Abstract
Pulsatile GNRH regulates the gonadotropin subunit genes in a differential manner, with faster frequencies favoring Lhb gene expression and slower frequencies favoring Fshb. Early growth response 1 (EGR1) is critical for Lhb gene transcription. We examined GNRH regulation of EGR1 and its two corepressors, Ngfi-A-binding proteins 1 and 2 (NAB1 and NAB2), both in vivo and in cultured rat pituitary cells. In rats, fast GNRH pulses (every 30 min) stably induced Egr1 primary transcript (PT) and mRNA 2-fold (P < 0.05) for 1–24 h. In contrast, slow GNRH pulses (every 240 min) increased Egr1 PT at 24 h (6-fold; P < 0.05) but increased Egr1 mRNA 4- to 5-fold between 4 and 24 h. Both GNRH pulse frequencies increased EGR1 protein 3- to 4-fold. In cultured rat pituitary cells, GNRH pulses (every 60 min) increased Egr1 (PT, 2.5- to 3-fold; mRNA, 1.5- to 2-fold; P < 0.05). GNRH pulses had little effect on Nab1/2 PT/mRNAs either in vivo or in vitro. We also examined specific intracellular signaling cascades activated by GNRH. Inhibitors of mitogen-activated protein kinase 8/9 (MAPK8/9 [also known as JNK]; SP600125) and MAP Kinase Kinase 1 (MAP2K1 [also known as MEK1]; PD98059) either blunted or totally suppressed the GNRH induction of Lhb PT and Egr1 PT/mRNA, whereas the MAPK14 (also known as p38) inhibitor SB203580 did not. In summary, pulsatile GNRH stimulates Egr1 gene expression and protein in vivo but not in a frequency-dependent manner. Additionally, GNRH-induced Egr1 gene expression is mediated by MAPK8/9 and MAPK1/3, and both are critical for Lhb gene transcription.
Keywords: Egr1, follicle-stimulating hormone, gonadotropin-releasing hormone, gonadotropins, luteinizing hormone, MAPK1/3(ERK), MAPK8/9(JNK), neuroendocrinology, pituitary hormones
GNRH pulse frequency does not differentially regulate Egr1 gene transcription or EGR1 protein, while Egr1 and Lhb gene transcriptions are regulated by both MAPK8/9(JNK) and MAPK1/3(ERK).
INTRODUCTION
The gonadotropins luteinizing hormone (LH) and follicle-stimulating hormone (FSH) are dimeric protein hormones composed of a common α subunit (CGA) and a unique β subunit [1]. The subunit genes are regulated by hypothalamic GNRH in both a coordinate and a differential manner. GNRH differentially regulates subunit mRNA synthesis via changes in pulse frequency, with faster-intermediate GNRH pulse frequencies (8- to 60-min pulse intervals) favoring Cga and Lhb, and slower-frequency pulses (≥120-min pulse intervals) favoring Fshb [2].
The signal transduction mechanisms responsible for interpreting GNRH pulse frequency and differentially regulating β-subunit gene expression are not well understood. The GNRH receptor (GNRHR) is a member of the G protein-coupled receptor family [3, 4]. Ligand-bound GNRHR activates several members of the G protein family, including Gq and G11. Activated Gq stimulates phospholipase C, resulting in increased inositol 1,4,5,-trisphosphate (IP3), elevated diacylglycerol levels, and activation of protein kinase C (PKC) [5, 6]. GNRHR activation also stimulates a transient increase in intracellular calcium (Ca2+) derived from IP3-induced release of Ca2+ from intracellular storage pools and from influx via L-type voltage-gated calcium channels, which can stimulate other Ca2+-sensitive protein kinases [5, 7]. Additionally, we and others have shown that GNRH stimulates activation of mitogen-activated protein kinase (MAPK) signaling cascades (MAPK1/3 [extracellular signal-regulated kinase, or ERK], MAPK8/9 [c-Jun N-terminal kinase, or JNK], and MAPK14 [p38]), and that members of this family are important in transducing GNRH pulse information in gonadotrophs [2].
GNRH-induced MAPK1/3 activation is via both PKC-dependent and independent mechanisms [2, 8]. We reported that GNRH pulses are more effective than continuous GNRH to stimulate sustained pituitary MAPK1/3 phosphorylation in rats, that MAPK1/3 phosphorylation is maximal after slow-frequency GNRH pulses [9, 10], and that inhibition of the pathway using a MAP Kinase Kinase 1 (MAP2K1, also known as MEK1) inhibitor blocked the GNRH-induced increase in Cga and Fshb mRNAs, but not Lhb mRNA, in primary pituitary cells [9]. GNRH also induces MAPK8/9 activation via a PKC-independent mechanism [11, 12]. Recently, we reported that MAPK8/9 blockade completely suppressed the GNRH-induced increase in Lhb transcription in perifused rat pituitary cells [13]. GNRH also increases MAPK14 activation via a PKC-dependent mechanism [14], but inhibition of MAPK14 activation had no effect on Lhb or Fshb transcriptional or gonadotropin secretory responses to pulsatile GNRH in rat pituitary cells [13].
The mechanism(s) by which MAPK1/3 and MAPK8/9 regulate β-subunit transcription have not been explored fully. MAPK1/3 and MAPK8/9 activation stimulates a number of transcription factors that are important in the regulation of the Lhb and Fshb subunit genes, including cFOS (FOS), cJUN (JUN), the ETS protein ELK1, and EGR1 [15]. The rodent Fshb proximal promoter contains a low-affinity activator protein-1 (AP1) half-site that binds JUN/FOS heterodimers and is important for maximal GNRH induction of the murine Fshb promoter in LβT2 cells [16]. This AP1 half-site is involved in MAPK1/3 activation of Fshb transcription, because treatment of LβT2 cells with a MAP2K1 inhibitor or cotransfection of a dominant/negative FOS expression vector reduced GNRH-stimulated Fshb promoter activity [16]. GNRH also regulates Fshb gene expression indirectly via changes in pituitary activin and follistatin (FST). Fast-frequency GNRH pulses selectively stimulate FST expression, reducing activin bioavailability and suppressing Fshb gene expression [2, 10, 17–20].
The rat Lhb promoter also contains a region that is highly homologous with a consensus AP1 site (−159/−153 bp [21, 22]), and mutation of this site diminishes Lhb promoter activity [23]. However, Lhb transcriptional responses to GNRH are primarily through actions on EGR1 and other transcription factors that bind to the proximal and distal GNRH-responsive regions [15]. EGR1 (also known as NGFI-A, Krox24, and zif268) is an immediate early gene of the zinc-finger subfamily and is expressed in many cell types during development and in differentiated cells in response to numerous types of signals and stress stimuli (for a review, see Thiel and Cibelli [24] and Knapska and Kaczmarek [25]). In the reproductive axis, EGR1 plays an essential role based on findings that Egr1 knockout mice are either completely infertile or subfertile, reflecting a lack of LHB synthesis in the gonadotroph (CGA and FSHB were unaffected [26, 27]). Two EGR1-binding sites have been identified in the proximal GNRH-responsive region of the Lhb promoter that are highly conserved across species [26, 28–31], and mutations within these EGR1-binding sites abrogate the GNRH induction of Lhb promoter reporter constructs in gonadotroph-derived cell lines [32–34]. Also, it has been observed that rat pituitary Egr1 mRNA expression is greatest during proestrus and is increased after ovariectomy (OVX), and the post-OVX increase can be blocked by estrogen [35], suggesting that GNRH plays a physiological role in regulating pituitary Egr1 expression.
Recent reports demonstrated that expression of Egr1 mRNA and protein and the two EGR1 corepressors Ngfi-A binding proteins (NAB1 and NAB2) are differentially regulated by GNRH pulse frequency in gonadotroph-derived cell lines. Kanasaki et al. [36] showed that EGR1 protein levels are 3-fold greater in LβT2 cells receiving GNRH pulses every 30 min vs. every 120 min. Similarly, Lawson et al. [37] reported that Egr1 mRNA increased rapidly and transiently in response to GNRH and was maximally stimulated by faster-frequency GNRH pulses (<60-min interpulse interval). In contrast, maximal Nab1 and Nab2 mRNA expressions were seen after slower-frequency GNRH pulses (≥60-min interpulse interval [37]). These findings suggest that regulating EGR1 synthesis and bioavailability plays a role in GNRH pulse frequency actions on the Lhb gene.
The aims of the present study were to investigate whether GNRH pulse frequency differentially regulates Egr1, Nab1, and Nab2 transcription in normal rat pituitary cells, and to assess the effects of MAPK8/9, MAPK1/3, and MAPK14 blockade on Egr1 and Nab gene expression.
MATERIALS AND METHODS
In Vivo Studies
Adult (225–250 g) Sprague Dawley rats (Harlan Sprague Dawley Inc., Indianapolis, IN) were used for all experiments. For in vivo studies, rats were housed in a light-controlled (lights on 0500–1700 h) and temperature-controlled (25°C) room and allowed access to food and water ad libitum. All surgeries were performed under isoflurane (2.5% isoflurane, balance O2; ISO-THESIA; Vetus Animal Heath, Burns Veterinary Supply Inc., Westbury, NY) anesthesia. At the completion of experiments, rats were euthanized by decapitation under anesthesia. The University of Virginia Animal Care and Use Committee approved all animal experimentation procedures described within this report.
To study the effects of GNRH pulse frequency, we used a GNRH-deficient (castrate and testosterone-replaced) rat model. Rats were castrated, and two 20-mm testosterone-containing silastic implants were inserted subcutaneously (serum testosterone levels were 4–5 ng/ml, 24 h after implant insertion) as described previously [20]. An indwelling right jugular cannula was also inserted at the time of castration for i.v. GNRH administration, and experiments began 24 h after castration. Rats (n = 5–7 per group) received 25-ng GNRH pulses (in 0.25 ml of 0.9% saline/0.1% bovine serum albumin) either every 30 or 240 min for 1–24 h. Controls were pulsed with vehicle only. Animals were killed 5 min after the last pulse, based on previous data in rats that β-subunit primary transcript (PT) responses to GNRH are maximal 5 min after a pulse [38]. Pituitaries were collected, snap frozen in liquid nitrogen, and stored at −70°C until RNA was extracted. In studies to determine the effects of GNRH pulse frequency on EGR1 protein, four to six rats per group received GNRH pulses (25 ng; i.v.) every 30 min or every 240 min for 8 h (controls were pulsed with vehicle only [10]).
In Vitro Studies
Pituitaries from adult female rats were pooled and dissociated in medium containing 0.35% collagenase, 0.1% hylauronidase, and 0.01% DNase. After dissociation, the cell suspension was aliquoted into culture wells (2 × 106 to 3 × 106 cells per well) containing 22-mm plastic coverslips coated with Matrigel (BD Biosciences, Bedford, MA). The cells were cultured for 48 h before beginning each experiment, and the media were augmented with testosterone (500 pg/ml) to allow Lhb mRNA expression in response to pulsatile GNRH [39, 40]. For perifusion studies, 48 h after plating cells, coverslips were inserted into custom-made chambers and allowed to equilibrate for 1 h before initiating treatment. The perifusion flow rate was 200 μl/min, and 100-μl pulses were administered during a 10-sec duration via autosyringe pumps. Studies were conducted as four separate experiments (12 chambers per experiment), with all treatment groups represented in each experiment (three chambers per treatment per experiment [39]).
Time course of Egr1 and Nab transcriptional responses to GNRH.
Cells received two prepulses of GNRH (1 nM; 5-min duration; or media pulses to controls) 2 h and 1 h before beginning each experiment. GNRH (1 nM) was administered to groups of cells for durations of 5, 10, 45, or 120 min. After GNRH treatment, cells were lysed and RNA extracted (n = 5–6 per group).
The role of MAPK8/9 (JNK) and MAPK14 (p38) in the regulation of Egr1 and Nab transcripts by pulsatile GNRH.
Perifused rat pituitary cells were given pulses of GNRH (peak chamber concentration, 200 pM; medium pulses to controls) every 60 min for 24 h [13]. GNRH pulses every 60 min were chosen because this intermediate pulse frequency stimulates both Lhb and Fshb expression in vitro [9]. For MAPK8/9 blockade studies, cells were perifused with medium containing the MAPK8/9-specific inhibitor SP600125 (SP; 20 μM; EMB Biosciences, San Diego, CA) or vehicle (inactive SP isoform; 20 μM; EMB Biosciences). For MAPK14 inhibitor studies, cells were treated with medium containing the MAPK14-specific blocker SB203580 (SB; 20 μM; EMB Biosciences) or vehicle (0.1% dimethyl sulfoxide [DMSO]). The SP and SB doses selected were based on previously published reports showing effective suppression of GNRH-induced activation of the MAPK8/9 or MAPK14 pathways within gonadotroph-derived cell lines [14, 41, 42]. Cells were recovered 10 min after the last pulse, and RNA was extracted.
The role of MAPK8/9 (JNK) and MAPK1/3 (ERK) in the regulation of Egr1 and Nab transcripts by pulsatile GNRH.
Rat pituitary cells were cultured as described above. Forty-eight hours after dissociation, cells were treated with the MAP2K-specific inhibitor PD98059 (PD; 50 μM; EMB Biosciences), the MAPK8/9-specific inhibitor SP600125 (20 μM), or vehicle (0.25% DMSO). All cells were also treated with 1 μM bromocriptine (EMB Biosciences) to suppress MAPK1/3 activational responses to the removal of dopamine in lactotropes [9]. The selected dose for PD was based on previous studies showing that 50 μM PD blocks the MAPK1/3 activational response to GNRH in rat pituitary cells in vitro [9, 43]. One hour later, cells received two prepulses of GNRH (1 nM; 5-min duration; or media pulses to controls) 2 h and 1 h before the final GNRH pulse. The final 1-nM GNRH pulse was administered for 10 min, which was optimal for both Lhb and Egr1 transcriptional responses. Cells were then lysed, and RNA and protein were extracted (n = 4 per group).
αT3 cells: time course of Egr1 and Nab mRNA expression in response to GNRH.
After initial results showed minimal Nab1/2 transcript responses to GNRH, which contrasts with earlier data in gonadotroph-derived cell lines [37], we used αT3 cells to assess the time course of Egr1 and Nab transcript expression in response to GNRH. αT3 cells were plated at a density of 1.0 × 106 cells onto 22-mm diameter coverslips coated with Matrigel diluted 1:3. Cells were incubated in Dulbecco modified Eagle medium augmented with 10% fetal bovine serum. Twenty-four hours later, the cells were transferred to serum-free medium for 16 h and then treated with 10 nM GNRH (or vehicle; 0-h time) for 10, 45, or 120 min. After GNRH, coverslips were rinsed twice in PBS, and cells were recovered and processed for RNA and protein.
RNA Preparation and Measurement of Egr1 and Nab Transcripts
Total pituitary RNA was extracted using the acid guanidinium method [44]. Residual genomic DNA was removed by treatment with 1 unit of RNase Free DNaseI per microgram of RNA (Roche Molecular Biochemicals, Indianapolis, IN) at 37°C for 1 h. RNA preparations were confirmed to be DNA free by PCR in the absence of a preceding RT reaction. Primary transcripts and mRNAs were measured by real-time PCR as described previously [10]. The PCR assay primers were: Lhb PT forward, 5′-AGAGGCTCCAGGTAAGATGGTA-3′; Lhb PT reverse, 5′-CTTTTGCATTCCAGGTCCTGGA-3′; Fshb PT forward, 5′-TTTCCCCAGGAGAGATAGCCAA-3′; Fshb PT reverse, 5′-GCAAACTGCTCTGTAAGTCAGA-3′; Egr1 PT/mRNA forward, 5′-AACAACCCTACGAGCACCTG-3′; Egr1-PT reverse, 5′-CCCCAGACATCCCTCTAACA-3′; Egr1 mRNA reverse, 5′-AGCGGCCAGTATAGGTGATG-3′; Nab1 mRNA forward, 5′-AAGGACAATGCCCTGCTG-3′; Nab1 PT forward, 5′-TCTGAGGGAATGTTACAGACTGA-3′; Nab1 PT/mRNA reverse, 5′-GGAGACAATTCATCTCTTTCACC-3′; Nab2 mRNA forward, 5′-CGTGAGGGCAAACAGCTTAG-3′; Nab2 PT forward, 5′-AAGCAGGCATTCTTTGGATG-3′; Nab2 PT/mRNA reverse, 5′-GTGCTCTCTCGGGCTACTTG-3′; Gnrhr mRNA forward, 5′-ATGCTGGAGAGTTCCTTTGC3′; and Gnrhr mRNA reverse, 3′-CCGTCGGCTAGGTAGATCAT-5′. Primers for Fst mRNA have been reported previously [19, 38]. To confirm amplification of a single product, the PCR reaction was followed by melt-curve analysis. To create a standard for each subscript, RNA from a pooled rat pituitary cDNA sample was amplified by PCR using the aforementioned primers and then subcloned into PGEM T-EZ (Promega, Madison, WI). The identity of all PCR products was confirmed by DNA sequencing. Each PCR reaction was optimized for annealing temperature and Mg2+ concentration to obtain a PCR efficiency of 90%–105%. Each sample was measured against a standard curve of 2.0E1 to 1.28E−4 pg of plasmid in 1:5 dilutions. All samples from within an experiment were measured in the same assay in triplicate. Mean intraassay CVs are Lhb PT (9.0%), Fshb PT (7.1%), Egr1 PT (9.3%), Egr1 mRNA (6.8%), Nab1 PT (7.9%), Nab1 mRNA (9.5%), Nab2 PT (12.8%), Nab2 mRNA (8.0%), Gnrhr mRNA (6.7%), and Fst mRNA (6.7%).
Protein Preparation and Western Blot Immunostaining
For the in vivo experiment, pituitary protein was prepared as described previously [10]. For some in vitro experiments, protein was precipitated from the phenol:isoamyl:chlorofom extract after lysis in acid guanidinium by 3 volumes of cold acetone. Precipitates were washed three times with 95% ethanol, 0.3 M guanidine hydrochloride, and 2.5% glycerol and then were solubilized directly into Laemmli sample buffer (2% SDS; 62.5 mM Tris, pH 6.8; 8.5% glycerol; and 1.5% β-mercaptoethanol). Pituitary protein lysates (20 μg for in vivo samples or 10% of total for in vitro samples) were resolved by 8% SDS-PAGE and then transferred to nitrocellulose filters. Blots were then immunostained for EGR1, phosphorylated p44/p42 ERK (MAPKs 3 and 1, respectively), or phosphorylated cJUN, with primary antibodies (rabbit) obtained from Cell Signaling Technologies (Beverly, MA). Protein loading was determined by immunostaining for GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit (Millipore, Billerica, MA). Immunoactivity was detected using SuperSignal West Pico chemiluminescent system (Pierce, Rockville, IL), followed by autoradiography. Protein bands were quantified by densitometry using TotalLab Software (Amersham Biosciences, Piscataway, NJ).
Analysis
All data were examined by ANOVA. Significant differences (P < 0.05) were determined posthoc by Duncan multiple range test. Prior to analyses, all measurements were transformed to the logarithmic scale to attain equal variation among treatments.
RESULTS
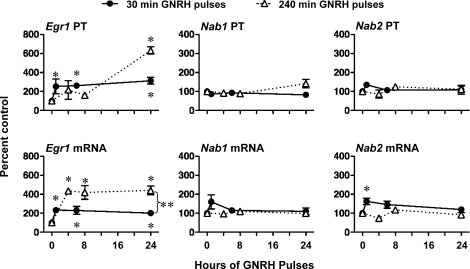
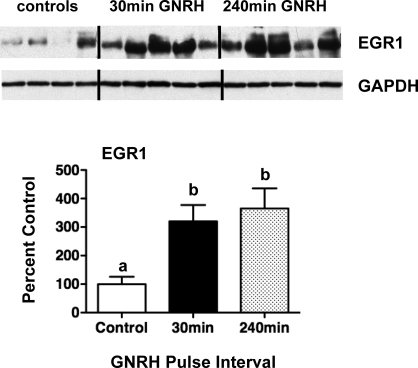
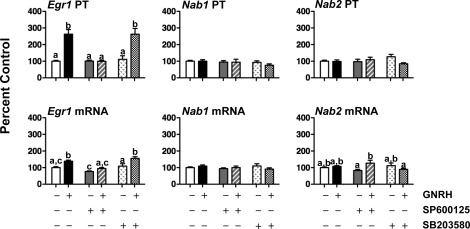
We have shown previously that Lhb transcription is maximally stimulated by faster-frequency (30 min) and Fshb by slower (240 min) GNRH pulses [20]. To examine potential links between frequency-dependent regulation of the Lhb gene and specific downstream transcriptional mediators, the effects of GNRH pulse frequency in vivo on Egr1, Nab1, and Nab2 PTs and mRNAs were determined (Fig. 1). GNRH pulses every 30 min increased Egr1 PT and mRNA 2- to 3-fold between 1 and 24 h (P < 0.05 vs. 0-h time point). In contrast, GNRH pulses every 240 min only increased Egr1 PT (6-fold; P < 0.05) after 24 h of pulses but increased Egr1 mRNA 4-fold between 4 and 24 h of treatment, and Egr1 mRNA levels were significantly greater at 24 h in rats receiving GNRH pulses every 240 min vs. 30 min (P < 0.05). Neither GNRH pulse frequency had much effect on Nab1 or Nab2 gene expression, although 30-min pulses induced a small increase in Nab2 mRNA at 1 h. The GNRH-induced increases in Egr1 gene expression had similar effects on EGR1 protein levels (Fig. 2). Although there was variability among samples, 8 h of GNRH pulses every 30 min or 240 min increased EGR1 protein levels to a similar degree (3- to 4-fold vs. controls; P < 0.05).
FIG. 1.
The effect of GNRH pulse frequency in vivo on Egr1, Nab1, and Nab2 PTs and mRNAs. Rats received 25-ng GNRH pulses either every 30 min or 240 min for 1–24 h (vehicle pulses to controls; n = 4–6 per group). Pituitaries were collected 5 min after the final pulse, and RNA was extracted. Primary transcripts or steady-state mRNAs were measured by quantitative real-time PCR. Data are expressed as percent 0-h controls ± SEM. Data were analyzed by two-way ANOVA with frequency and time as main effects. Basal levels (femtograms of plasmid/nanograms of RNA) for Egr1 PT and mRNA are 0.35 ± 0.06 and 34.0 ± 1.4; Nab1 PT and mRNA are 0.07 ± 0.01 and 0.8 ± 0.1; and Nab2 PT and mRNA are 0.70 ± 0.07 and 14.8 ± 1.01. *Significant difference (P < 0.05) vs. controls (0 h). **Significant difference between GNRH pulse-frequency paradigms at 24 h.
FIG. 2.
The effects of GNRH pulse frequency on EGR1 protein in vivo. Top: Representative Western blots of pituitary protein from rats pulsed with GNRH either every 30 min or 240 min for 8 h (n = 4–5 per group). Blots were immunostained for EGR1 and GAPDH. Protein amounts are 20 μg per lane. Bottom: Changes in EGR1 densitometry normalized to GAPDH for protein loading and expressed as percent 0-h (±SEM) controls. Points with different letters are significantly different (P < 0.05).
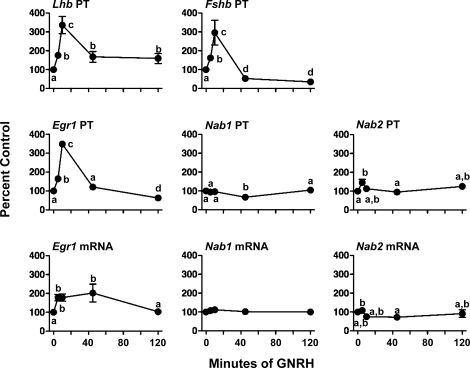
Figure 3 shows the time course of β-subunit, Egr1, and Nab gene expression responses to GNRH in cultured rat pituitary cells. GNRH rapidly increased Lhb PT (1.8-fold 5 min after GNRH pulse), peaked at 10 min (3-fold), then declined but remained elevated vs. controls at 45 min and 120 min (P < 0.05 vs. 0-h time controls). Fshb PT levels also increased rapidly after GNRH (1.6-fold at 5 min), with maximal (3-fold; P < 0.05) increases seen at 10 min before declining to <50% of controls between 45 min and 120 min (P < 0.05 vs. 0 min). Egr1 PT increased to a peak (3.5-fold; P < 0.05) 10 min after GNRH, returned to control levels at 45 min, and was suppressed by 40% after 120 min (P < 0.05). Egr1 mRNA increased approximately 2-fold between 5 min and 45 min after GNRH (P < 0.05), then returned to basal levels at 120 min. Nab1 PT and mRNA changed little during the GNRH treatment time course, although Nab1 PT was transiently lower (30% vs. control; P < 0.05) 45 min after GNRH. Nab2 PT increased 1.5-fold 5 min after GNRH before returning to basal after 10 min of treatment (P < 0.05), whereas Nab2 mRNA showed little change.
FIG. 3.
The effects of GNRH on Lhb and Fshb PTs, and Egr1, Nab1, and Nab2 PTs and mRNAs in cultured rat pituitary cells. Rat pituitary cells received two prepulses of GNRH (1 nM; media pulses to controls; 60-min interval). One hour later, cells were treated with 1 nM GNRH for 5–120 min (media to 0-h time point). Data are presented as percent (±SEM) of 0-h control (n = 4–5 per group). Groups marked with different letters are statistically different (P < 0.05). Basal levels (femtograms of plasmid/nanograms of RNA) for Lhb PT and Fshb PT are 0.59 ± 0.02 and 57.6 ± 2.9; Egr1 PT and mRNA are 4.7 ± 0.9 and 230.2 ± 22.6; Nab1 PT and mRNA are 0.15 ± 0.01 and 2.4 ± 0.2; and Nab2 PT and mRNA are 0.93 ± 0.06 and 24.2 ± 2.6.
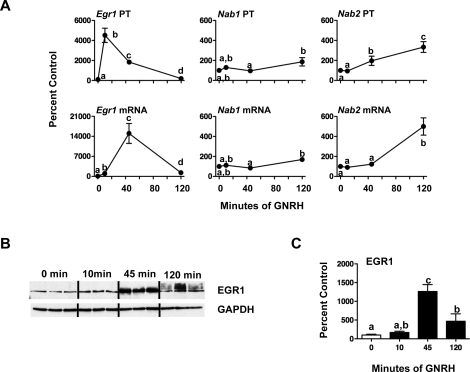
In gonadotroph-derived cell lines, GNRH has been reported to induce large increases in Egr1 mRNA [36, 37], which contrasts with our observations either in vivo or in cultured primary pituitary cells. To confirm that this difference reflects cell type, we investigated Egr1, Nab1, and Nab2 gene responses to GNRH in αT3 cells (Fig. 4A). GNRH increased both Egr1 PT and mRNA, with maximal responses seen at 10 min (45-fold) for PT and 45 min (150-fold) for mRNA (P < 0.05 vs. 0-h time). Changes in Nab1 PT and mRNA after GNRH were modest and only significant at 120 min (1.9- and 1.7-fold, respectively; P < 0.05), whereas Nab2 PT increased 2- to 3.5-fold 45–120 min after GNRH, and mRNA increased 5-fold at 120 min (P < 0.05). The large changes in Egr1 gene expression reflected similar increases in EGR1 protein levels; GNRH increased EGR1 protein 13- and 4.7-fold at 45 and 120 min, respectively (P < 0.05; Fig. 4C).
FIG. 4.
The regulation of Egr1, Nab1, and Nab2 transcripts and EGR1 protein in αT3 cells. A) Gonadotroph-derived αT3 cells were treated with 10 nM GNRH for 10–120 min. After completing the study, cells were recovered, and PT/mRNA levels were determined. Data are presented as percent (±SEM) control (n = 6 per group). Groups marked with different letters are statistically different (P < 0.05). Basal levels (femtograms of plasmid/nanograms of RNA) for Egr1 PT and mRNA are 0.06 ± 0.01 and 3.4 ± 0.3; Nab1 PT and mRNA are 0.021 ± 0.002 and 7.4 ± 0.7; and Nab2 PT and mRNA are 0.004 ± 0.001 and 1.2 ± 0.1. B) Representative Western blots of protein from gonadotroph-derived αT3 cells treated as in A (n = 3 of 6 per group). Blots were immunostained for EGR1 and GAPDH. Protein amounts are 10% of culture lysate. C) Changes in EGR1 densitometry normalized to GAPDH for protein loading and expressed as percent 0-h (±SEM) controls. Points with different letters are significantly different (P < 0.05).
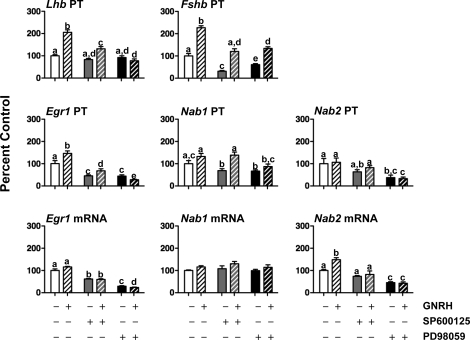
We have reported that the MAPK8/9 inhibitor SP600125, but not the MAPK14 inhibitor SB203580, abolished the Lhb transcriptional response to pulsatile GNRH in perifused rat pituitary cells [13]. To determine whether blockade of Lhb PT reflects altered pituitary Egr1 gene expression, GNRH pulses (every 60 min for 24 h) with or without SP or SB were given to perifused rat pituitary cells, and Egr1, Nab1, and Nab2 PTs and mRNAs were measured (Fig. 5). Egr1 PT and mRNA increased 2.5-fold and 1.5-fold, respectively, in GNRH-treated cells (P < 0.05 vs. controls). The SP blocked the Egr1 PT and mRNA responses to pulsatile GNRH, but SB had no effect. Nab1 and Nab2 PTs and mRNAs were not different from vehicle controls after either GNRH or either inhibitor treatment, but Nab2 mRNA was greater in SP plus GNRH-treated cells vs. SP-only cells (P < 0.05). GNRH regulation of the Gnrhr gene has been reported to be MAPK8/9 dependent, because GNRH-induced Gnrhr promoter activity was blocked in αT3 cells stably expressing a dominant-negative MAP2K4 (also known as MKK4/JNKK [45]). To determine whether the effects of MAPK8/9 blockade on Egr1 gene expression are indirect (i.e., via downregulation of Gnrhr), we measured Gnrhr mRNA. Twenty-four hours of GNRH pulses increased Gnrhr mRNA 2.5-fold (P < 0.05 vs. vehicle controls), but neither MAPK8/9 nor MAPK14 blockade altered basal or GNRH-induced increases in Gnrhr mRNA (data not shown).
FIG. 5.
The effects of MAPK8/9 and MAPK14 blockade on Egr1, Nab1, and Nab2 PTs and mRNAs. Cultured rat pituitary cells received pulses of GNRH (200 pM; media pulses to controls; 60-min interval) in the presence of the MAPK8/9 blocker (SP600125; 20 μM), MAPK14 blocker (SB203580; 20 μM), or vehicle for 24 h. After completing the study, pituitary cells were recovered, and Egr1, Nab1, and Nab2 PTs and mRNAs were measured (n = 6 per group). Data are presented as percent (±SEM) control. Groups with different letters are statistically different (P < 0.05). Basal levels (femtograms of plasmid/nanograms of RNA) for Egr1 PT and mRNA are 2.7 ± 0.2 and 82.0 ± 5.8; Nab1 PT and mRNA are 0.09 ± 0.01 and 1.1 ± 0.1; and Nab2 PT and mRNA are 0.8 ± 0.1 and 29.0 ± 1.2.
Evidence in gonadotroph-derived cell lines [36, 46–49], and recently in pituitary-specific MAPK1/3 (ERK2/1) knockout mice [50], suggests that MAPK1/3 regulation of EGR1 is fundamental for Lhb gene expression. To determine the importance of MAPK1/3 in the regulation of Egr1 gene expression, and hence Lhb, we examined the effects of GNRH pulses with or without the MAP2K1 inhibitor PD in primary rat pituitary cells. For comparison, we also measured the effects of GNRH with or without the MAPK8/9 inhibitor SP. GNRH (three pulses, every 60 min, cells recovered 10 min after the last pulse) increased Lhb PT 2-fold and Egr1 PT 1.5-fold (P < 0.05; Fig. 6). MAP2K1 blockade by PD reduced both basal Egr1 PT and mRNA and completely abrogated the GNRH-induced increases in Lhb and Egr1 PT. MAPK8/9 blockade by SP also suppressed both basal Egr1 PT and mRNAs but, in contrast to our data in perifused rat pituitary cells after 24 h of treatment (Fig. 5), short-term SP only partially suppressed the GNRH induction of Lhb and Egr1 PTs. As seen earlier, GNRH had little effect on Nab gene expression in rat pituitary cells, although GNRH did increase Nab2 mRNA 1.5-fold (P < 0.05). Nonetheless, both inhibitors suppressed basal Nab1 PT and GNRH-induced increases in Nab2 mRNA. PD also suppressed basal Nab2 PT and mRNA (P < 0.05).
FIG. 6.
The effects of MAPK8/9 and MAPK1/3 blockade on Egr1, Nab1, and Nab2 PTs and mRNAs. Rat pituitary cells were pretreated with: MAPK8/9 blocker (SP600125; 20 μM), MAP2K1 blocker (PD98059; 50 μM), or vehicle for 1 h. Then, cells received two prepulses of GNRH (1 nM; media pulses to controls; 60-min interval). One hour later, cells were treated with 1 nM GNRH for 10 min (media to controls). After completing the study, pituitary cells were recovered, and Lhb PT, Fshb PT, and Egr1, Nab1, and Nab2 PTs and mRNAs were measured (n = 4 per group). Data are presented as percent (±SEM) control. Groups marked with different letters are statistically different (P < 0.05). Basal levels (femtograms of plasmid/nanograms of RNA) for Lhb and Fshb PTs are 0.25 ± 0.01 and 63.2 ± 7.0; Egr1 PT and mRNA are 3.3 ± 0.5 and 153.1 ± 11.2; Nab1 PT and mRNA are 0.13 ± 0.02 and 3.5 ± 0.1; and Nab2 PT and mRNA are 0.60 ± 0.14 and 25.4 ± 1.2.
Previously, we found that both MAPK8/9 and MAP2K1 blockades influence Fshb gene expression after 24 h in perifused rat pituitary cells [9, 13], so we also examined the effects of SP and PD on Fshb PT in this short-term treatment model. GNRH pulses increased Fshb PT 2.3-fold. Both SP and PD suppressed basal Fshb PT (P < 0.05 vs. controls), but GNRH induction was not impaired. The suppression of basal Fshb PT by MAP2K1 blockade may be indirect, at least in part, because PD increased basal Fst mRNA 1.5-fold vs. vehicle controls (P < 0.05), whereas SP had no effect on either basal or GNRH-induced Fst mRNA (data not shown).
Because MAPK1/3 appeared to play a major role in Egr1 gene expression, and we have previously shown that androgens increase pituitary MAPK1/3 activation [43], we determined whether the levels of testosterone (500 pg/ml) added to cultured rat pituitary cells (required to allow Lhb gene responses to pulsatile GNRH [40]) affect Egr1 expression. After 24 h of androgen (testosterone or dihydrotestosterone) treatment, Egr1 PT and mRNA levels were no different compared with cells in steroid-free media (data not shown).
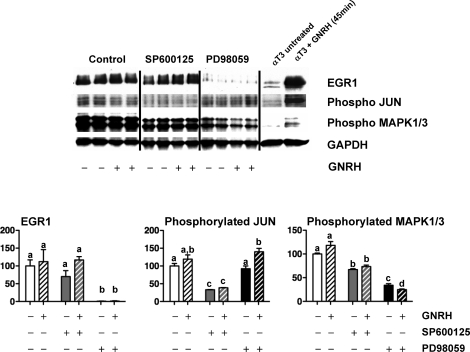
Finally, to determine both efficacy and specificity of the inhibitors, we examined the effects of MAPK8/9 and MAP2K1 blockade on basal and responses to GNRH (cells recovered 10 min after GNRH pulse; Fig. 7) for EGR1, phosphorylated p42/p44 MAPK1/3, and phosphorylated JUN protein levels. Significant stimulatory responses to GNRH were not seen (Fig. 7), which was anticipated based on previous time course studies showing that 10 min after GNRH is a little early to observe effective increases in EGR1 expression or MAPK1/3 and JUN phosphorylation [37, 41]. The SP tended to decrease basal EGR1 protein, but it did not reach significance. In contrast, PD reduced EGR1 protein to nearly undetectable levels (P < 0.05). The PD suppressed MAPK1/3 phosphorylation 65%–75% (P < 0.05). Of note, SP also partially suppressed MAPK1/3 phosphorylation (approximately 25%–30%; P < 0.05). The cross-reactivity of SP to inhibit MAPK1/3 has been noted previously [51]. Of note, SP specifically reduced cJUN phosphorylation by approximately 60% (P < 0.05), whereas PD had no effect.
FIG. 7.
The effects of MAPK8/9 and MAPK1/3 blockade on EGR1, phosphorylated cJUN, and phosphorylated MAPK1/3. Top: Representative Western blots of protein from rat pituitary cells that were treated as in Figure 6 (n = 4 per group). Blots were immunostained for EGR1, phosphorylated cJUN, phosphorylated MAPK1/3, and GAPDH. Untreated αT3 protein lysates or αT3 cells plus 50 nM GNRH for 45 min were included as negative and positive controls, respectively. Protein amounts are 10% of culture lysate. Bottom: Changes in EGR1, phosphorylated cJUN, and phosphorylated MAPK1/3 densitometry normalized to GAPDH for protein loading and expressed as percent (±SEM) control. Points with different letters are significantly different (P < 0.05).
DISCUSSION
These results expand our previous investigations into the mechanisms responsible for differential regulation of Lhb and Fshb transcription by GNRH pulses. We reported previously that in vivo GNRH pulses every 30 min preferentially increased Lhb PT levels and also stimulated a transient increase in FSHB transcription, which declined to basal levels coincidentally with an increase in Fst mRNA. In contrast, GNRH pulses every 240 min maximally increased Fshb PT, which was correlated with the suppression of Fst mRNA but had either a modest or no effect on Lhb PT [10, 20]. These and other data suggest that pulsatile GNRH acts on the gonadotroph both directly and indirectly to regulate Fshb transcription, but its actions on Lhb remain unclear. Because EGR1 has been shown to be critical for Lhb transcriptional responses to GNRH, we examined whether GNRH pulse frequency regulates EGR1 and its corepressors NAB1 and NAB2.
In rat pituitary cells, we found that Egr1 responded to GNRH with a rapid but short-duration burst of transcription, and Egr1 mRNA increased during 45 min; in perifused rat pituitary cells, Egr1 expression continued to respond to pulsatile GNRH (every 60 min) for at least 24 h. In vivo, fast-frequency GNRH pulses (every 30 min) maintained a rise in both Egr1 PT and steady-state mRNA levels during 24 h, whereas slow-frequency GNRH pulses (every 240 min) only increased Egr1 transcription after 24 h. Surprisingly, a GNRH-induced increase in Egr1 mRNA was seen earlier (beginning at 4 h), suggesting that slow-frequency GNRH pulses increase Egr1 mRNA via a nontranscriptional mechanism, such as increased mRNA stability. The regulation of Egr1 mRNA by RNA stability has been reported in several cell types [52–56]. The half-life of the Egr1 mRNA is approximately 30 min, but it increased to 70 min when protein synthesis was inhibited with cyclohexamide, indicating posttranscriptional regulation of Egr1 mRNA by a labile protein [52]. The Egr1 mRNA has a lengthy 3′ untranslated region (3′ UTR; >1000 bp) containing several elements that are known to regulate mRNA stability, such as adenosine/uracil (AU)-rich elements, polypyrimidine tracts, cold shock domain sequences (CDSs), and alternative polyadenlyaltion sites [57, 58]. Chauvin and Nilson [58] have reported that although GNRH does not affect Egr1 mRNA stability per se in LβT2 cells, the 3′ UTR-enhanced GNRH induction of a luciferase reporter and treatment with a CDS siRNA attenuated GNRH induction of Lhb mRNA.
GNRH pulses stimulated an increase in EGR1 protein, but an effect of pulse frequency was not seen. However, we cannot exclude the possibility that posttranslational effects of GNRH pulse frequency on EGR1 may play a role in Lhb gene regulation. EGR1 is regulated by a number of mechanisms that can alter transactivational activity and/or protein turnover. EGR1 is Ser/Thr phosphorylated by a number of kinases, most notably casein kinase II [59, 60], PKC [60], and AKT1 [61]. Inhibitors of protein phosphatases 1 and 2A also increase EGR1 phosphorylation [62]. The function of phosphorylated EGR1 is uncertain and has been reported to either enhance [62, 63] or inhibit [59, 60] EGR1′s ability to bind and transactivate DNA directly or via other transcription factors. Lysine residues on EGR1 are also posttranslationally modified by acetylation, sumoylation, and/or ubiquitination. EGR1 acetylation increases EGR1 protein stability [64], and sumoylated EGR1 induces the tumor suppressor phosphatase and tensin homologue [61]. Multiubiquitinated EGR1 associates with proteosome complex 8 and is targeted for degradation via the ubiquitin-dependent proteosome pathway, and proteosome blockade results in increased EGR1 accumulation [65]. Recently, Walsh and Shupnik [66] have suggested that ubiquitination and degradation of EGR1 by the proteasome are critical for Lhb transcription. When proteasome activity was inhibited in LβT2 cells, GNRH induction of an Lhb promoter reporter was suppressed, and both Egr1 mRNA and protein levels accumulated to high levels. They hypothesized that proteasome activity in the gonadotroph allows for degradation and clearing of transcription factors—in this case EGR1—from the Lhb promoter to initiate the next round of transcription. Perhaps slow-frequency GNRH pulses do not stimulate ubiquitination and/or proteasome-directed degradation of EGR1, resulting in high levels of Egr1 mRNA and protein and relatively low levels (vs. fast frequency) of Lhb transcription.
The mechanism(s) by which GNRH regulates Egr1 expression are not fully known. The rodent Egr1 promoter has five serum response elements (SREs; a distal group of three and a proximal group of two), a cAMP response element (CRE), and an AP1 site [46, 67]. The distal group of SREs contributes the majority of the Egr1 promoter responsiveness to GNRH [46], but mutation of the CRE or expression of a dominant-negative CRE-binding protein (CREB) also reduced GNRH induction of Egr1 expression [46, 49]. The role of the AP1 site in GNRH regulation of the Egr1 promoter has not been examined. The SREs bind serum response factor and recruit binding of ternary complex factors, such as the Ets protein, ELK1 [24]. Expression of a dominant-negative ELK1 blocks induction of EGR1 protein by a GNRH agonist in αT3 cells [49]. Also of note, ELK1 is a substrate for all three MAPK pathways [68], and as such connects MAPK signaling to SRE-mediated transcription of the Egr1 gene.
Several laboratories have reported that the Egr1 transcriptional response to GNRH is PKC dependent [32, 34, 69]. Further, Egr1 promoter activity or EGR1 protein levels in αT3 and LβT2 cells can be suppressed by MAPK1/3 blockade [36, 46, 49]. Recently, Bliss et al. [50] reported that pituitary-specific Mapk1/3 (ERK2/ERK1) double-knockout mice result in female, but not male, infertility, with reduced basal and GNRH-stimulated Egr1 gene expression and a loss of LHB biosynthesis. Prior to the development of this mouse model, a regulatory link between Lhb and the MAPK1/3 pathway was not well established in primary pituitary cells. In fact, we and others found that MAP2K1 blockade did not suppress Lhb mRNA expression or promoter activity in response to GNRH [9, 70]. However, in the present study we report that PD98059 completely blocked GNRH induction of Lhb PT, Egr1 PT and mRNA, and EGR1 protein in primary rat pituitary cells. The explanations for the differences between this and our former studies are uncertain but may be related to the efficacy of blockade during the treatment duration (3 h vs. 24 h) or Lhb gene expression markers measured (PT vs. steady-state mRNA). In previous studies, we measured the effects of MAP2K1 blockade on Lhb mRNA expression in perifused rat pituitary cells given GNRH pulses every 60 min for 24 h. It is possible that the actions of PD on Lhb expression are transient, and/or that MAP2K1 blockade can be compensated for by another signaling pathway(s), after an extended duration of GNRH pulse treatment. In previous experiments, we determined the effects of PD plus GNRH on steady-state Lhb mRNA by Northern dot blotting vs. measuring Lhb PT using quantitative real-time PCR. Thus, PD may suppress Lhb transcription but not significantly deplete the pool of steady-state mRNA, which has a much longer half-life [71].
We have also reported that MAPK8/9 blockade by either the inhibitor SP600125 or the dominant negatives of MAP2K4 and/or 7 (also known as MKK4/7) inhibited the Lhb transcriptional response to GNRH in both perifused rat pituitary cells (24-h treatment duration) and LβT2 cells (6-h treatment duration [13]), whereas MAPK14 blockade had no effect. Here, we report that GNRH induction of Egr1 PT and mRNA in perifused rat pituitary cells is also MAPK8/9, but not MAPK14, dependent; however, in shorter-duration (3 h) studies, SP was only partially effective in suppressing GNRH-induced increases in Lhb and Egr1 PTs. We acknowledge that blocking MAPK8/9 activity with chemical inhibitors is not completely specific, and kinase inhibition by SP is not limited to MAPK8/9 [51]. Indeed, we observed that 3 h of SP treatment also partially suppressed MAPK1/3 activation (Fig. 7), although it was quite specific for inhibiting cJUN phosphorylation. However, the present results and our earlier data in LβT2 cells using dominant-negative MAP2K4 and MAP2K7 proteins [13] support a role for GNRH-induced MAPK8/9 activation in Egr1-dependent Lhb transcription. Furthermore, MAPK8/9 has been implicated in the regulation EGR1 in several other cell types [72–76].
The ability of EGR1 to transactivate target genes may be altered by expression of its corepressors NAB1 or NAB2. NAB1 is constitutively expressed in most cell types [24, 77], whereas NAB2 is induced by the same stimuli as EGR1, but responses are often delayed [78]. The human NAB2 promoter contains 11 EGR1-binding sites and is strongly stimulated by EGR1, suggesting that EGR1 induction of NAB2 is a negative-feedback circuit to regulate EGR1 activity [79]. In LβT2 cells, a single GNRH pulse increased Nab1 and Nab2 mRNAs 2- to 4-fold 1–4 h after pulse [37]. We saw similar responses to GNRH for Nab2 gene expression in αT3 cells, whereas Nab1 was largely unaffected. In contrast to LβT2 or αT3 cells, Nab1 and Nab2 transcripts showed little change after GNRH either in vivo or in cultured rat pituitary cells, indicating that GNRH does not significantly regulate EGR1 bioavailability via the NAB proteins in normal rat pituitary cells.
In conclusion, these findings reveal that pulsatile GNRH increases Egr1 transcription both in vivo and in cultured rat pituitary cells, but not in a frequency-dependent manner and not to the degree seen in gonadotroph-derived αT3 cells. Additionally, GNRH induction of Egr1 transcription and mRNA expression is dependent on MAPK8/9 and MAPK1/3 but not on MAPK14. These results suggest that both MAPK8/9 and MAPK1/3 are critical regulators of Lhb gene expression, in part by mediating the Egr1 transcriptional response to pulsatile GNRH.
Acknowledgments
The authors would like to thank the University of Virginia Center for Research in Reproduction Ligand Preparation and Assay Core for conducting the radioimmunoassay, Dr. Pamela Mellon for generating the αT3 cell line, and Drs Heidi Walsh and Margaret Shupnik for their assistance with these cells.
Footnotes
Supported by U.S. Public Health Service grant no. HD-33039 to J.C.M., and the National Institute of Child Health and Human Development/National Institutes of Health through a cooperative agreement (U54-HD28934, Ligand Assay and Analysis Core, Molecular Core) as part of the Specialized Cooperative Centers Program in Reproductive Research to J.C.M. and D.J.H.
REFERENCES
- Gharib SD, Wierman ME, Shupnik MA, Chin WW.Molecular biology of the pituitary gonadotropins. Endocrine Rev 1990; 11: 177–199. [DOI] [PubMed] [Google Scholar]
- Burger LL, Haisenleder DJ, Dalkin AC, Marshall JC.Regulation of gonadotropin subunit gene transcription. J Mol Endocrinol 2004; 33: 559–584. [DOI] [PubMed] [Google Scholar]
- Grosse R, Schmid A, Schoneberg T, Herrlich A, Muhn P, Schultz G, Gudermann T.Gonadotropin-releasing hormone receptor initiates multiple signaling pathways by exclusively coupling to G(q/11) proteins. J Biol Chem 2000; 275: 9193–9200. [DOI] [PubMed] [Google Scholar]
- Liu F, Usui I, Evans LG, Austin DA, Mellon PL, Olefsky JM, Webster NJ.Involvement of both Gq/11 and Gs proteins in gonadotropin-releasing hormone receptor-mediated signaling in LβT2 cells. J Biol Chem 2002; 277: 32099–32108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojilkovic SS, Catt KJ.Novel aspects of GnRH-induced intracellular signaling and secretion in pituitary gonadotrophs. J Neuroendocrinol 1995; 7: 739–757. [DOI] [PubMed] [Google Scholar]
- Ando H, Hew CL, Urano A.Signal transduction pathways and transcription factors involved in the gonadotropin-releasing hormone-stimulated gonadotropin subunit gene expression. Comp Biochem Physiol B Biochem Mol Biol 2001; 129: 525–532. [DOI] [PubMed] [Google Scholar]
- Naor Z.Signal transduction mechanisms of Ca2+ mobilizing hormones: the case of gonadotropin-releasing hormone. Endocr Rev 1990; 11: 326–353. [DOI] [PubMed] [Google Scholar]
- Dobkin-Bekman M, Naidich M, Pawson AJ, Millar RP, Seger R, Naor Z.Activation of mitogen-activated protein kinase (MAPK) by GnRH is cell-context dependent. Mol Cell Endocrinol 2006; 252: 184–190. [DOI] [PubMed] [Google Scholar]
- Haisenleder DJ, Cox ME, Parsons SJ, Marshall JC.Gonadotropin-releasing hormone pulses are required to maintain activation of mitogen-activated protein kinase: role in stimulation of gonadotrope gene expression. Endocrinology 1998; 139: 3104–3111. [DOI] [PubMed] [Google Scholar]
- Burger LL, Haisenleder DJ, Aylor KA, Marshall JC.Regulation of intracellular signaling cascades by GnRH pulse frequency in the rat pituitary: roles for CaMK II, ERK and JNK activation. Biol Reprod 2008; 79: 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D, Bonfil D, Chuderland D, Kraus S, Seger R, Naor Z.Activation of MAPK cascades by GnRH: ERK and JNK are involved in basal and GnRH-stimulated activity of the glycoprotein hormone LHβ-subunit promoter. Endocrinology 2002; 143: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Mulvaney JM, Roberson MS.Divergent signaling pathways requiring discrete calcium signals mediate concurrent activation of two mitogen-activated protein kinases by gonadotropin-releasing hormone. J Biol Chem 2000; 275: 14182–14189. [DOI] [PubMed] [Google Scholar]
- Haisenleder DJ, Burger LL, Walsh HE, Stevens J, Aylor KW, Shupnik MA, Marshall JC.Pulsatile gonadotropin-releasing hormone stimulation of gonadotropin subunit transcription in rat pituitaries: evidence for the involvement of Jun N-terminal kinase but not p38. Endocrinology 2008; 149: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson MS, Zhang T, Li HL, Mulvaney JM.Activation of the p38 mitogen-activated protein kinase pathway by gonadotropin-releasing hormone. Endocrinology 1999; 140: 1310–1318. [DOI] [PubMed] [Google Scholar]
- Ferris HA, Shupnik MA.Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GnRH1. Biol Reprod 2006; 74: 993–998. [DOI] [PubMed] [Google Scholar]
- Coss D, Jacobs SB, Bender CE, Mellon PL.A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J Biol Chem 2004; 279: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Guendner MJ, Halvorson LM, Jameson JL.Transcriptional activation of the follicle-stimulating hormone β-subunit gene by activin. Endocrinology 1995; 136: 1885–1891. [DOI] [PubMed] [Google Scholar]
- Bilezikjian LM, Blount AL, Leal AM, Donaldson CJ, Fischer WH, Vale WW.Autocrine/paracrine regulation of pituitary function by activin, inhibin and follistatin. Mol Cell Endocrinol 2004; 225: 29–36. [DOI] [PubMed] [Google Scholar]
- Kirk SE, Dalkin AC, Yasin M, Haisenleder DJ, Marshall JC.Gonadotropin-releasing hormone pulse frequency regulates expression of pituitary follistatin messenger ribonucleic acid: a mechanism for differential gonadotrope function. Endocrinology 1994; 135: 876–880. [DOI] [PubMed] [Google Scholar]
- Burger LL, Dalkin AC, Aylor KW, Haisenleder DJ, Marshall JC.GnRH pulse frequency modulation of gonadotropin subunit gene transcription in normal gonadotropes-assessment by primary transcript assay provides evidence for roles of GnRH and follistatin. Endocrinology 2002; 143: 3243–3249. [DOI] [PubMed] [Google Scholar]
- Clayton RN, Lalloz MR, Salton SR, Roberts JL.Expression of luteinising hormone-beta subunit chloramphenicol acetyltransferase (LH-beta-CAT) fusion gene in rat pituitary cells: induction by cyclic 3′-adenosine monophosphate (cAMP). Mol Cell Endocrinol 1991; 80: 193–202. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK.Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001; 20: 2438–2452. [DOI] [PubMed] [Google Scholar]
- Ferris HA, Walsh HE, Stevens J, Fallest PC, Shupnik MA.Luteinizing hormone beta promoter stimulation by adenylyl cyclase and cooperation with gonadotropin-releasing hormone 1 in transgenic mice and LβT2 cells. Biol Reprod 2007; 77: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Thiel G, Cibelli G.Regulation of life and death by the zinc finger transcription factor Egr1. J Cell Physiol 2002; 193: 287–292. [DOI] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L.A gene for neuronal plasticity in the mammalian brain: Zif268/Egr1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol 2004; 74: 183–211. [DOI] [PubMed] [Google Scholar]
- Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J.Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr1). Science 1996; 273: 1219–1221. [DOI] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P.Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr1)-targeted mice. Mol Endocrinol 1998; 12: 107–122. [DOI] [PubMed] [Google Scholar]
- Halvorson LM, Ito M, Jameson JL, Chin WW.Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem 1998; 273: 14712–14720. [DOI] [PubMed] [Google Scholar]
- Kaiser UB, Halvorson LM, Chen MT.Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (Egr1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-β gene promoter: an integral role for SF-1. Mol Endocrinol 2000; 14: 1235–1245. [DOI] [PubMed] [Google Scholar]
- Call GB, Wolfe MW.Species differences in GnRH activation of the Lhb promoter: role of Egr1 and Sp1. Mol Cell Endocrinol 2002; 189: 85–96. [DOI] [PubMed] [Google Scholar]
- Horton CD, Halvorson LM.The cAMP signaling system regulates Lhb gene expression: roles of early growth response protein-1, SP1 and steroidogenic factor-1. J Mol Endocrinol 2004; 32: 291–306. [DOI] [PubMed] [Google Scholar]
- Wolfe MW, Call GB.Early growth response protein 1 binds to the luteinizing hormone-beta promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol Endocrinol 1999; 13: 752–763. [DOI] [PubMed] [Google Scholar]
- Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y.Activation of luteinizing hormone beta gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem 1999; 274: 13870–13876. [DOI] [PubMed] [Google Scholar]
- Tremblay JJ, Drouin J.Egr1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol Cell Biol 1999; 19: 2567–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade JP, Carter DA.Cyclical expression of Egr1/NGFI-A in the rat anterior pituitary: a molecular signal for ovulation? J Neuroendocrinol 2000; 12: 671–676. [DOI] [PubMed] [Google Scholar]
- Kanasaki H, Bedecarrats GY, Kam KY, Xu S, Kaiser UB.Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LβT2 cells. Endocrinology 2005; 146: 5503–5513. [DOI] [PubMed] [Google Scholar]
- Lawson MA, Tsutsumi R, Zhang H, Talukdar I, Butler BK, Santos SJ, Mellon PL, Webster NJ.Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop involving early growth response-1 and Ngfi-A binding protein 1 and 2. Mol Endocrinol 2007; 21: 1175–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkin AC, Burger LL, Aylor KW, Haisenleder DJ, Workman LJ, Cho S, Marshall JC.Regulation of gonadotropin subunit gene transcription by gonadotropin-releasing hormone: measurement of primary transcript ribonucleic acids by quantitative reverse transcription-polymerase chain reaction assays. Endocrinology 2001; 142: 139–146. [DOI] [PubMed] [Google Scholar]
- Haisenleder DJ, Burger LL, Aylor KW, Dalkin AC, Marshall JC.GnRH stimulation of gonadotropin subunit transcription: evidence for the involvement of calcium/calmodulin-dependent kinase II activation in rat pituitaries. Endocrinology 2003; 144: 2768–2774. [DOI] [PubMed] [Google Scholar]
- Yasin M, Dalkin AC, Haisenleder DJ, Marshall JC.Testosterone is required for gonadotropin-releasing hormone stimulation of luteinizing hormone-beta messenger ribonucleic acid expression in female rats. Endocrinology 1996; 137: 1265–1271. [DOI] [PubMed] [Google Scholar]
- Xie J, Bliss SP, Nett TM, Eersole BJ, Sealfon SC, Roberson MS.Transcript profiling of immediate early genes reveals a unique role of ATF-3 in mediating activation of the glycoprotein hormone alpha-subunit promoter by GnRH. Mol Endocrinol 2005; 19: 2624–2638. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fortin J, Lamba P, Bonomi M, Persani L, Roberson MS, Bernard DJ.Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity. Endocrinology 2008; 149: 5577–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haisenleder DJ, Burger LL, Aylor KW, Dalkin AC, Walsh HE, Shupnik MA, Marshall JC.Testosterone stimulates follicle-stimulating hormone beta transcription via activation of extracellular signal-regulated kinase: evidence in rat pituitary cells. Biol Reprod 2005; 72: 523–529. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N.Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Ellsworth BS, White BR, Burns AT, Cherrington BD, Otis AM, Clay CM.c-Jun N-terminal kinase activation of activator protein-1 underlies homologous regulation of the gonadotropin-releasing hormone receptor gene in alpha T3–1 cells. Endocrinology 2003; 144: 839–849. [DOI] [PubMed] [Google Scholar]
- Duan WR, Ito M, Park Y, Maizels ET, Hunzicker-Dunn M, Jameson JL.GnRH regulates early growth response protein 1 transcription through multiple promoter elements. Mol Endocrinol 2002; 16: 221–233. [DOI] [PubMed] [Google Scholar]
- Liu F, Austin DA, Mellon PL, Olefsky JM, Webster NJ.GnRH activates ERK1/2 leading to the induction of c-fos and LHbeta protein expression in LbetaT2 cells. Mol Endocrinol 2002; 16: 419–434. [DOI] [PubMed] [Google Scholar]
- Maudsley S, Naor Z, Bonfil D, Davidson L, Karali D, Pawson AJ, Larder R, Pope C, Nelson N, Millar RP, Brown P.Proline-rich tyrosine kinase 2 mediates gonadotropin-releasing hormone signaling to a specific extracellularly regulated kinase-sensitive transcriptional locus in the luteinizing hormone beta-subunit gene. Mol Endocrinol 2007; 21: 1216–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer SI, Willars GB, Nishida E, Thiel G.Elk-1, CREB, and MKP-1 regulate Egr-1 expression in gonadotropin-releasing hormone stimulated gonadotrophs. J Cell Biochem 2008; 105: 1267–1278. [DOI] [PubMed] [Google Scholar]
- Bliss SP, Miller A, Navratil AM, Xie J, McDonough SP, Fisher PJ, Landreth GE, Roberson MS.ERK signaling in the pituitary is required for female but not male fertility. Mol Endocrinol 2009; 23: 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P.The selectivity of protein kinase inhibitors: a further update. Biochem J 2007; 408: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein SH, Kharbanda SM, Sherman ML, Sukhatme VP, Kufe DW.Posttranscriptional regulation of the zinc finger-encoding EGR-1 gene by granulocyte-macrophage colony-stimulating factor in human U-937 monocytic leukemia cells: involvement of a pertussis toxin-sensitive G protein. Cell Growth Differ 1991; 2: 273–278. [PubMed] [Google Scholar]
- Long KD, Salbaum JM.Evolutionary conservation of the immediate-early gene ZENK. Mol Biol Evol 1998; 15: 284–292. [DOI] [PubMed] [Google Scholar]
- Baek SJ, Wilson LC, Hsi LC, Eling TE.Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem 2003; 278: 5845–5853. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Hood DA.Regulation of Egr-1, SRF, and Sp1 mRNA expression in contracting skeletal muscle cells. J Appl Physiol 2004; 97: 2207–2213. [DOI] [PubMed] [Google Scholar]
- Simon P, Schott K, Williams RW, Schaeffel F.Posttranscriptional regulation of the immediate-early gene EGR1 by light in the mouse retina. Eur J Neurosci 2004; 20: 3371–3377. [DOI] [PubMed] [Google Scholar]
- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, Le Beau MM, Adamson ED.A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 1988; 53: 37–43. [DOI] [PubMed] [Google Scholar]
- Chauvin T, Nilson J.Posttranscriptional regulation of Egr1 in L beta T2 cells is influenced by CSDA. In: Abstracts of the 40th Annual Meeting of the Society for the Study of Reproduction, July 21–25, 2007, San Antonio, Texas. Biol Reprod 2007; Special Issue: Abstract 93. [PubMed]
- Jain N, Mahendran R, Philp R, Guy GR, Tan YH, Cao X.Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells. J Biol Chem 1996; 271: 13530–13536. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Weitzmann MN, Kimble RB, Rizzo M, Zahner M, Milbrandt J, Ross FP, Pacifici R.Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp-1. J Clin Invest 1998; 102: 1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, Ke Y, Baron V, Mercola D, Feng GS, Adamson E, Mustelin T.PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J 2009; 28: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Mahendran R, Guy GR, Tan YH.Protein phosphatase inhibitors induce the sustained expression of the Egr-1 gene and the hyperphosphorylation of its gene product. J Biol Chem 1992; 267: 12991–12997. [PubMed] [Google Scholar]
- Huang RP, Adamson ED.The phosphorylated forms of the transcription factor, Egr-1, bind to DNA more efficiently than non-phosphorylated. Biochem Biophys Res Commun 1994; 200: 1271–1276. [DOI] [PubMed] [Google Scholar]
- Yu J, de Belle I, Liang H, Adamson ED.Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Mol Cell 2004; 15: 83–94. [DOI] [PubMed] [Google Scholar]
- Bae MH, Jeong CH, Kim SH, Bae MK, Jeong JW, Ahn MY, Bae SK, Kim ND, Kim CW, Kim KR, Kim KW.Regulation of Egr-1 by association with the proteasome component C8. Biochim Biophys Acta 2002; 1592: 163–167. [DOI] [PubMed] [Google Scholar]
- Walsh HE, Shupnik MA.Proteasome regulation of dynamic transcription factor occupancy on the GnRH-stimulated luteinizing hormone beta-subunit promoter. Mol Endocrinol 2009; 23: 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtgen JL, Campbell CJ, Braddock M.Full promoter sequence of human early growth response factor-1 (Egr1): demonstration of a fifth functional serum response element. DNA Seq 2000; 10: 429–432. [DOI] [PubMed] [Google Scholar]
- Yang SH, Whitmarsh AJ, Davis RJ, Sharrocks AD.Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J 1998; 17: 1740–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorson LM, Kaiser UB, Chin WW.The protein kinase C system acts through the early growth response protein 1 to increase LHβ gene expression in synergy with steroidogenic factor-1. Mol Endocrinol 1999; 13: 106–116. [DOI] [PubMed] [Google Scholar]
- Weck J, Fallest PC, Pitt LK, Shupnik MA.Differential gonadotropin-releasing hormone stimulation of rat luteinizing hormone subunit gene transcription by calcium influx and mitogen-activated protein kinase-signaling pathways. Mol Endocrinol 1998; 12: 451–457. [DOI] [PubMed] [Google Scholar]
- Paul SJ, Ortolano GA, Haisenleder DJ, Stewart JM, Shupnik MA, Marshall JC.Gonadotropin subunit messenger RNA concentrations after blockade of gonadotropin-releasing hormone action: testosterone selectively increases follicle-stimulating hormone beta-subunit messenger RNA by posttranscriptional mechanisms. Mol Endocrinol 1990; 4: 1943–1955. [DOI] [PubMed] [Google Scholar]
- Uchida C, Gee E, Ispanovic E, Haas TL.JNK as a positive regulator of angiogenic potential in endothelial cells. Cell Biol Int 2008; 32: 769–776. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Ashouri J, Wolter S, Doerrie A, Dittrich-Breiholz O, Schneider H, Wagner EF, Troppmair J, Mackman N, Kracht M.Transcriptional regulation of EGR-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J Biol Chem 2008; 283: 12120–12128. [DOI] [PubMed] [Google Scholar]
- Shin SY, Lee JH, Min B, Lee YH.The translation inhibitor anisomycin induces Elk-1-mediated transcriptional activation of egr-1 through multiple mitogen-activated protein kinase pathways. Exp Mol Med 2006; 38: 677–685. [DOI] [PubMed] [Google Scholar]
- Gururajan M, Chui R, Karuppannan AK, Ke J, Jennings CD, Bondada S.c-Jun N-terminal kinase (JNK) is required for survival and proliferation of B-lymphoma cells. Blood 2005; 106: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Salman H, Danilenko M, Studzinski GP.Cooperation between antioxidants and 1,25-dihydroxyvitamin D3 in induction of leukemia HL60 cell differentiation through the JNK/AP-1/Egr-1 pathway. J Cell Physiol 2005; 204: 964–974. [DOI] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, Milbrandt J.Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci U S A 1995; 92: 6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Berk BC.NAB2: a transcriptional brake for activated gene expression in the vessel wall? Am J Pathol 1999; 155: 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbrink J, Gerlinger M, Johnson JP.Egr1 induces the expression of its corepressor Nab2 by activation of the Nab2 promoter thereby establishing a negative feedback loop. J Biol Chem 2005; 280: 42785–42793. [DOI] [PubMed] [Google Scholar]