Abstract
Previous genome-wide screens identified >100 host genes affecting tombusvirus replication using yeast model host. One of those factors was Nsr1p (nucleolin), which is an abundant RNA binding shuttle protein involved in rRNA maturation and ribosome assembly. We find that over-expression of Nsr1p in yeast or in Nicotiana benthamiana inhibited the accumulation of tombusvirus RNA by ~10-fold. Regulated over-expression of Nsr1p revealed that Nsr1p should be present at the beginning of viral replication for efficient inhibition, suggesting that Nsr1p inhibits an early step in the replication process. In vitro experiments revealed that Nsr1p binds preferably to the 3' UTR in the viral RNA. The purified recombinant Nsr1p inhibited the in vitro replication of the viral RNA in a yeast cell-free assay when pre-incubated with the viral RNA before the assay. These data support the model that Nsr1p/nucleolin inhibits tombusvirus replication by interfering with the recruitment of the viral RNA for replication.
INTRODUCTION
RNA viruses, which have small genomes with limited coding potential, depend on recruited host factors during the replication process. Therefore, virus - host interaction is critical for successful viral infections as well as for triggering anti-viral responses in the host. Recent genome-wide screens with several RNA viruses revealed rather complex interactions involving several hundred host genes (Cherry et al., 2005; Hao et al., 2008; Jiang et al., 2006; Krishnan et al., 2008; Kushner et al., 2003; Panavas et al., 2005b; Serviene et al., 2006; Serviene et al., 2005; Tai et al., 2009). While many of the identified genes are important for RNA virus replication, other host genes were found inhibitory by reducing the accumulation of the viral RNA. The identified inhibitory genes could be part of the innate immune responses of the host.
Tomato bushy stunt virus (TBSV) has emerged as one of highly suitable model virus systems to study RNA virus replication and host - virus interaction due to the recent development of the highly tractable yeast as a model host (Nagy, 2008; Panavas and Nagy, 2003b) and cell-free approaches (Panaviene et al., 2004; Pogany and Nagy, 2008; Pogany et al., 2008). Replication of a short TBSV replicon (rep)RNA, which is a 621 nt defective interfering RNA carrying four noncontiguous regions from the genomic (g)RNA, in yeast requires the co-expression of the viral p33 and p92pol replication proteins, which form the membrane-associated viral replicase (Panaviene, Panavas, and Nagy, 2005; Panaviene et al., 2004). Systematic, genome-wide and proteomics approaches have led to the identification of more than 200 host proteins/genes affecting TBSV replication/recombination or interacting with the viral replication proteins/viral RNA (Jiang et al., 2006; Li et al., 2008; Li et al., 2009; Panavas et al., 2005b; Serva and Nagy, 2006; Serviene et al., 2006; Serviene et al., 2005). A rapidly progressing research area after the systematic genome-wide screens is the dissection of the functions of the identified host factors during virus replication. Five of the identified host factors are part of the viral replicase complex, facilitating the assembly of the replicase, regulating the ratio of plus- versus minus-strand RNA synthesis, enhancing the stability of the viral replication proteins or their intracellular transportations and insertions into subcellular membranes (Jonczyk et al., 2007; Li et al., 2008; Li et al., 2009; Pathak, Sasvari, and Nagy, 2008; Pogany et al., 2008; Serva and Nagy, 2006; Wang and Nagy, 2008; Wang, Stork, and Nagy, 2009). Other host proteins tested in more detail affected viral RNA degradation and viral recombination (Cheng et al., 2007; Cheng, Serviene, and Nagy, 2006; Jaag and Nagy, 2009) or had only indirect effect on TBSV repRNA accumulation (Jaag, Stork, and Nagy, 2007). Importantly, the relevance of several host genes identified in yeast has also been confirmed in the natural plant host as well (Jaag and Nagy, 2009; Wang and Nagy, 2008; Wang, Stork, and Nagy, 2009). These discoveries justify the use of yeast model host for replication studies with TBSV.
In this paper, we further characterize the inhibitory role of the previously identified nucleolin (Nsr1p in yeast) in TBSV replication (Panavas et al., 2005b). Nucleolin/Nsr1p is an abundant, ubiquitously expressed protein, which is involved in ribosome biogenesis (Mongelard and Bouvet, 2007). Nucleolin also affects transcription of rDNA, processing and modification of rRNA and nuclear - cytosolic transport of ribosomal protein and ribosomal subunits by shuttling between the nucleus and the cytoplasm (Tuteja and Tuteja, 1998). Nucleolin is found in various cell compartments and it is especially abundant in the nucleolus.
Nuceolin/Nsr1 has three well-defined domains: the N-terminal domain with alternating acidic and basic stretches is involved in rDNA transcription by interacting with rDNA repeats and histone H1 as well as in nuclear localization. The central portion is the RNA-binding domain carrying RRM (RNA recognition motif) repeats, whereas the C-terminal part contains the glycine-arginine-rich (GAR) domain. The GAR domain is involved in interaction with the ribosomal proteins and it was suggested to affect ribosomal assembly and transport (Tuteja and Tuteja, 1998).
The analysis of nucleolin functions is challenging due to the broad range of mechanisms performed by nucleolin, which affect DNA and RNA metabolism, and its presence in various subcellular locations (Mongelard and Bouvet, 2007). In addition to binding to RNA/DNA and its role in proper folding of pre-rRNA, nucleolin also interacts with many proteins during ribosome assembly and it is involved in regulating the RNA polymerase I-based transcription. Arabidopsis has two nucleolin genes, but only AtNuc-L1 is expressed ubiquitously under normal growth conditions (Kojima et al., 2007; Pontvianne et al., 2007). The nucleolin gene from pea was able to complement nsr1Δ yeast by rescuing the reduced level of rRNA (Reichler et al., 2001), suggesting that the plant nucleolin has similar functions to the yeast NSR1.
Here, we confirm that Nsr1p/nucleolin is an inhibitor of TBSV replication. Over-expression of the yeast Nsr1p in yeast or the Arabidopsis nucleolin in Nicotiana benthamiana reduced the accumulation of tombusvirus RNA and inhibited the in vitro activity of the tombusvirus replicase. We found that Nsr1p binds to the upstream portion of the 3'UTR in (+)repRNA in vitro. Overall, these data suggest that Nsr1p could inhibit TBSV RNA replication by inhibiting the recruitment of the viral RNA for replication.
RESULTS
Over-expression of Nsr1p inhibits TBSV repRNA replication in yeast
To test the effect of Nsr1p on TBSV repRNA accumulation in yeast, we over-expressed Nsr1p either as an N-terminal 6xHis-tagged Nsr1p or the C-terminal FLAG-tagged Nsr1p from a high copy number plasmid together with p33 and p92pol replication proteins and the TBSV repRNA (Fig. 1A). The accumulation of repRNA was measured via Northern blotting 24 hours after induction of TBSV repRNA replication via the galactose-inducible GAL1 promoter. These experiments revealed that the C-terminal FLAG-tagged Nsr1p inhibited repRNA accumulation by 10-fold (Fig. 1A, lanes 4–6), while the inhibitory effect of 6xHis-tagged Nsr1p was less (by ~60%, lanes 1–3). Also, over-expression of the 6xHis-tagged Nsr1p inhibited repRNA accumulation in nsr1Δ yeast by ~3-fold (Fig. 1A, lanes 14–16), when compared with TBSV repRNA accumulation in nsr1Δ yeast. These experiments also confirmed that TBSV repRNA accumulation is 3-fold higher in nsr1Δ yeast (Fig. 1A, lanes 10–13) than in the parental BY4741 that expresses Nsr1p from the native promoter (lanes 7–9). Altogether, these data firmly established that Nsr1p is a potent inhibitor of TBSV repRNA accumulation in yeast.
Fig. 1.
Inhibition of tombusvirus RNA accumulation by over-expression of Nsr1p in yeast. (A) Nsr1p as a FLAG or 6xHis fusion protein and the TBSV DI-72 repRNA were expressed from the GAL1 promoter, whereas 6xHis-p33 and 6xHis-p92 were expressed from the ADH1 and CUP1 promoters, respectively, by simultaneous induction with galactose and copper ions in the parental BY4741 or nsr1Δ yeast strains. The total RNA samples were obtained after 24 hour culturing at 29 ºC. The accumulation of repRNA was estimated using Northern blotting. The 18S rRNA was used as a loading control. Each experiment was repeated twice. (B) The level of transcription of TBSV DI-72 repRNA from the GAL1 promoter in BY4741 strain expressing Nsr1-FLAG or a short peptide (pYES), but not expressing p92, was estimated by Northern blotting. The two bands represent the ribozyme cleaved and uncleaved TBSV repRNA transcripts. The quantified data are based on the cleaved bands, but the total level (cleaved plus uncleaved) of repRNA transcripts show similar values (not shown). Note that the replicating TBSV repRNA in yeast co-expressing p33/p92 was used as a size marker. The replicating repRNA reaches more than 1,000-fold higher accumulation level. (C) Decreased replicase activity from yeast over-expressing Nsr1p. The top panel shows a replicase activity assay with membrane-enriched preparations obtained from yeast expressing high level of Nsr1p in the parental BY4741 or nsr1Δ yeast strains. The membrane-enriched fraction contains the endogenous repRNA template that is used during the in vitro replicase assay in the presence of 32P-UTP and the other unlabeled rNTPs. Note that the in vitro activities of the tombusviral replicase were normalized based on p33 levels (see bottom panel). The bottom panel also shows the level of FLAG or 6xHis-tagged Nsr1p expression based on Western blotting with the mixture of anti-FLAG and anti-6xHis antibodies.
Since Nsr1p is mostly a nuclear protein, it is possible that it could affect the plasmid-based transcription of the TBSV repRNA or the cleavage at the 3’ end by the ribozyme, which have been engineered to launch TBSV repRNA replication with the authentic 3’ end from the expression plasmid in the yeast model host (Panavas and Nagy, 2003b; Panaviene et al., 2004). Over-expression of Nsr1p, however, did not affect significantly the amount of repRNA transcripts made from the GAL1 promoter/expression plasmid in the absence of the viral replication proteins (Fig. 1B, lanes 2–4 versus 5–7). Also, the amount of p33 made in yeast over-expressing Nsr1p was comparable to that obtained in the parental yeast expressing native level of Nsr1p (not shown). These data suggest that over-expression of Nsr1p does not affect the amount of plasmid-born repRNA, its processing by the rybozyme or the expression of the viral replication proteins.
To test if Nsr1p can affect the activity of the tombusvirus replicase, we isolated membrane-bound replicase preparations from the above yeast strains, followed by in vitro replicase assay with the co-purified repRNA (Panaviene, Panavas, and Nagy, 2005; Panaviene et al., 2004). As expected, we found that the tombusvirus replicase activity was ~3-fold lower when obtained from yeast over-expressing the FLAG-tagged Nsr1p (Fig. 1C, lanes 4–6) when compared with the preparation obtained from the parental BY4741 (lanes 1–3). On the contrary, the replicase preparation obtained from nsr1Δ yeast (lanes 7–9) was almost twice as active as the control preparation. Altogether, the in vitro data support the model that Nsr1p inhibits TBSV repRNA accumulation by inhibiting the viral replicase.
Expression of the plant nucleolin inhibits TBSV replication in Nicotiana benthamiana host
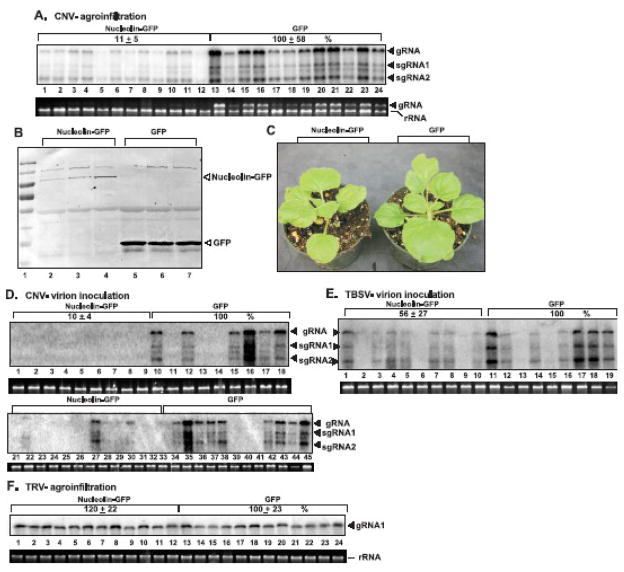
To test if the plant nucleolin, the homolog of yeast Nsr1p, might have similar inhibitory function against TBSV, we expressed the Arabidopsis thaliana nucleolin (AtNuc-L1) tagged with GFP (Kojima et al., 2007) in N. benthamiana leaves via agroinfiltration (Jaag and Nagy, 2009). The genomic RNA of Cucumber necrosis virus (CNV), a very close relative of TBSV, was co-expressed with AtNuc-L1-GFP via agroinfiltration in the same leaves. Leaf samples taken 2.5 days latter were analyzed via Northern blotting to estimate the level of CNV RNA accumulation (Fig. 2A). Interestingly, expression of AtNuc-L1-GFP in N. benthamiana leaves inhibited the accumulation of CNV RNA by ~10-fold when compared with the control that expressed GFP in leaves (Fig. 2 A, lanes 1–12 versus 13–24). The agroinfiltrated leaves expressed the GFP control at a higher level than AtNuc-L1-GFP (Fig. 2B). Over-expression of AtNuc-L1-GFP had only mild effect on the uninoculated agro-infiltrated leaves during these experiments, reducing the overall size of the leaves and causing minor growth inhibition of the plants (Fig. 2C).
Fig. 2.
Inhibition of tombusvirus genomic RNA accumulation by transient expression of Arabidopsis nucleolin in N. benthamiana leaves. (A) Leaves of N. benthamiana were co-agroinfiltrated to express Nucleolin-GFP (AtNuc-L1-GFP) and CNV genomic RNA from expression plasmids. Total RNA samples were prepared from randomly chosen areas of the infiltrated leaves 2.5 days post-infiltration, followed by Northern blotting to detect the accumulation level of CNV gRNA. rRNA was used as a loading control. Agroinfiltrated leaves co-expressing CNV and GFP were used as controls. (B) Western blotting shows the accumulation level of AtNuc-L1-GFP in the agroinfiltrated N. benthamiana leaves 2.5 days post-infiltration in comparison with the accumulation level of GFP. (C) Transient expression of AtNuc-L1-GFP in agroinfiltrated N. benthamiana leaves did not cause significant growth inhibition 2.5 days post-infiltration in comparison with plant expressing GFP. (D) Leaves of N. benthamiana were agroinfiltrated to express AtNuc-L1-GFP or GFP, followed by inoculation with inoculum containing CNV (mutant 20KSTOP) virions. Total RNA samples were prepared from randomly chosen areas of the infiltrated leaves 4 days post-infiltration, followed by Northern blotting to detect the accumulation level of CNV gRNA. See panel A for details. Note that variation in CNV RNA levels in different samples within the same set of experiment is likely due to uneven distribution of 'infection foci" (those areas in the leaf where the virus was able to start infection) and the random sampling approach. The experiments were repeated and the averages were calculated based on 18–42 samples/experiment. Two panels are shown for CNV due to the low level accumulation. (E) Leaves of N. benthamiana were agroinfiltrated to express AtNuc-L1-GFP or GFP, followed by inoculation with inoculum containing TBSV virions. See panel D for details. (F) Leaves of N. benthamiana were co-agroinfiltrated to express AtNuc-L1-GFP and TRV genomic RNA1/2. See panel A for details.
We found similar ~10-fold inhibition of CNV RNA accumulation when CNV replication was initiated by sap-inoculation with CNV virions, which represents one of the natural ways for CNV to spread, in N. benthamiana leaves agroinfiltrated 2 days earlier with a DNA construct expressing AtNuc-L1-GFP (Fig. 2D). The inhibitory effect of AtNuc-L1-GFP was less pronounced against infections started with TBSV virions (Fig. 2E). It is possible that higher level expression of AtNuc-L1-GFP is required against TBSV than CNV infection. It is also possible that the bigger inhibitory effect of nucleolin over-expression on CNV was due to the lack of expression of p20 suppressor of gene silencing from the CNV genome, while the TBSV expressed the p19 suppressor of gene silencing. This difference may make CNV more sensitive to nucleolin levels than TBSV. Overall, these data demonstrate that the plant nucleolin can inhibit the accumulation of tombusvirus genomic RNAs in an experimental host, even when the infection is initiated with the highly infectious virions.
To test if AtNuc-L1 can also inhibit a distantly related plant RNA virus, namely Tobacco rattle virus (TRV), which belongs to a different supergroup, we agroinfiltrated N. benthamiana leaves to co-express TRV RNAs and AtNuc-L1-GFP. Northern blot analysis of TRV RNA1 levels revealed the lack of inhibition of TRV accumulation by AtNuc-L1-GFP (Fig. 2F, lanes 1–12 versus 13–24). Thus, nucleolin has different effects on tombus- versus tobraviruses, which belong to different supergroups of RNA viruses.
Nsr1p inhibits the early steps in TBSV replication
After confirming the relevance of nucleolin/Nsr1p in inhibition of tombusvirus RNA replication in yeast as well as in a plant host, our goal was to dissect what steps of TBSV replication could be inhibited by this host protein. Tombusvirus replication is a complex process that consists of at least six defined steps after translation of the viral RNA (Nagy and Pogany, 2006). The early steps include selection of the viral RNA by selective binding of the viral p33 to the p33RE cis-acting element in the (+)RNA (Monkewich et al., 2005; Pogany, White, and Nagy, 2005), followed by recruitment of the viral RNA/replication protein complex to the site of replication (peroxisomal or ER membranes), and the assembly of the viral replicase into special membranous spherules. This is followed by the late steps of replication, such as minus- and plus-strand synthesis, release of the newly synthesized (+)RNA progeny from the replicase and the final disassembly of the replicase complex (Nagy and Pogany, 2006).
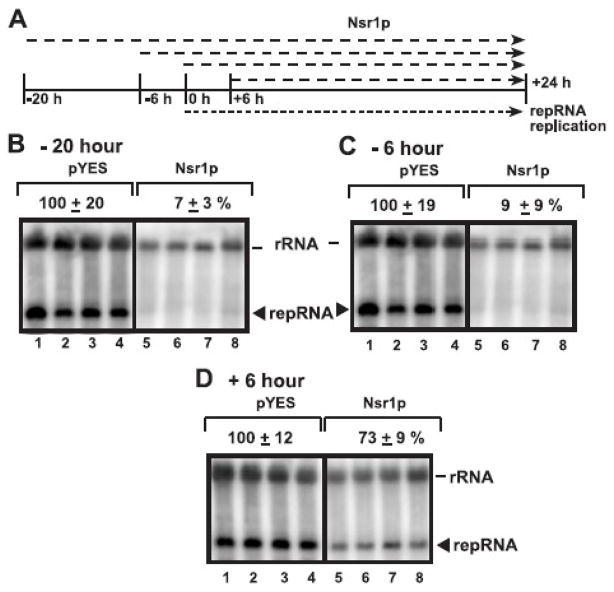
To test if Nsr1p could inhibit early or late steps of TBSV replication, we started the over-expression of Nsr1p from the galactose-inducible GAL1 promoter at various time points when compared with initiating TBSV replication from the copper-inducible CUP1 promoter (chosen as 0 hr time point, Fig. 3A). Over-expression of Nsr1p starting from 20 hr or 6 hr prior to launching TBSV replication resulted in ~10–15-fold inhibition of TBSV repRNA accumulation (Fig. 3B–C). This level of inhibition is higher than that obtained when Nsr1p was over-expressed from 0 hr time point (Fig. 1A, lanes 4–6). However, over-expression of Nsr1p 6 hours after launching TBSV replication (Fig. 3D) resulted in only a moderate level (by ~30 %) inhibition of TBSV repRNA accumulation. Altogether, these data support the model that Nsr1p inhibits TBSV replication most efficiently at the early time points.
Fig. 3.
The effect of time of expression of Nsr1p on inhibition of tombusvirus RNA accumulation in yeast. (A) A scheme showing the time of expression of 6xHis-Nsr1p in comparison with TBSV replication in yeast. 6xHis-Nsr1p was expressed form the GAL1 promoter, whereas repRNA replication was launched from the CUP1 promoter in the parental BY4741 yeast strain. (B-C-D) The yeast transformants were pre- grown in SC-ULH media with 2% glucose for 24 hr at 29°C, then transferred to a media with 2% galactose (starting OD600 was ~0.3) and further cultured at 29°C. Copper sulfate (50 μM) was added at different time points, such as 20 hr or 6 hr after or 6 hr prior to the addition of galactose containing medium to initiate repRNA replication. The accumulation of repRNA was estimated using Northern blotting after 24 hours of culturing of yeast in the presence of copper ions. See further details in the legend to Fig. 1A.
Nsr1p binds to the 3’ UTR of the TBSV (+)RNA in vitro
To identify the target of Nsr1p during TBSV repRNA replication that leads to inhibition of replication, we tested if the purified recombinant Nsr1p could bind to p33 and p92pol replication proteins and/or the viral (+)RNA. Although we could not detect interaction between Nsr1p and the viral replication proteins in vitro (not shown), we observed that Nsr1p bound to the 32P-labeled DI-72 (+) repRNA in a UV-cross-linking assay (Fig. 4A, lane 2). Deletion of the known RNA-binding domain in the recombinant Nsr1p (mutant GST-ΔRBD, lane 1) (Bouvet et al., 2001) abolished the ability of Nsr1p to bind to the repRNA. The purified GST was incapable of binding to the repRNA under the conditions used, suggesting that the recombinant GST-Nsr1 was responsible for RNA binding.
Fig. 4.
In vitro binding of recombinant Nsr1p to the (+)repRNA. (A) Top panel: UV-cross-linking assay with 2 μg of purified recombinant GST-ΔRBD (Nsr1p missing the central RNA binding domains), GST-Nsr1 or GST and ~5nM 32P-labeled DI-72 (+)repRNA. Bottom panel: Coomassie blue staining of the SDS-PAGE shown in the top panel, showing the purified recombinant GST-ΔRBD, GST-Nsr1 and GST proteins from E. coli. The fusion proteins were purified using GST affinity chromatography. (B) A gel mobility shift assay showing interactions between the recombinant GST-Nsr1 and 32P-labeled TBSV DI-72 (+)repRNA. The in vitro binding was analyzed in 4% non-denaturing polyacrylamide gel. The unbound, free RNA probe and the shifted (bound) RNA/protein complexes are marked on the right. GST-Nsr1 and GST were used in increasing amounts (400, 800 and 1600 ng protein/per lane).
To confirm the results from the above UV-cross-linking experiments, we performed gel mobility-shift experiments with purified recombinant GST-Nsr1 and 32P-labeled DI-72 (+) repRNA. This experiment revealed that Nsr1p bound to the viral RNA (Fig. 4B). Since the extent of the band shift increased with increasing amounts of GST-Nsr1, it is likely that more than one Nsr1p molecules can bind to the same viral RNA molecule in vitro.
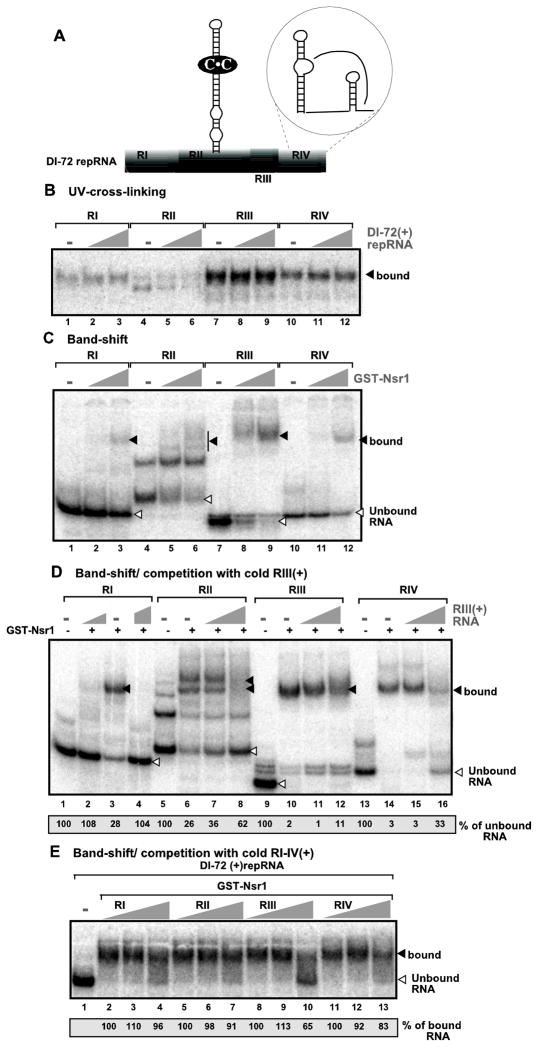
To test if there is a specific binding site for Nsr1p in DI-72(+) repRNA, we separately used the four segments of DI-72(+), known as RI-RIV (Fig. 5A) (White and Morris, 1994), as 32P-labeled probes in UV cross-linking experiments. This analysis revealed that Nsr1p bound preferably to RIII(+), moderately to RIV(+) and to a lesser extent to RI(+) and RII(+) (Fig. 5B). Gel mobility-shift assays confirmed that Nsr1p binding to RIII(+) was the most efficient (Fig. 5C, lanes 8–9). However, RII(+) and RIV(+) also bound to Nsr1p (lanes 6 and 12), while binding of RI(+) was the least efficient (lane 3).
Fig. 5.
Nsr1p binds to RIII in (+)repRNA in vitro. (A) Schematic representation of the various regions in DI-72(+) repRNA. The 169 nt long RI(+) represents the 5' UTR; the 239 nt long RII(+) is derived from the p92 ORF, whereas the 82nt long RIII(+) represents a short segment of the very 3' end of p19/p22 ORF and the 5' portion of 3' UTR and the 131 nt long RIV(+) is from the very 3' end of the genomic RNA. RII(+) contains the p33 recognition element (p33RE), which is a stem-loop structure with a C•C mismatch, required for RNA recruitment, while RIV(+) contains the replication silencer element and the genomic promoter (circled), required together with p33RE for the assembly of the viral replicase. (B) UV-cross-linking assay with 2 μg of purified GST-Nsr1 and ~5nM 32P-labeled RI(+), RII(+), RIII(+) or RIV(+) of DI-72 repRNA. See further details in Fig. 4A. We performed the UV-cross-linking assay in the absence (lanes 1, 4, 7, 10) or presence of 5 and 50 nM cold competitor DI-72 (+)RNA. (C) A gel mobility shift assay showing interactions between the recombinant GST-Nsr1 and 32P-labeled RI(+), RII(+), RIII(+) or RIV(+). GST-Nsr1 was used in increasing amounts (0, 800 and 1600 ng protein/per lane). See further details in Fig. 4B. (D) Band shift experiments with cold competitor RIII(+) RNA. The gel mobility shift assay was performed with 32P-labeled RI(+), RII(+), RIII(+) or RIV(+) and recombinant GST-Nsr1 (2 μg / per lane) in the absence or presence of 50 and 500 nM cold competitor RIII(+) RNA. We quantified the unbound RNA and the values show the % of the control samples (no GST-Nsr1 and competitor RNA, such as lanes 1, 5, 9 and 13). (E) Gel mobility shift assay with cold competitor RI, RII, RIII and RIV RNAs. The assay was performed with 32P-labeled DI-72 (+)RNA and recombinant GST-Nsr1 (2 μg / per lane) in the absence or presence of 50 and 500 nM cold competitor RI(+), RII(+), RIII(+) or RIV(+) RNAs. We quantified the bound RNA and the values show the % of the control samples.
Template-competition experiments with cold RIII(+) confirmed that the cold template competed the least efficiently against the 32P-labeled RIII(+) probe (Fig. 5D, lane 11–12), while it competed efficiently against the RI(+) probe (lanes 2 and 4). Again, RII(+) and RIV(+) showed moderate level of competition based on the amount of released probes when excess amount of cold competitor was used.
Finally, we used the cold RI(+), RII(+), RIII(+) and RIV(+) competitors separately against the 32P-labeled full-length DI-72(+) RNA probe in a gel mobility-shift assay (Fig. 5E). Only RIII(+) competed efficiently with the labeled DI-72(+) RNA for binding to Nsr1p (Fig. 5E, lane 10), confirming that RIII(+) is the preferred site for Nsr1p binding. RIII(+) does not contain the previously identified nucleolin-recognition element, which is a stem-loop structure with the loop containing UCCCGA sequence (Bouvet et al., 2001). Thus, it is likely that RIII(+) contains a not yet defined sequence/structure recognized by Nsr1p.
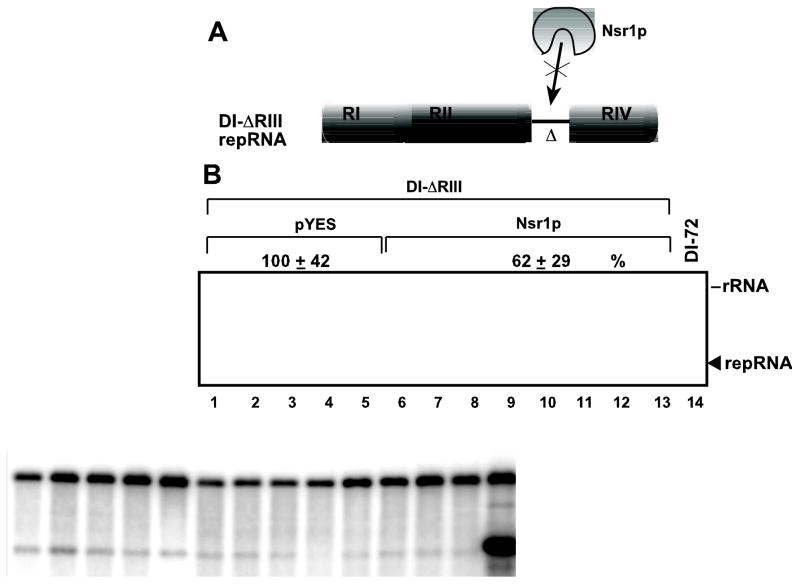
RIII is mostly derived from the 3’ UTR of the TBSV genome, but its function is not essential and the currently known role is on the minus-strand serving as a replication enhancer (Panavas and Nagy, 2003a; Ray and White, 2003). Accordingly, deletion of RIII in DI-72 leads to vastly reduced replication (Ray and White, 2003). Since Nsr1p binds preferably to RIII(+), we reasoned that deletion of RIII should make the repRNA insensitive to expression of excess amount of Nsr1p. Indeed, the low level accumulation of ΔRIII repRNA, lacking RIII, was inhibited only slightly by the over-expression of Nsr1p (Fig. 6, lane 6–13), which inhibits DI-72 repRNA accumulation by 10-fold (Fig. 1A, lanes 4–6). This further supports that RIII in the tombusvirus genome is the main target of Nsr1p.
Fig. 6.
Accumulation of tombusvirus RNA lacking RIII sequence is only moderately inhibited by over-expression of Nsr1p in yeast. (A) Schematic representation of the repRNA used and the model for Nsr1p-based inhibition of repRNA accumulation. (B) To study the effect of over-expression of Nsr1p on the accumulation of DI-ΔRIII RNA, BY4741 yeast were transformed with three plasmids, such as pHisGBK-His33/DI-ΔRIII -Gal [co-expressing CNV p33 from the ADH1 promoter and DI-ΔRIII RNA from the GAL1 promoter], pGAD-His92-CUP1 (containing the CNV p92pol gene behind the CUP1 promoter), and pYES-NSR1 (expressing N-terminally 6xHis-tagged Nsr1p from the GAL1 promoter) (lanes 10–13). Alternatively, we also used pYES-C-FLAG-NSR1 (expressing C-terminal FLAG-tagged Nsr1p from the GAL1 promoter) (lanes 6–9) or the empty plasmid pYES-NT-C expressing a short peptide as a control (lanes 1–5). The accumulation of repRNA was estimated using Northern blotting. See Fig. 1A for further details.
The recombinant Nsr1p inhibits the tombusvirus replicase in vitro
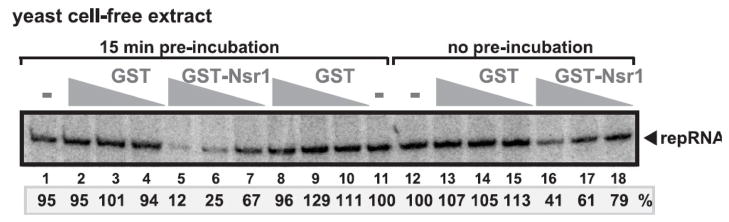
To test if the purified recombinant GST-Nsr1p can inhibit the tombusvirus replicase, we used a tombusvirus replicase assay based on a yeast cell-free extract containing subcellular membranes. This extract is capable of supporting authentic TBSV replication in vitro (Pogany and Nagy, 2008), due to the requirement of viral RNA recruitment and replicase assembly in the membranous fraction in vitro. Programming the cell-free extract with DI-72(+) repRNA leads to asymmetrical replication, resulting in small amount of (-)RNA intermediate and abundant (+)RNA progeny (Pogany and Nagy, 2008). Interestingly, addition of increasing amounts of purified GST-Nsr1 to the cell-free extract led to ~90% inhibition of TBSV repRNA replication when the (+)repRNA was pre-incubated with GST-Nsr1 prior to adding to the cell-free extract (Fig. 7, lanes 5–7). On the other hand, adding GST-Nsr1 and DI-72(+) repRNA simultaneously to the cell-free extract resulted in less inhibition (by 60%, lanes 16–18) when compared with the GST control (lanes 13–15). These results suggest that Nsr1p inhibits an early step in TBSV replication, likely the recruitment of the (+)repRNA into replication (see Discussion).
Fig. 7.
Inhibition of replication of the TBSV repRNA in the cell-free yeast extract. The panel shows a denaturing PAGE analysis of the 32P-labeled RNA products obtained when the 621 nt DI-72(+) repRNA added to the cell-free extract in the absence of GST-Nsr1 (lanes 1, 11, 12) or in the presence of the purified recombinant GST-Nsr1 or GST as shown. The cell-free extract was obtained from yeast expressing p33 and p92pol replication proteins. The amount of RNA synthesis by the replicase assembled in the yeast cell-free extract was compared to control samples, which did not contain recombinant GST-Nsr1 or GST proteins. The 200 ng repRNA and the recombinant proteins (400, 800 and 1600 ng per sample) were pre-incubated for 15 min (lanes 2–10) or without pre-incubation (lanes 13–18), and then mixed with the replicase mixture to perform in vitro TBSV replication.
Lack of changes in subcellular localization of nucleolin during tombusvirus replication in plants
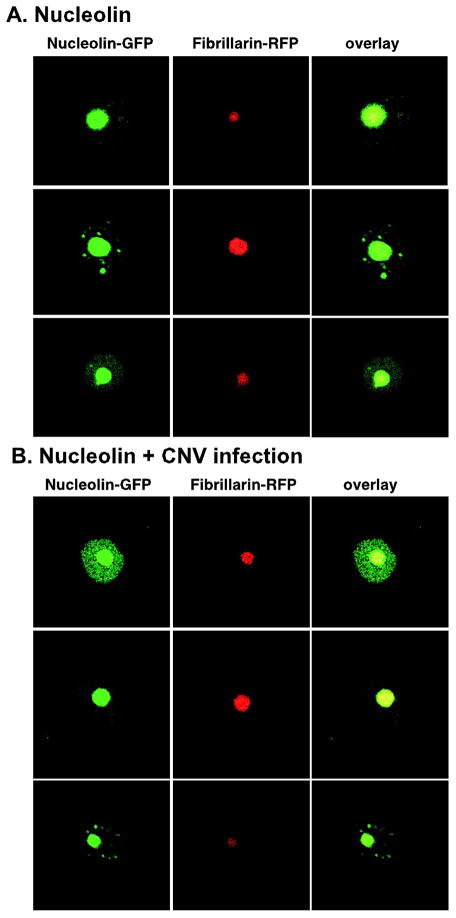
To test if the subcellular localization of nucleolin changes during replication of tombusviruses, we transiently expressed the AtNuc-L1-GFP fusion protein via agroinfiltration in transgenic plants expressing fibrillarin-RFP, a nucleolar marker protein (Kanneganti et al., 2007). Confocal laser microscopy revealed mostly nucleolar localization of AtNuc-L1 in both CNV infected and control plant cells (Fig. 8). In addition, we also observed a small portion of AtNuc-L1 in the nucleus in both experiments. Overall, the subcellular distribution of AtNuc-L1-GFP was comparable in CNV infected and control plant cells, suggesting that tombusvirus replication did not lead to nucleus-to-cytosol re-distribution of nucleolin. Additional detailed experiments will address if fraction of nucleolin might be redistributed at early time points of CNV infection to the cytosol or if the viral RNA might be redirected to the nucleus.
Fig. 8.
Nucleosomal and nucleolar localization of nucleolin in N. benthamiana cells expressing AtNuc-L1-GFP and infected with CNV. (A) Nucleosomal and nucleolar localization of nucleolin in the absence of CNV replication. The A. thaliana fibrillarin1-RFP expressed transgenically was used as a nucleolar marker. Each experiment was repeated and 20 or more cells were analyzed. (B) Similar localization of AtNuc-L1-GFP in transgenic fibrillarin1-RFP N. benthamiana cells infected with CNV.
DISCUSSION
Host proteins could affect viral replication in various ways. Those host proteins, which facilitate or regulate virus replication, are called host factors. Previous works have identified several host factors for TBSV, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), HSP70 heat shock protein, eEF1A translation elongation factor, and Cdc34 E2 ubiquitin ligase, which are part of the viral replicase complex together with p33 and p92pol replication proteins. These host factors have been shown to regulate the ratio of plus- versus minus-strand synthesis; participate in the assembly of the replicase; promote insertion of viral proteins into subcellular membranes; enhance the stability of the viral replication proteins or affect their intracellular transportations (Li et al., 2008; Li et al., 2009; Pathak, Sasvari, and Nagy, 2008; Pogany et al., 2008; Serva and Nagy, 2006; Wang and Nagy, 2008; Wang, Stork, and Nagy, 2009). The detailed functions of additional host factors identified during previous genome-wide screens (Jiang et al., 2006; Panavas et al., 2005b; Serviene et al., 2006; Serviene et al., 2005) are not yet known.
The second group of host proteins inhibits tombusvirus replication and they might be components of the host innate immunity. The best-characterized example is Xrn1p 5’-3’ exoribonuclease (Xrn4p in plants/mammals), which is involved in degradation of tombusvirus RNA, including partially degraded viral RNAs generated by endoribonucleases (Cheng et al., 2007; Cheng, Serviene, and Nagy, 2006; Jaag and Nagy, 2009). In the absence of Xrn1p/Xrn4p, tombusvirus RNA accumulation increased several fold and novel viral recombinant RNAs or variants emerged rapidly in yeast and in plants. Thus, in addition to inhibiting tombusvirus RNA accumulation, Xrn1p also affects the rate of virus evolution, suggesting complex interactions between host proteins and plant viruses (Nagy, 2008).
Another member of this group of inhibitory host factors is Nsr1p/nucleolin characterized in this work, which has been identified during the genome-wide screen of yeast strains for affecting TBSV repRNA accumulation (Panavas et al., 2005b). Deletion of NSR1 increased TBSV RNA accumulation by 2-fold, suggesting that this host protein has anti-TBSV activity as confirmed in this paper. Accordingly, over-expression of Nsr1p in yeast led to ~10-fold inhibition of repRNA accumulation (Fig. 1A). Interestingly, the over-expression was the most effective when it occurred before or at the beginning of TBSV repRNA replication, suggesting that Nsr1p could inhibit an early step in the viral replication process. This step could be the viral RNA recruitment step, since preincubation of repRNA and the purified recombinant Nsr1p inhibited the activity of the in vitro assembled tombusvirus replicase more efficiently than adding Nsr1p directly to the cell-free extract. Indeed, we found strong interaction between the RNA-binding domain of Nsr1p and repRNA, while we could not demonstrate direct interaction between Nsr1p and p33 or p92 replication proteins in this work (not shown) or during previous proteomics screens (Li et al., 2008).
Another piece of evidence for the role of Nsr1p in inhibition of repRNA recruitment is the ability of Nsr1p to bind to RIII(+) sequence in the repRNA. This region is not known to play a role in the assembly of the tombusvirus replicase (Panaviene, Panavas, and Nagy, 2005). It is more likely that binding of Nsr1p to RIII(+) could lead to sequestration of the viral RNA, inhibiting its recruitment by the p33 replication protein, which binds to RII(+) (Pogany, White, and Nagy, 2005). Also, over-expression of Nsr1p prior to the viral repRNA in yeast was the most effective in inhibiting repRNA accumulation. This fits well with the model that high concentration of nucleolin could sequester the TBSV repRNA, especially at the early stage of infection when the viral RNA is present in limiting amounts.
In addition to the broad range of activities of nucleolin in the host cell, it is also involved in replication/pathogenesis of various RNA and DNA viruses. Similar to the findings in this paper that Nsr1p/nucleolin can be inhibitory to tombusvirus replication, nucleolin has also been found to act as an inhibitor of DNA replication of simian virus 40 (SV40) virus. It has been shown that nucleolin inhibited the unwinding of SV40 origin (Daniely and Borowiec, 2000). However, in several other cases, nucleolin has been shown to stimulate viral infections. For example, nucleolin has been shown to interact with the 3’ UTR of poliovirus (PV) and stimulate an early step of PV replication in vitro (Waggoner and Sarnow, 1998). Nucleolin was also shown to relocalize from the nucleolus to the cytoplasm in PV infected cells, suggesting the existence of virus-induced mechanism to redistribute certain nuclear proteins in infected cells. Interestingly, the 5’ UTR of PV also binds to nucleolin and this interaction affects the IRES-mediated translation of the poliovirus RNA both in vivo and in vitro (Izumi et al., 2001). The NS5B RdRp protein of hepatitis C virus interacts with nucleolin, which could be relevant for virus replication (Kusakawa et al., 2007). The NS1 protein of influenza A virus, a negative-strand RNA virus, binds to nucleolin and colocalizes with nucleolin in the nucleolus, possibly affecting cellular events, such as shut down of host protein synthesis (Murayama et al., 2007). Herpes simplex virus 1 affects the subcellular localization of nucleolin in order to regulate rRNA levels and ultimately to alter cellular metabolism (Bertrand and Pearson, 2008). Nucleolin is also involved in the budding of retrovirus virions from the infected cells by interacting with the gag protein and the RNA packaging signal (Ueno et al., 2004). Over expression of the C-terminal portion of nucleolin inhibited the assembly of retrovirus virions, suggesting that nucleolin – gag interaction is critical during the virion assembly process (Bacharach et al., 2000).
Based on data presented here, it seems that the yeast Nsr1p and the Arabidopsis nucleolin play comparable inhibitory roles in tombusvirus replication, thus adding another example that host factors affecting TBSV repRNA accumulation in yeast are also effective against the fully infectious tombusvirus genomic RNA in plants. Further experiments will be conducted to see if nucleolin/Nsr1p acts alone against tombusviruses or it is part of a larger innate immunity system of the host.
MATERIALS AND METHODS
Yeast and Escherichia coli plasmids
To study the effect of over-expression of Nsr1p protein on viral RNA replication, we transformed Saccharomyces cerevisiae parental strain (BY4741) or nsr1Δ strain from the YKO library (Open Biosystems) with three plasmids: pHisGBK-His33/DI-72 [co-expressing CNV p33 from the ADH1 promoter and DI-72 (+) RNA from the GAL1 promoter] (Jiang et al., 2006), pGAD-His92-CUP1 (containing the CNV p92pol gene behind the CUP1 promoter) (Li et al., 2008), and pYES-C-FLAG-NSR1 (expressing C terminal FLAG-tagged NSR1) or pYES-Nsr1 or empty plasmid pYES-NT-C (Invitrogen) as a control.
To study the effect of Nsr1p expression at different time points on tombusvirus RNA replication, we transformed the S. cerevisiae parental strain (BY4741) with three plasmids: pHisGBK-His33/DI72-CUP1 [co-expressing CNV p33 from the ADH1 promoter and DI-72 (+)RNA from the CUP1 promoter], pGAD-His92-CUP1 (Li et al., 2008) and pYES-Nsr1.
To obtain pYES-NSR1, the full-length NSR1 sequence was amplified by PCR with primers #1947 (CGCGGGATCCATGGCTAAGACTACTAAAG) and #1948 (CGCGCTCGAGTCAATCAAATGTTTTCTTTGAACC) from a yeast genomic DNA preparation. The PCR product was treated with BamHI and XhoI and ligated to pYES-NT-C, which was also treated with the same enzymes. The expression plasmid pYES-C-FLAG-NSR1 was prepared by PCR using primers #1951 (CgcgAAGCTTACCATGGCTAAGACTACTAAAG) and #2832 (CGACCTCGAGTCACTTATCGTCGTCATCCTTGTAATCATCAAATGTTTTCTTTGAAC-C) and the yeast genomic DNA as template. The PCR product was inserted between HindIII and XhoI sites (engineered in the Nagy lab) in pYES-NT-C (Invitrogen).
Plasmid pGWB5 expressing the Arabidopsis nucleolin (AtNuc-L1p) from the 35S promoter was the generous gift of Dr. K. Nakamura (Kojima et al., 2007). GFP expression plasmid pGD was used as a control (Goodin et al., 2002). The CNV expression plasmid pGD-CNV and pGD-p19 were described (Cheng et al., 2007; Jaag and Nagy, 2009). The TRV plasmids pTRV1 and pTRV2 were described (Liu, Schiff, and Dinesh-Kumar, 2002).
To generate the E. coli expression plasmids for Nsr1p and its deletion derivative NSR1ΔRBD lacking the central RNA-binding domain with the two RBD repeats, we introduced the C-terminal portion of Nsr1 and an extra XhoI restriction site into pGEX-2T plasmids at the BamHI and EcoRI sites by using PCR and primers #1972 (CgcgGGATCCGACTTCTCTTCTCCAAGACC) and #2040 (CGCGGAATTCCTCGAGTCAATCAAATGTTTTCTTTGAACC). Then, to obtain pGEX-NSR1, the full-length sequence of NSR1 (primers #1947 and #1948) was inserted between the BamHI and XhoI sites of modified pGEX-2T plasmid. Plasmid pGEX-NSR1ΔRBD was obtained by ligating together the PstI-treated DNA sequence representing the N-terminal part of NSR1 gene generated by PCR using primers #1947 and #1975 (CgcgCTGCAGAGTAGCTGGTTCTTCGG) and the C-terminal part with primers #1978 (CgcgCTGCAGGACTTCTCTTCTCCAAGACC) and #1948. The ligated PCR products were then inserted between the BamHI and XhoI sites of modified pGEX-2T plasmid.
Yeast transformation and culturing
Yeast transformation was done by using the standard lithium acetate-single-stranded DNA-polyethylene glycol method, and transformants were selected by complementation of auxotrophic markers, ULH− media lacking uracine, leucine and histidine as described before (Panaviene et al., 2004). The transformed yeast cells were grown at 29°C for 24 hours in SC media (synthetic media, SC-ULH−) and 2% galactose as the carbon source and 50 μM copper sulfate to express p92 and DI-72 RNA.
Expression and purification of recombinant Nsr1p protein
We used pGEX-NSR1 and pGEX-NSR1ΔRBD plasmids to express the GST tagged protein in E. coli. Purification of recombinant NSR1 protein was performed as described with slight modification (Rajendran and Nagy, 2006). Briefly, E. coli Epicurion BL21-CodonPlus RIL (Stratagene) cells were pelleted from 25 ml culture media was resuspended in 1x PBS buffer (with 0.7% beta-mercaptoethanol) and sonicated and centrifuged to remove cell debris. The supernatant was loaded on GST resin column in PBS buffer, and then the GST fusion protein was eluted in 0.32% glutathione in PBS. Similarly expressed and purified GST protein from pGEX-2T plasmid was used as a control in the RNA binding assay.
RNA analysis and Northern blotting
Total RNA isolation and Northern blot analysis were done as described (Panaviene et al., 2004). Briefly, pelleted yeast cells were resuspended in RNA extraction buffer [50 mM sodium acetate, pH 5.2, 10 mM EDTA, 1% sodium dodecyl sulfate (SDS)] and the same volume of phenol. Samples were vortexed for ~1 min at room temperature, followed by incubation for 4 min at 65 °C and on ice for ~ 1 min. Then, the total RNA was precipitated with ethanol. The obtained total RNA samples were separated by 1.5% agarose gel electrophoresis and were transferred to a Hybond-XL membrane (GE Healthcare). Northern blotting was done as described (Li et al., 2009). Briefly, the blotted total RNA samples fixed on the membrane were hybridized with a mixture of two 32P-labeled probes to detect DI-72 (+)RNA and the 18S rRNA. Hybridization signals were detected using a Typhoon 9400 imaging scanner (GE Healthcare) and quantified by ImageQuant software.
Protein extraction and western blotting
Total protein extraction from yeast and Western blot were performed as described previously (Panaviene et al., 2004). Briefly, the yeast pellets were resuspended in 0.1M NaOH, followed by vortexing for 30s and shaking for another 10 min. Then, the samples were centrifuged at 15,000 x g for 5 min at 4 ºC and the pellet was resuspend in 1x SDS-PAGE buffer. The protein samples were electrophoresed in 0.1% SDS-8% PAGE gel, and transferred to a PVDF membrane (Bio-Rad). Nonspecific binding was blocked with 5% nonfat dry milk solution. The primary antibody was anti-His antibody (GE Healthcare), and the secondary antibody was anti-mouse IgG alkaline phosphotase (Sigma).
Total protein from plant leaf samples was extracted from 30 mg plant leaf tissue. The plant tissue was grinded with a pestle in a microcentrifuge tube in 30 μl buffer A (50mM Tris-HCl, 10mM KCl, 15mM MgCl2, 2mM EDTA, 20% Glycerol), followed by centrifugation at 400 x g for 5 min at 4°C. The supernatant was mixed with 0.5 volume of 3x SDS loading buffer and heated at 85°C for 15 min, followed by electrophoresis in 0.1% SDS-9% PAGE. Western blot analysis was done using anti-GFP as the primary antibody and anti-chicken as the secondary antibody.
Transformation of Agrobacterium, agroinfiltration and inoculation of plants
The procedure used was as described (Cheng et al., 2007). Briefly, expression plasmids pGWB5, pGD-GFP and pGD-p19 and pGD-CNV or pGD-TRV1 / pGD-TRV2 were transformed into Agrobacterium C58C1. Tranformants were selected in LB medium containing 50 μg/ml kanamycin, 100 μg/ml rifampicin and 5 μg/ml tetracycline. The transformed agrobacteria were grown in LB media containing the antibiotics and 20 μM acetosyringone at 29°C until the OD600 reached 1.0. The bacterial cells were pelleted and resuspended in MMA media [10 mM MES (pH 5.6), 10 mM MgCl2, 200 μM acetosyringone) and incubated for 2–4 hrs on the bench. We used the obtained Agrobacteria culture of 0.8~1.0 OD600 for agro-infiltration. Agrobacteria carrying pGWB5 (or pGD-GFP as a control), pGD-P19 and pGD-CNV were mixed in a ratio of 5:5:1 prior to infiltration to N. benthamiana leaves. For the TRV experiment, the agrobacteria cultures containing pGWB5 (or pGD-GFP), pGD-p19 and pGD-TRV1 / pGD-TRV2 were mixed in a ratio of 5:5:1:1 before infiltration.
For the analysis of agroinfiltrated leaf tissues, we randomly chose the same sized leaf areas and excised 30 mg of leaf tissue to extract total RNA (Cheng et al., 2007; Jaag and Nagy, 2009). Then, the leaf samples were grinded in liquid nitrogen, followed by shaking for 5 min at room temperature in 200 μl of RNA extraction buffer [50 mM NaOAc (pH5.2), 10 mM EDTA, 1% SDS] and 200 μl water-saturated phenol and then additional incubation for 4 min at 65 °C. The RNA was precipitated by ethanol. The obtained total RNA samples were analyzed by Northern blotting as described (Jaag and Nagy, 2009).
To test the effect of nucleolin on CNV infections started via rub-inoculation, agrobacterium strains carrying pGWB5 and pGD-p19 were mixed in a ratio of 1:1 prior to agroinfiltration into N. benthamiana leaves. Two days after agroinfiltration, the infiltrated leaves were inoculated with sap preparation containing CNV virions. The plant sap preparation was obtained from CNV/20KSTOP gRNA transcript-inoculated N. benthamiana plants in 0.02 M sodium-acetate pH 5.3 as described (Cheng, Serviene, and Nagy, 2006). The infiltrated leaves were also inoculated with the sap containing TBSV virions prepared from TBSV infected N. benthamiana leaves.
RNA probes and competitors used for RNA-protein interactions
To study the binding of Nsr1p to the full-length DI-72 (+)RNA and its four different regions, we PCR amplified DI-72SXP (White and Morris, 1994) or its portions using primers described in (Rajendran and Nagy, 2003). The RNA transcripts were synthesized on the PCR templates using T7-based transcription in the presence or absence of 32P-UTP to generate labeled probes or cold transcripts, which were used as competitors during RNA-protein interactions. The amounts of transcripts were quantified by UV spectrophotometer (Beckman).
Nsr1p - viral RNA interactions in vitro
The UV crosslinking assay was performed according to (Hirose and Harada, 2008). The reaction mixture was 12 μl containing 2 μg purified GST-Nsr1 protein, 10 ng (about 5nM) 32P-UTP labeled RNA probe, 10mM Hepes, pH 7.9; 100 mM KCl; 1mM MgCl2; 10% glycerol; 0.5% NP40; 2 μg tRNA; and 0.2 μg Heparin. In the competition assay, we used cold RNA transcripts as competitors in 5 nM or 50 nM amounts. The reaction mixtures were incubated at room temperature for ~20 min to allow the formation of RNA-protein complexes. To crosslink RNA and protein, we transferred the reaction mixture to a 96-well plate on ice, then irradiation was done at 254 nm for 10 min using an UV Stratalinker 1800 (Stratagene). Then, the unprotected RNAs were digested by 1mg/ml RNase A for 10 min at 37ºC. Samples were mixed with 0.5 volume 3x SDS loading dye and boiled for 10 min. Analysis was performed using SDS-PAGE and phosphoimaging.
For the band shift (gel mobility shift) assays, the reaction mixtures were set up as described above for UV crosslinking, except that the 32P-UTP labeled RNA probes were diluted ~50 times and 10U of RNase inhibitor was also included. Analysis was performed using 4% nondenaturing PAGE and phosphoimaging. For the template competition assay, we used the cold RNA transcripts as competitors in 0, 0.05, and 0.5 μM concentration.
In vitro replicase assays
One of the replicase assays was based on the membrane-enriched fraction of yeast as described earlier (Panaviene et al., 2004). Yeast co-transformed with pGAD-His92-CUP1 / pHisGBK-His33/DI-72 and one of the following: pYES-NSR1, pYES-C-FLAG-NSR1 or empty plasmid pYES-NT-C (used as control) was pre-grown in Sc-ULH− media containing 2% glucose at 29°C for 24 hr, then switched to 2% galactose for 4–5 hr before adding 50 μM copper sulfate to the media. After culturing for 22 hrs in the presence of copper sulfate, the yeast cells were harvest by centrifugation (Panaviene et al., 2004). The membrane enriched fraction for each strain was prepared by disrupting the cells in an ice cold extraction buffer (200 mM sorbitol, 50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, 1% yeast protease inhibitor mix; Sigma), followed by centrifugation 100 x g for 5 min at 4°C to remove cell debris. Then, the enriched membrane fraction was obtained by centrifugation at 21,000 x g for 10 min. Before the replicase reaction, we performed Western blotting for estimating p33 levels in order to normalize the amount of p33 in each sample. The in vitro replicase reactions were set up according to Panaviene et al. (2004). The RdRp products were analyzed by electrophoresis on 5% PAGE containing 8 M urea and phosphoimaging.
To test the effect of Nsr1p on the activity of the in vitro assembled tombusvirus replicase, a yeast cell free extract was prepared as described previously (Pogany and Nagy, 2008). Note that this essay is based on the CNV replication proteins, which show 100% (indistinguishable) activity with the repRNA when compared with the TBSV replication proteins in yeast or in vitro. Briefly, yeast cells expressing p33/p92 from plasmids pGAD-His92 (Panaviene et al., 2004) and pHisGBK-His33 (Panavas et al., 2005a) were cultured in LH− media containing glucose for 24 hr, followed by pelleting and resuspension in buffer A. After breaking the cells gently by glass beads, the cell debris was removed by centrifugation at 500 x g to obtain the cell free extract. The in vitro replication assays were also performed as described (Pogany and Nagy, 2008). The replication mixture (total of 20 μl) contained 1 μl cell free extract, 50 mM HEPES-KOH, pH 7.4, 150 mM potassium acetate, 5 mM magnesium acetate, 0.2 M sorbitol, and 0.4 μl actinomycin D (5 mg/ml), 2 μl of 150 mM creatine phosphate; 2 μl of 10 mM ATP, CTP, and GTP and 0.25 mM UTP; 0.3 μl of 32P-UTP, 0.2 μl of 10-mg/ml creatine kinase, 0.2 μl of RNase inhibitor, 0.2 μl of 1 M dithiothreitol, and 0.2 μg DI-72 (+)RNA transcript. The reaction mixture was incubated at 25°C for 3 hr and terminated by adding 110 μl stop buffer (1% SDS and 0.05 M EDTA, pH 8.0), followed by phenol-chloroform extraction, isopropanol-ammonium actetate precipitation, and analysis with electrophoresis on 5% PAGE containing 8 M urea and phosphoimaging.
Confocal microscopy-based observation of nucleolin localization
Transgenic N. benthamiana expressing fibrillarin-RFP was kindly provided by Dr. Goodin. Transient expression of pGWB5 (to express GFP tagged nucleolin), pGD-P19 and pGD-CNV via agroinfiltration was done as described above. The confocal laser microscopy was performed on an Olympus FV1000 (Olympus America Inc., Melville, NY). The images were acquired using sequential line-by-line mode in order to reduce excitation and emission crosstalk (Wang and Nagy, 2008). The primary objective used was water-immersion PLAPO60XWLSM (Olympus). Image acquisition was conducted at a resolution of 512 × 512 pixels and a scan-rate of 10 μs/pixel. Image acquisition was performed by using Olympus Fluoview software version 1.5.
Acknowledgments
The authors thank Drs. Judit Pogany and Daniel Barajas. The authors are grateful to Dr. K. Nakamura for sharing plasmid pGWB5 and to Dr. M. Goodin for the At-Fibrillarin1-RFP transgenic N. benthamiana and anti-GFP and anti-chicken antibodies. This work was supported by the National Science Foundation (IOB-0517218), NIH-NIAID and the Kentucky Tobacco Research and Development Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacharach E, Gonsky J, Alin K, Orlova M, Goff SP. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J Virol. 2000;74(23):11027–39. doi: 10.1128/jvi.74.23.11027-11039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L, Pearson A. The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J Gen Virol. 2008;89(Pt 5):1142–51. doi: 10.1099/vir.0.83573-0. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Allain FH, Finger LD, Dieckmann T, Feigon J. Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J Mol Biol. 2001;309(3):763–75. doi: 10.1006/jmbi.2001.4691. [DOI] [PubMed] [Google Scholar]
- Cheng CP, Jaag HM, Jonczyk M, Serviene E, Nagy PD. Expression of the Arabidopsis Xrn4p 5'-3' exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology. 2007;368(2):238–48. doi: 10.1016/j.virol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Cheng CP, Serviene E, Nagy PD. Suppression of viral RNA recombination by a host exoribonuclease. J Virol. 2006;80(6):2631–40. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19(4):445–52. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Borowiec JA. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol. 2000;149(4):799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31(3):375–83. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454(7206):890–3. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Harada F. Mouse nucleolin binds to 4.5S RNAh, a small noncoding RNA. Biochem Biophys Res Commun. 2008;365(1):62–8. doi: 10.1016/j.bbrc.2007.10.117. [DOI] [PubMed] [Google Scholar]
- Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 2001;76(1):17–29. doi: 10.1016/s0168-1702(01)00240-4. [DOI] [PubMed] [Google Scholar]
- Jaag HM, Nagy PD. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology. 2009;386(2):344–52. doi: 10.1016/j.virol.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Jaag HM, Stork J, Nagy PD. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology. 2007;368(2):388–404. doi: 10.1016/j.virol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J Virol. 2006;80(15):7394–404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk M, Pathak KB, Sharma M, Nagy PD. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362(2):320–30. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Bai X, Tsai CW, Win J, Meulia T, Goodin M, Kamoun S, Hogenhout SA. A functional genetic assay for nuclear trafficking in plants. Plant J. 2007;50(1):149–58. doi: 10.1111/j.1365-313X.2007.03029.x. [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Saez-Vasquez J, Echeverria M, Nakagawa T, Ishiguro S, Nakamura K. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49(6):1053–63. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455(7210):242–5. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakawa T, Shimakami T, Kaneko S, Yoshioka K, Murakami S. Functional interaction of hepatitis C Virus NS5B with Nucleolin GAR domain. J Biochem. 2007;141(6):917–27. doi: 10.1093/jb/mvm102. [DOI] [PubMed] [Google Scholar]
- Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A. 2003;100(26):15764–9. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. Cdc34p Ubiquitin-Conjugating Enzyme Is a Component of the Tombusvirus Replicase Complex and Ubiquitinates p33 Replication Protein. J Virol. 2008;82(14):6911–26. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385(1):245–60. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31(6):777–86. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17(2):80–6. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Monkewich S, Lin HX, Fabian MR, Xu W, Na H, Ray D, Chernysheva OA, Nagy PD, White KA. The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication. J Virol. 2005;79(8):4848–58. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama R, Harada Y, Shibata T, Kuroda K, Hayakawa S, Shimizu K, Tanaka T. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem Biophys Res Commun. 2007;362(4):880–5. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol. 2008;46:217–42. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344(1):211–20. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005a;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T, Nagy PD. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J Virol. 2003a;77(1):258–69. doi: 10.1128/JVI.77.1.258-269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Nagy PD. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003b;314(1):315–25. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Brasher J, Nagy PD. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A. 2005b;102(20):7326–31. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3'-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005;79(16):10608–18. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Serva S, Nagy PD. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J Virol. 2004;78(15):8254–63. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379(2):294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Pogany J, Nagy PD. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J Virol. 2008;82(12):5967–80. doi: 10.1128/JVI.02737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci U S A. 2008;105(50):19956–61. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol. 2005;79(8):4859–69. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18(2):369–79. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran KS, Nagy PD. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J Virol. 2003;77(17):9244–58. doi: 10.1128/JVI.77.17.9244-9258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran KS, Nagy PD. Kinetics and functional studies on interaction between the replicase proteins of Tomato Bushy Stunt Virus: requirement of p33:p92 interaction for replicase assembly. Virology. 2006;345(1):270–9. doi: 10.1016/j.virol.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Ray D, White KA. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J Virol. 2003;77(1):245–57. doi: 10.1128/JVI.77.1.245-257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichler SA, Balk J, Brown ME, Woodruff K, Clark GB, Roux SJ. Light differentially regulates cell division and the mRNA abundance of pea nucleolin during de-etiolation. Plant Physiol. 2001;125(1):339–50. doi: 10.1104/pp.125.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serva S, Nagy PD. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol. 2006;80(5):2162–9. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J Virol. 2006;80(3):1231–41. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 2005;102(30):10545–50. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5(3):298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33(6):407–36. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- Ueno T, Tokunaga K, Sawa H, Maeda M, Chiba J, Kojima A, Hasegawa H, Shoya Y, Sata T, Kurata T, Takahashi H. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type-1 (HIV-1) Microbiol Immunol. 2004;48(2):111–8. doi: 10.1111/j.1348-0421.2004.tb03496.x. [DOI] [PubMed] [Google Scholar]
- Waggoner S, Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J Virol. 1998;72(8):6699–709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Nagy PD. Tomato bushy stunt virus Co-Opts the RNA-Binding Function of a Host Metabolic Enzyme for Viral Genomic RNA Synthesis. Cell Host Microbe. 2008;3(3):178–87. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wang RY, Stork J, Nagy PD. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol. 2009;83(7):3276–87. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Morris TJ. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J Virol. 1994;68(1):14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]