar conservation within the lipid modification and glycosylation domains, containing only zero to three amino acid differences across the entire protein. Conversely, N. meningitidis strains appear to have acquired a wide range of sequence changes within AniA (Table 3, Supplementary Fig. S1). It was found that 13 out of 41 N. meningitidis strains were missing the catalytic domains, while 46 of 46 meningococcal AniA sequences show either small deletions or complete absence of the glycosylation domain (Ku et al., 2009). Previous reports have demonstrated a high degree of sequence diversity within AniA among meningococcal strains, in addition to strains that do not express aniA at all (Stefanelli et al., 2008; Thomson et al., 2008). One of the fully sequenced strains, 053442, was lacking aniA in its entirety (Table 2). In contrast, commensal Neisseria generally had high conservation across the entire protein sequence, with the exception of the proline-rich glycosylation domain and the N terminus. N. lactamica and N. cinerea were nearly identical to N. gonorrhoeae at the N terminus, while the other commensals had a number of insertions within this region. The essential signal peptidase II cleavage sequence, both type I and type II copper centres, and the important active site residues Asp141 and His 284, which are required for nitrite reduction capability in other CuNirs, were conserved across all Neisseria species (Supplementary Fig. S1) (Boulanger et al., 2000; Kataoka et al., 2000).
Table 3.
Conservation of specific protein domains within AniA among Neisseria species
+, Present; +/−, small deletions; −, missing, relative to N. gonorrhoeae FA 1090.
| Species and strain | N-terminal lipid modification domain (amino acids 1–100) | Catalytic domain (amino acids 101–350) | Glycosylation domain (amino acids 351–394) |
|---|---|---|---|
| N. gonorrhoeae* | + | + | + |
| N. meningitidis† | + | + | +/− |
| N. meningitidis‡ | +/− | + | +/− |
| N. meningitidis§ | +/− | + | − |
| N. meningitidis|| | +/− | − | − |
| N. meningitidis¶ | − | − | − |
| N. lactamica ATCC 23970 | + | + | +/− |
| N. cinerea ATCC 14685 | + | + | − |
| N. subflava NJ9703 | + | + | − |
| N. mucosa ATCC 25996 | + | + | − |
| N. flavescens NRL 30031/H210 | + | + | − |
| N. sicca ATCC 29256 | + | + | − |
*Eleven sequenced strains in NCBI and five strains whose aniA was sequenced in this laboratory.
†MC58, three strains whose aniA was sequenced as reported by Ku et al. (2009), and two strains whose aniA was sequenced in this laboratory.
‡Z2491 and three strains whose aniA was sequenced in this laboratory.
§Twenty-two strains whose aniA was sequenced as reported by Ku et al. (2009).
||FAM18 and 13 strains whose aniA was sequenced as reported by Ku et al. (2009).
¶Genome-sequenced strain 053442.
Biochemical analysis of nitrite reductase activity
The specific activity of anaerobically induced AniA in N. gonorrhoeae strain F62 has been previously reported to be 241 nanomoles nitrite reduced per minute per OD600 unit, with no more than a twofold difference in AniA activity across nine gonococcal isolates (Barth & Clark, 2008). We expanded our analysis to examine AniA activity in the commensal Neisseria species (Table 4). The commensal Neisseria grew anaerobically in the presence of 10, 25 and 50 mM nitrite for all strains tested, except RUN5084 (N. cinerea), which failed to grow at the highest nitrite concentration.
Table 4.
Nitrite and nitric oxide reductase specific activities in Neisseria species
| Species | Strain | Nir activity* | Nor activity† | Nir : Nor ratio |
|---|---|---|---|---|
| N. gonorrhoeae | Laboratory strain F62 | 241±3 | 88±18 | 2.74 |
| N. meningitidis | RUN 5645 | 94±2 | 240±15 | 0.39 |
| N. meningitidis | RUN 5647 | 84±7 | 256±10 | 0.33 |
| N. lactamica | RUN 5071 | 57±3 | 75±15 | 0.76 |
| N. lactamica | RUN 5106 | 87±10 | 27±10 | 3.22 |
| N. cinerea | RUN 5066 | 149±2 | 74±22 | 2.01 |
| N. cinerea | RUN 5084 | 120±8 | 138±4 | 0.87 |
| N. mucosa | RUN 5100 | 295±14 | 86±10 | 3.43 |
| N. mucosa | RUN 5101 | 279±11 | 131±6 | 2.13 |
| N. subflava | RUN 5075 | 124±1 | 181±9 | 0.69 |
| N. flava | RUN 5068 | 94±3 | 186±22 | 0.51 |
| N. perflava | RUN 5073 | 121±6 | 171±23 | 0.71 |
| N. sicca | RUN 5074 | 99±9 | 136±3 | 0.73 |
*Values represents the mean±se of three independent determinations; units are nanomoles NO2 reduced per minute per OD600 unit.
†Values represents the mean±se of three independent determinations; units are nanomoles NO reduced per minute per OD600 unit.
The majority of commensal strains displayed a lower level of Nir activity than that seen in N. gonorrhoeae F62, ranging from 57 to 149 nanomoles NO2 reduced per minute per OD600 unit (Table 4). N. mucosa strains RUN5100 and RUN5101 were the exceptions, having Nir activities of 295 and 279 nanomoles NO2 reduced per minute per OD600 unit, respectively, which are more similar to levels seen in N. gonorrhoeae F62 (Table 4). The largest difference in AniA activity was observed in N. lactamica strain RUN5071, which had a 4.2-fold lower Nir activity than N. gonorrhoeae F62. The two N. meningitidis strains used in this study, RUN5645 and RUN5647, were able to grow under anaerobic conditions when supplemented with nitrite (data not shown), representing a subtype of meningococcal strains that are nitrite tolerant. Meningococcal Nir activity was approximately 2.5- to threefold lower than that of gonococcal strain F62 (84–94 nanomoles NO2 reduced per minute per OD600 unit) (Table 4).
Role of AniA in neisserial growth and survival
It would appear that AniA does not play a major role in survival for N. meningitidis considering that 32 % (13 out of 41) of sequenced strains were shown to contain a frame-shift mutation in aniA, resulting in premature termination of translation, and that strain 053442 contains a deletion of the entire aniA gene (Ku et al., 2009). Commensal strains share a niche within the human host similar to that of the meningococcus, and a significant proportion of these strains have acquired mutations within aniA. Sequencing of aniA in additional commensal strains might indicate a range of AniA mutations akin to that observed in the meningococcus. Of the 16 gonococcal strains that have had aniA sequenced, all contain a fully intact nitrite reductase gene (Table 3), suggesting that AniA may be more important for survival of the organism within the urogenital tract.
Genomic analysis of the neisserial nitric oxide reductase
The gonococcal nitric oxide reductase, NorB, was originally identified due to its high sequence similarity to the nitric oxide reductase of R. eutropha. NorB is a single subunit qNOR that accepts electrons directly from quinols (Householder et al., 2000). This reductase has been shown to be required for anaerobic growth but not anaerobic survival under denitrifying conditions (Householder et al., 2000). Sequence analysis indicates that the entire NorB protein is highly conserved across all Neisseria species (Supplementary Table S2, Supplementary Fig. S2). The amino acid sequence of NorB from 12 sequenced gonococcal strains contains zero to two amino acid changes as compared with N. gonorrhoeae FA 1090, while NorB orthologues from other Neisseria species display 96–99 % identity. The high degree of sequence conservation across the genus suggests that NorB is essential for growth or survival of the Neisseria species in the various ecological niches within the human host.
Biochemical analysis of nitric oxide reductase activity
Previous studies have reported the Nor activity in anaerobically induced N. gonorrhoeae F62 to be 88 nanomoles NO reduced per minute per OD600 unit. Other gonococcal isolates showed no more than a twofold difference, having similar activities in the range of 88–155 nmoles NO reduced per minute per OD600 (Barth & Clark, 2008). N. meningitidis strains displayed an approximately 2.5- to threefold higher Nor activity (240 and 256 nanomoles NO reduced per minute per OD600 unit) compared with F62. The commensal species displayed greater variation in activity (27–186 nanomoles NO reduced per minute per OD600 unit); however, the observed activities were more similar to those of gonococci (Table 4). Thus, the nitric oxide reductase is functional across all species examined in the genus Neisseria, with activities that vary over a threefold range relative to N. gonorrhoeae.
Genomic analysis of the neisserial nitrous oxide reductase
The last enzymic step of the denitrification pathway is catalysed by the nitrous oxide reductase and results in the production of dinitrogen gas. The Nos system is composed of the catalytic subunit encoded by nosZ, a regulatory protein encoded by nosR, and a number of accessory/assembly proteins encoded by the nosDFYL genes (Holloway et al., 1996; Schwartz et al., 2003; Velasco et al., 2004). Denitrifying bacteria usually maintain a conserved organization of the Nos regulon having the order nosRZDFYL (Philippot, 2002).
blast analysis using the blastp algorithm revealed differences among Neisseria species in regards to organization and completeness of the Nos genes (Table 2; specific gene identification numbers are given in Supplementary Tables S3 and S4). The organization of the Nos regulon in Neisseria is generally well conserved with respect to maintaining the order nosRZDFYL, with the exception of N. mucosa, which has Nos genes at three distinct loci consisting of nosRZ, nosDFYL and nosX. The commensal strains N. subflava and N. flavescens have an organization similar to that of R. eutropha, with the full complement of coding regions contained at one locus (nosRZDFYLX), while in N. gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea and N. mucosa, nosX is located distantly from the rest of the regulon, ranging from 45 to 1201 ORFs away.
Of the sequenced Neisseria species, only N. lactamica, N. cinerea, N. subflava, N. flavescens and N. sicca contain an intact Nos system (Table 2, Supplementary Fig. S3). N. mucosa contains a premature stop codon within nosZ and a deletion in nosF. Both of the pathogenic Neisseria have multiple coding regions disrupted. Interestingly, however, they have eliminated the Nos regulon via independent mechanisms. N. gonorrhoeae contains premature stop codons within nosR, nosZ, nosD and nosX, while N. meningitidis has a 4 kb deletion that extends from the middle of nosR into the middle of nosD, completely eliminating the catalytic subunit nosZ (Table 2). Furthermore, nosY and nosX each contain premature stop codons in meningococci.
NO steady states established during denitrification
Previous studies examined NO levels in bacteria capable of complete denitrification. Paracoccus denitrificans, Pseudomonas stutzeri and Achromobacter cycloclastes have been shown to produce a steady-state NO level in the range of 1–65 nM (Goretski et al., 1990). N. gonorrhoeae, although only encoding the internal reactions of the denitrification pathway, also established an NO steady-state level in the presence of nitrite or a long-term NO donor (Cardinale & Clark, 2005). In the presence of nitrite, the NO steady-state level was directly dependent on the Nir activity : Nor activity ratio, while in the presence of an exogenous NO source the NO steady-state level was dependent on the input NO concentration.
With the addition of a long-term NO donor, which mimics the local environment during an active host response, all strains of Neisseria species were efficient at metabolizing NO and establishing an NO steady-state level that reduced input concentrations by 80–98 % (31–157 nM NO) (Table 5). Thus, all Neisseria strains tested have the potential to reduce in vivo NO concentrations to levels that would result in the anti-inflammatory signalling events of NO predominating (Stefano et al., 2000).
Table 5.
NO steady-state concentrations established by anaerobically grown Neisseria species in the presence of nitrite or DETA/NO
| Species | Strain | NO steady state with NO2* | NO steady state with DETA/NO |
|---|---|---|---|
| N. gonorrhoeae | Laboratory strain F62 | 1166±70 | 129±5 |
| N. meningitidis | RUN 5645 | 73±12 | 276±24 |
| N. meningitidis | RUN 5647 | 289±20 | 150±14 |
| N. lactamica | RUN 5071 | 2274±131 | 107±10 |
| N. lactamica | RUN 5106 | 271±15 | 68±18 |
| N. cinerea | RUN 5066 | 4810±128 | 157±69 |
| N. cinerea | RUN 5084 | 1142±22 | 31±12 |
| N. mucosa | RUN 5100 | 0† | 80±14 |
| N. mucosa | RUN 5101 | 0† | 44±15 |
| N. subflava | RUN 5075 | 256±22 | 74±54 |
| N. flava | RUN 5068 | 195±35 | 153±38 |
| N. perflava | RUN 5073 | 406±70 | 157±23 |
| N. sicca | RUN 5074 | 13±9 | 140±21 |
*Steady-state NO concentrations measured as nmol NO l−1 (mean±se).
†N. mucosa strains RUN 5100 and RUN 5101 brought NO levels to zero during each independent trial in the presence of 2 mM nitrite.
The NO steady-state concentration was determined for each Neisseria species in the presence of 2 mM nitrite. With the addition of nitrite, each Neisseria species, except N. mucosa, generated an NO steady-state level that remained consistent for several hours (data not shown). It appears that the NO steady-state levels established by Neisseria strains form three distinct groups (Table 5): (1) strains that established high NO steady-state levels similar to gonococcal strain F62 (e.g. N. cinerea RUN5084, N. lactamica RUN5071), (2) strains that established moderate to low NO steady-state levels, similar to a class of gonococcal strains auxotrophic for arginine, hypoxanthine and uracil (AHU; e.g. N. meningitidis RUN5647, N. subflava RUN5075) (Barth & Clark, 2008), and (3) strains that established very low NO steady-state levels or brought the NO concentration to zero, similar to a second class of gonococcal AHU strains (e.g. N. sicca and both N. mucosa strains, RUN5100 and RUN5101). These data show that there were not only significant differences in the NO steady-state level between species but also examples of differences within the same species.
Regulation of the neisserial denitrification pathway
Extensive research has shown that regulation of the genes involved in denitrification and/or adaptation to anaerobic growth in the neisserial species involves the transcriptional regulators FNR and NsrR, and the two-component system NarQP (for a detailed review regarding these neisserial regulators, see Clark et al., 2009). FNR is the only regulator in the pathogenic Neisseria known to respond directly to anaerobic environmental conditions, and does so through a labile, oxygen-sensitive [4Fe–4S] cluster coordinated by four cysteine residues (Clark et al., 2009; Overton et al., 2003). FNR activation is a requirement for efficient transcription of aniA, and fnr mutations in both the meningococcus and the gonococcus result in the inability of these strains to reduce nitrite (Clark et al., 2009; Householder et al., 1999). Sequence alignments show that FNR is very highly conserved across all members of the neisserial genus, with the most distant FNR orthologues, expressed by N. mucosa and N. sicca, displaying 95 % similarity to the FNR sequence of N. gonorrhoeae FA1090 (Supplementary Table S5, Supplementary Fig. S4A). All neisserial species contain the four Fe–S coordinating cysteines (Overton et al., 2003). Furthermore, the amino acid sequence is conserved in and around areas where mutations have been found to increase the oxygen stability of gonococcal FNR (L22H and D148A) (Householder, 2000; Overton et al., 2003). This suggests that FNR likely functions in a similar, oxygen-dependent manner in all neisserial strains. The high level of conservation of this anaerobic regulator across the genus also implies that anaerobiosis is a physiologically significant state not only for the pathogenic Neisseria species but also for commensal Neisseria species (Clark et al., 1987; Rock et al., 2005).
NsrR is an NO-sensing rrf2-type transcriptional repressor, and although the discovery of this protein has been somewhat recent, extensive research has characterized its regulatory role in several bacterial species (Bodenmiller & Spiro, 2006; Overton et al., 2006; Clark et al., 2009; Filenko et al., 2007; Gilberthorpe et al., 2007; Isabella et al., 2008; Nakano et al., 2006; Rock et al., 2007). In N. gonorrhoeae and N. meningitidis, NsrR negatively regulates aniA and norB, and derepression occurs in the presence of NO (Clark et al., 2009; Isabella et al., 2008, 2009). Sequence alignments of the NsrR protein across the neisserial genus illustrate that this regulator is well conserved, with the most distant NsrR orthologue, encoded by N. mucosa, displaying 88 % similarity to that of N. gonorrhoeae FA1090 (see Supplementary Table S5, Supplementary Fig. S4B). Furthermore, most of the sequence differences are contained within the C-terminal end of the protein, which is not predicted to play a role in protein function (Isabella et al., 2009). In the gonococcus, NsrR has been shown to contain three cysteine residues that coordinate a [2Fe–2S] cluster involved in NO sensing (Isabella et al., 2009). All neisserial strains analysed here show conservation of these key cysteine residues (Supplementary Fig. S4B). In N. gonorrhoeae, N. lactamica and N. cinerea, the amino acid at position 93, which is located between the first two cysteines involved in Fe–S coordination (Cys90 and Cys97), is proline, whereas in N. meningitidis MC58, N. mucosa, N. subflava, N. flava and N. sicca, the amino acid at position 93 is a negatively charged Glu or Asp residue. Since these amino acids are within the Fe–S cluster coordination pocket of NsrR (Isabella et al., 2009), these amino acid differences may affect the chemical reactivity of the [2Fe–2S] cluster towards NO, and thus potentially change the sensitivity of the protein to its ligand. Regardless of this, both meningococcal and gonococcal NsrRs have been shown to be capable of responding to NO (Isabella et al., 2008; Rock et al., 2007).
The NarQP two-component regulatory system is involved in regulation of the neisserial denitrification pathway, and is known to activate aniA in both N. gonorrhoeae and N. meningitidis (Clark et al., 2009; Householder et al., 1999). The response regulator NarP is highly conserved in the neisserial genus, with the most distant NarP orthologue, expressed by N. mucosa, displaying 90 % similarity to gonococcal FA1090 NarP (Supplementary Table S5, Supplementary Fig. S4C). Interestingly, N. mucosa contains two copies of narP, which are highly similar to each other but not adjacent to one other on the chromosome. N. mucosa is also the only neisserial species to contain a functional Nar complex. This second NarP gene product could be involved in regulation of a subset of genes involved in nitrate utilization and/or reduction, though this strain apparently contains only one orthologue of NarQ (Supplementary Fig. S4D). All neisserial NarP orthologues contain the conserved aspartate residue presumed to be the phosphorylation target for the sensor kinase NarQ (Householder, 2000; Whitehead & Cole, 2006). Despite the similarity among neisserial NarP orthologues, large differences exist within the sequence of NarQ (Supplementary Table S5, Supplementary Fig. S4D).
NarQ is the sensor kinase of the NarQP two-component system and is anchored in the bacterial inner membrane (Szurmant et al., 2007). In E. coli, NarQ senses the presence of nitrite and relays this signal, via phosphorylation, to NarP, its cognate response regulator (Hartig et al., 1999). Contrary to what is observed in E. coli, the gonococcal NarQ homologue appears to be insensitive to nitrite and constitutively active, thus leading to constitutive phosphorylation of NarP (Clark et al., 2009; Isabella et al., 2008; Overton et al., 2006). Nitrite sensing in the gonococcus, at least at the level of aniA regulation, has apparently been taken over by NsrR in a complex regulatory mechanism whereby the product of nitrite reduction, NO, causes derepression by NsrR, which in turn allows access of NarP to RNA polymerase (RNAP), thus leading to activation (Clark et al., 2009; Overton et al., 2006). To date, no ligand has been found for gonococcal NarQ (Clark et al., 2009; Overton et al., 2006). Contrary to this finding, aniA expression has been shown to be nitrite-, but not NO-dependent in N. meningitidis, suggesting that meningococcal NarQ still responds directly to nitrite (Rock et al., 2007).
NarQ is not as well conserved as the response regulator NarP, suggesting that environmental sensing is different among strains and thus elicits a different regulatory response. The most divergent NarQ sequence, encoded by N. mucosa, contains 73 % similarity and only 60 % identity to that of the N. gonorrhoeae FA1090 NarQ protein (Supplementary Table S5). Many sensor kinase proteins contain periplasmic sensing domains on their N terminus (Szurmant et al., 2007; Wolanin et al., 2002). There exists the possibility that sequence differences in the N terminus of neisserial NarQ affect the ability to sense and respond and/or halt a response to nitrite.
A conserved domain search of neisserial NarQ proteins revealed a composition of four key domains, including HAMP, GAF, HisKA_3 and HATPase_c domains, all of which are predicted to be located on the cytoplasmic face of the inner membrane (Supplementary Table S6) (Marchler-Bauer et al., 2009; Szurmant et al., 2007). The HAMP domain, through rotation of a four α-helical bundle, is implicated in transduction of a perceived environmental signal (Szurmant et al., 2007). The HATPase_c and HisKA_3 domains form the catalytic core of the protein, where ATP is hydrolysed and kinase activity is utilized in the transfer of phosphate to the response regulator, respectively (Szurmant et al., 2007; Wolanin et al., 2002). The GAF domain is thought to play a role in environmental sensing at the cytoplasmic face of the inner membrane (Szurmant et al., 2007). Interestingly, whereas the HAMP, HATPase_c and HisKA_3 domains are very well conserved across the neisserial genus, the GAF domain is particularly divergent (Supplementary Table S6, Supplementary Fig. S4D) (Marchler-Bauer et al., 2009). GAF domains from N. gonorrhoeae and N. lactamica NarQ display the weakest conservation compared with other neisserial species (Supplementary Table S6, Supplementary Fig. S4D). The divergence within this GAF domain may affect the structure of NarQ in such a way as to alter its signal perception properties. In the case of N. gonorrhoeae, the GAF domain may have evolved this way to maintain NarQ in a constitutively active state, perhaps due to the fact that the gonococcus would not be expected to encounter much nitrite in its own environmental niche. The NarQ proteins from other neisserial species show greater conservation of this GAF domain, and due to this fact, may be capable of sensing nitrite, as has been observed in the meningococcus (Rock et al., 2007).
Concluding remarks
Examination of the neisserial denitrification pathway has resulted in a number of interesting observations. Some portions of the pathway, such as NO reduction, are very highly conserved across this genus (NorB, NsrR), while others, including nitrite reduction, have a moderate to high degree of divergence (AniA, NarQ). These evolutionary differences are likely to be due to the different ecological niches that these bacteria inhabit. Regulation of neisserial denitrification is highly conserved, with the exception of the N terminus and GAF domain within NarQ, which may cause differences in nitrite-sensing capability. The nitrous oxide reductase operon represents an example of extreme divergence among Neisseria species, with less than half of the species containing what appears to be a functional Nos system. Interestingly, both of the pathogenic species and a number of commensal species have used divergent mechanisms to disrupt genes in the Nos operon, suggesting that inactivation of this branch of the denitrification pathway provides some benefit. Measurement of nitrite and nitric oxide reductase activities in various Neisseria species suggests a similar potential for the generation and degradation of NO, which is important for establishment of an NO steady-state level. This study represents only a survey of neisserial denitrification, and examination of additional strains within a specific Neisseria species may reveal larger differences between the pathogenic and commensal Neisseriaceae.
The physiological relevance of denitrification can be considered by measuring nitrogen oxide levels that can serve as sources of input for the pathway. Dietary nitrate is absorbed in the gastrointestinal tract, leading to increases in plasma nitrate levels (Olin et al., 2001). The salivary glands actively take up nitrate in a plasma concentration-dependent manner, and this is reduced to nitrite by anaerobic bacteria within the oral cavity, generating nitrite levels ranging from 10 μM to >1 mM (Lundberg et al., 2004). High nitrite concentrations, in concert with a microaerobic niche, may afford Neisseria species that reside in the nasopharynx opportunities to utilize denitrification as a route for electron transfer. N. gonorrhoeae primarily colonizes the genital mucosa, a niche that may encounter an intermittent supply of nitrite through the flushing of urine. In addition, both anatomical sites are competent at host-generated NO production, providing an additional electron acceptor for energy production (Stefano et al., 2000; Bogdan, 2001). The NO steady-state levels produced by each Neisseria species are contingent on nitrite and NO inputs, as well as bacterial metabolism, so that the steady-state level of NO will differ depending on anatomical site, bacterial population size and immune system state of activation. Future studies aim to further elucidate the involvement of neisserial denitrification in bacterial metabolism as well as in the host response.
Acknowledgments
This study was supported by Public Health Service grant R01 AI 11709 from the National Institutes of Health. Additionally, V. M. I. was supported by NIH grant T32 AI07362.
Abbreviations
DETA/NO, 2,2′-(hydroxynitrosohydrazono) bis-ethanimine
Footnotes
Four supplementary figures and six supplementary tables are available with the online version of this paper. The supplementary figures show amino acid alignments of AniA across Neisseria species, amino acid alignments of NorB across Neisseria species, alignments of the nitrous oxide reductase complex proteins among Neisseria species and amino acid alignments of the denitrification pathway regulators. The supplementary tables list AniA/NirK amino acid alignments across Neisseria species, NorB sequence conservation among Neisseria species, gene identifications for the denitrification reductases within Neisseria species and the full denitrifier Ralstonia eutropha, gene identifications for the nitrous oxide reductase regulon within Neisseria species and Ralstonia eutropha, denitrification pathway regulators among Neisseria species and domain homologies for NarQ.
References
- Baek, S. H., Rajashekara, G., Splitter, G. A. & Shapleigh, J. P. (2004). Denitrification genes regulate Brucella virulence in mice. J Bacteriol 186, 6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, K. & Clark, V. L. (2008). Differences in nitric oxide steady states between arginine, hypoxanthine, uracil auxotrophs (AHU) and non-AHU strains of Neisseria gonorrhoeae during anaerobic respiration in the presence of nitrite. Can J Microbiol 54, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, U. (1961). Reduction of nitrate and nitrite by Neisseria. . Z Hyg Infektionskr 148, 45–50. [PubMed] [Google Scholar]
- Blakebrough, I. S., Greenwood, B. M., Whittle, H. C., Bradley, A. K. & Gilles, H. M. (1982). The epidemiology of infections due to Neisseria meningitidis and Neisseria lactamica in a northern Nigerian community. J Infect Dis 146, 626–637. [DOI] [PubMed] [Google Scholar]
- Bodenmiller, D. M. & Spiro, S. (2006). The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188, 874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan, C. (2001). Nitric oxide and the immune response. Nat Immun 2, 907–916. [DOI] [PubMed] [Google Scholar]
- Boulanger, M. J. & Murphy, M. E. (2002). Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: a new class of copper-containing nitrite reductases. J Mol Biol 315, 1111–1127. [DOI] [PubMed] [Google Scholar]
- Boulanger, M. J., Kukimoto, M., Nishiyama, M., Horinouchi, S. & Murphy, M. E. (2000). Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J Biol Chem 275, 23957–23964. [DOI] [PubMed] [Google Scholar]
- Cardinale, J. A. (1999). Structural and functional analysis of aniA, the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae. PhD thesis, University of Rochester, Rochester, NY.
- Cardinale, J. A. & Clark, V. L. (2005). Determinants of nitric oxide steady-state levels during anaerobic respiration by Neisseria gonorrhoeae. Mol Microbiol 58, 177–188. [DOI] [PubMed] [Google Scholar]
- Clark, V. L., Campbell, L. A., Palermo, D. A., Evans, T. M. & Klimpel, K. W. (1987). Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infect Immun 55, 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, V. L., Isabella, V. M., Barth, K. & Overton, T. (2009). Regulation and function of the Neisserial denitrification pathway: life with limited oxygen. In Neisseria: Molecular Mechanisms of Pathogenesis. Edited by C. A. Genco & L. Wetzler. Norwich, UK: Horizon Scientific Press.
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta, M. L. B. T., Shao J., Ketterer C., Steichen, C., Jennings, M. P. & Apicella, M. A. (2008). Transcriptional profiling of Neisseria gonorrhoeae biofilm indicates that biofilms grow using anaerobic or microaerobic metabolism. Abstract P033, p114, in 16th International Pathogenic Neisseria Conference, September 7–12 2008, Rotterdam.
- Fenderson, F. F., Kumar, S., Adman, E. T., Liu, M. Y., Payne, W. J. & LeGall, J. (1991). Amino acid sequence of nitrite reductase: a copper protein from Achromobacter cycloclastes. Biochemistry 30, 7180–7185. [DOI] [PubMed] [Google Scholar]
- Filenko, N., Spiro, S., Browning, D. F., Squire, D., Overton, T. W., Cole, J. & Constantinidou, C. (2007). The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol 189, 4410–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberthorpe, N. J., Lee, M. E., Stevanin, T. M., Read, R. C. & Poole, R. K. (2007). NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-γ-stimulated J774.2 macrophages. Microbiology 153, 1756–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, R., Goldschneider, I., Lepow, M. L., Draper, T. F. & Randolph, M. (1978). Carriage of Neisseria meningitidis and Neisseria lactamica in infants and children. J Infect Dis 137, 112–121. [DOI] [PubMed] [Google Scholar]
- Goretski, J., Zafiriou, O. C. & Hollocher, T. C. (1990). Steady-state nitric oxide concentrations during denitrification. J Biol Chem 265, 11535–11538. [PubMed] [Google Scholar]
- Hartig, E., Schiek, U., Vollack, K. U. & Zumft, W. G. (1999). Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J Bacteriol 181, 3658–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, D. A. & Ruskin, J. (1981). Are the “nonpathogenic” neisseriae pathogenic? Am J Clin Pathol 75, 739–743. [DOI] [PubMed] [Google Scholar]
- Holloway, P., McCormick, W., Watson, R. J. & Chan, Y. K. (1996). Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J Bacteriol 178, 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Householder, T. C. (2000). Regulation of gene expression in Neisseria gonorrhoeae: aniA and norB, examples from the denitrification pathway. PhD thesis, University of Rochester, Rochester, NY.
- Householder, T. C., Belli, W. A., Lissenden, S., Cole, J. A. & Clark, V. L. (1999). cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J Bacteriol 181, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Householder, T. C., Fozo, E. M., Cardinale, J. A. & Clark, V. L. (2000). Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect Immun 68, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella, V., Wright, L. F., Barth, K., Spence, J. M., Grogan, S., Genco, C. A. & Clark, V. L. (2008). cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154, 226–239. [DOI] [PubMed] [Google Scholar]
- Isabella, V. M., Lapek, J. D., Jr, Kennedy, E. M. & Clark, V. L. (2009). Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol 71, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani, T., Watanabe, H., Arima, K. & Beppu, T. (1981). A blue protein as an inactivating factor for nitrite reductase from Alcaligenes faecalis strain S-6. J Biochem 89, 463–472. [DOI] [PubMed] [Google Scholar]
- Kataoka, K., Furusawa, H., Takagi, K., Yamaguchi, K. & Suzuki, S. (2000). Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J Biochem 127, 345–350. [DOI] [PubMed] [Google Scholar]
- Kellogg, D. S., Jr, Peacock, W. L., Jr, Deacon, W. E., Brown, L. & Pirkle, D. I. (1963). Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol 85, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, J. S. (1988). Historical perspectives and identification of Neisseria and related species. Clin Microbiol Rev 1, 415–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, J. S. & Clark, V. L. (1984). Anaerobic growth of Neisseria gonorrhoeae coupled to nitrite reduction. Infect Immun 46, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, S. C., Schulz, B. L., Power, P. M. & Jennings, M. P. (2009). The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem Biophys Res Commun 378, 84–89. [DOI] [PubMed] [Google Scholar]
- Lauer, B. A. & Fisher, C. E. (1976). Neisseria lactamica meningitis. Am J Dis Child 130, 198–199. [DOI] [PubMed] [Google Scholar]
- Lissenden, S., Mohan, S., Overton, T., Regan, T., Crooke, H., Cardinale, J. A., Householder, T. C., Adams, P., O'Conner, C. D. & other authors (2000). Identification of transcription activators that regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol Microbiol 37, 839–855. [DOI] [PubMed] [Google Scholar]
- Lundberg, J. O., Weitzberg, E., Cole, J. A. & Benjamin, N. (2004). Nitrate, bacteria and human health. Nat Rev Microbiol 2, 593–602. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., Fong, J. H., Geer, L. Y., Geer, R. C., Gonzales, N. R. & other authors (2009). CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37, D205–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies, J., Jose, J. & Meyer, T. F. (1997). The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol Gen Genet 256, 525–532. [DOI] [PubMed] [Google Scholar]
- Morse, S. A. & Knapp, J. A. (1987). Neisserial infections. In Diagnostic Procedures for Bacterial Infections, pp. 407–432. Edited by B. B. Wentworth. Washington, DC: American Public Health Association.
- Nakano, M. M., Geng, H., Nakano, S. & Kobayashi, K. (2006). The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J Bacteriol 188, 5878–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olin, A. C., Aldenbratt, A., Ekman, A., Ljungkvist, G., Jungersten, L., Alving, K. & Toren, K. (2001). Increased nitric oxide in exhaled air after intake of a nitrate-rich meal. Respir Med 95, 153–158. [DOI] [PubMed] [Google Scholar]
- Overton, T., Reid, E. G., Foxall, R., Smith, H., Busby, S. J. & Cole, J. A. (2003). Transcription activation at Escherichia coli FNR-dependent promoters by the gonococcal FNR protein: effects of a novel S18F substitution and comparisons with the corresponding substitution in E. coli FNR. J Bacteriol 185, 4734–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton, T. W., Whitehead, R., Li, Y., Snyder, L. A., Saunders, N. J., Smith, H. & Cole, J. A. (2006). Coordinated regulation of the Neisseria gonorrhoeae-truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ–NarP. J Biol Chem 281, 33115–33126. [DOI] [PubMed] [Google Scholar]
- Philippot, L. (2002). Denitrifying genes in bacterial and archaeal genomes. Biochim Biophys Acta 1577, 355–376. [DOI] [PubMed] [Google Scholar]
- Potter, L., Angove, H., Richardson, D. & Cole, J. (2001). Nitrate reduction in the periplasm of Gram-negative bacteria. Adv Microb Physiol 45, 51–112. [DOI] [PubMed] [Google Scholar]
- Rock, J. D., Mahnane, M. R., Anjum, M. F., Shaw, J. G., Read, R. C. & Moir, J. W. (2005). The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol Microbiol 58, 800–809. [DOI] [PubMed] [Google Scholar]
- Rock, J. D., Thomson, M. J., Read, R. C. & Moir, J. W. (2007). Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J Bacteriol 189, 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, E., Henne, A., Cramm, R., Eitinger, T., Friedrich, B. & Gottschalk, G. (2003). Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. J Mol Biol 332, 369–383. [DOI] [PubMed] [Google Scholar]
- Short, H. B., Clark, V. L., Kellogg, D. S., Jr & Young, F. E. (1982). Anaerobic survival of clinical isolates and laboratory strains of Neisseria gonorrhoea: use in transfer and storage. J Clin Microbiol 15, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanelli, P., Colotti, G., Neri, A., Salucci, M. L., Miccoli, R., Di Leandro, L. & Ippoliti, R. (2008). Molecular characterization of nitrite reductase gene (aniA) and gene product in Neisseria meningitidis isolates: is aniA essential for meningococcal survival? IUBMB Life 60, 629–636. [DOI] [PubMed] [Google Scholar]
- Stefano, G., Gouman, Y., Bilfinger, T. V., Welters, I. & Cadet, P. (2000). Basal nitric oxide limits immune, nervous, and cardiovascular excitation: human endothelia express a mu opiate receptor. Prog Neurobiol 60, 513–530. [DOI] [PubMed] [Google Scholar]
- Stevanin, T. M., Moir, J. W. & Read, R. C. (2005). Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun 73, 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin, T. M., Laver, J. R., Poole, R. K., Moir, J. W. & Read, R. C. (2007). Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect 9, 981–987. [DOI] [PubMed] [Google Scholar]
- Szurmant, H., White, R. A. & Hoch, J. A. (2007). Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol 17, 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares, P., Pereira, A. S., Moura, J. J. & Moura, I. (2006). Metalloenzymes of the denitrification pathway. J Inorg Biochem 100, 2087–2100. [DOI] [PubMed] [Google Scholar]
- Thomson, M. J., Stevanin, T. M. & Moir, J. W. (2008). Measuring nitric oxide metabolism in the pathogen Neisseria meningitidis. Methods Enzymol 437, 539–560. [DOI] [PubMed] [Google Scholar]
- Tunbridge, A. J., Stevanin, T. M., Lee, M., Marriott, H. M., Moir, J. W., Read, R. C. & Dockrell, D. H. (2006). Inhibition of macrophage apoptosis by Neisseria meningitidis requires nitric oxide detoxification mechanisms. Infect Immun 74, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst, N. E., Picardo, K. F., Iglewski, B. H. & Haidaris, C. G. (2007). Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun 75, 3780–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedros, N. A. (1984). Genus I. Neisseria. In Bergey's Manual of Systematic Bacteriology, pp. 290–296. Edited by N. R. Krieg and J. G. Holt. Baltimore, MD: Williams & Wilkins.
- Velasco, L., Mesa, S., Xu, C. A., Delgado, M. J. & Bedmar, E. J. (2004). Molecular characterization of nosRZDFYLX genes coding for denitrifying nitrous oxide reductase of Bradyrhizobium japonicum. Antonie Van Leeuwenhoek 85, 229–235. [DOI] [PubMed] [Google Scholar]
- Whitehead, R. N. & Cole, J. A. (2006). Different responses to nitrate and nitrite by the model organism Escherichia coli and the human pathogen Neisseria gonorrhoeae. Biochem Soc Trans 34, 111–114. [DOI] [PubMed] [Google Scholar]
- Wolanin, P. M., Thomason, P. A. & Stock, J. B. (2002). Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3, REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]