Abstract
Bone homeostasis is tightly regulated by matrix-producing osteoblasts and bone-resorbing osteoclasts. During osteoclast development, mononuclear preosteoclasts derived from myeloid cells fuse together to form multinucleated, giant cells. Previously, we reported that the d2 isoform of the vacuolar (H+) ATPase V0 domain (Atp6v0d2) plays an important role in osteoclast maturation and bone formation. To understand how Atp6v0d2 controls osteoclast maturation, we have performed a yeast two-hybrid screen using full-length Atp6v0d2 as the bait, and identified adhesion-regulating molecule 1 protein (Adrm1) as a potential functional partner of Atp6v0d2. The interaction between Atp6v0d2 and Adrm1 was confirmed in yeast and in vivo using immunoprecipitation assays. We also show that Adrm1 is required for cell migration and osteoclast maturation.
Keywords: Osteoclast, Atp6v0d2, Adrm1, cell fusion, differentiation, yeast-two hybrid screen
Introduction
Bone homeostasis is maintained through a dynamic balance between bone-resorbing osteoclasts and matrix-producing osteoblasts. Disruption of this balance may lead to bone diseases, such as osteoporosis. Osteoclasts—giant multinucleated cells (MNCs) derived from hematopoietic macrophage/monocyte lineage precursor cells—are formed as mononuclear preosteoclasts fuse together. Bone resorption is multistep process, which includes secretion of such lysosomal enzymes as tartrate-resistant acid phosphatase (TRAP), cathepsin K, and several matrix metalloproteases, as well as extracellular acidification at the ruffled border [1, 2, 3]. This extracellular acidification is mediated by vacuolar type H+-ATPase complex (V-ATPase). V-ATPases are composed of at least 13 distinct subunits, and several of these subunits have multiple isoforms [4]. The expression of these isoforms is regulated in a cell type- and tissue-specific manner. Two isoforms of V-ATPase V0 subunit d, d1 and d2, have been identified in mice and humans. Both of these isoforms are more abundantly expressed in osteoclasts than in other tissues [5, 6, 7]. In particular, the expression of Atp6v0d2 is more specific than that of Atp6v0d1 during osteoclast differentiation, which is thought to highlight the importance of Atp6v0d2 for osteoclastogenesis, although the exact function of Atp6v0d2 has not been elucidated [8]. Previously, we reported that genetic inactivation of Atp6v0d2 in mice results in markedly increased bone mass due to defective osteoclasts. Interestingly, Atp6v0d2 deficiency did not affect the differentiation of osteoclasts or V-ATPase activity in this cell population. Rather, Atp6v0d2 was required for efficient osteoclast maturation [9].
In this study, we have further investigated the functions of Atp6v0d2 during osteoclast differentiation by searching for Atp6v0d2-interacting proteins. We screened a human bone marrow cDNA-derived two-hybrid library using Atp6v0d2 as a bait, and identified adhesion-regulating molecule 1 (Adrm1) as an Atp6v0d2-binding partner. Adrm1 was previously identified due to its upregulated expression levels in metastatic tumor cells, and its involvement in cell adhesion and cell migration [10, 11, 12, 13]. Although initially described as a cell membrane glycoprotein, Adrm1 is not glycosylated, is intracellularly localized, and probably has no direct role in cell adhesion [11, 14, 15]. Furthermore, recent studies suggested that Adrm1 is a proteasome subunit that functions as novel ubiquitin receptor [16, 17, 18, 19]. To elucidate the role of Adrm1 during osteoclast differentiation, we performed retrovirus-mediated RNA interference (RNAi) knockdown of Adrm1 expression during in vitro osteoclast differentiation. We also examined the effects of Adrm1 on fusion efficiency, cell adhesion, and cell migration.
Materials and Methods
Cell culture
Murine preosteoclasts were prepared from the bone marrow cells of 6- to 8-week-old mice as described previously [9]. To generate osteoclasts, bone marrow-derived monocytes/macrophages (BMMs; 1 × 104 cells/well in 96-well plates) were cultured with macrophage-colony stimulating factor (M-CSF, 50 ng/ml) and soluble receptor of nuclear factor-κB (RANKL, 150 ng/ml) for 3–4 days.
Yeast two-hybrid screen
The yeast two-hybrid Matchmaker 3 system (Clontech Laboratories, Palo Alto, California, USA) was used to screen for potential ATP6v0d2-interacting proteins. A human bone marrow cDNA library cloned into pGAD-Rec such that it contained sequences encoding an amino-terminal GAL4 activation domain and a hemagglutinin tag was pretransformed into the Y187 yeast strain. This library was then screened after mating the transformants with AH109 yeast containing the human ATP6v0d2 bait construct pGBKT7-d2, which encoded full-length human ATP6v0d2 fused to a GAL4 DNA-binding domain. The mated yeast cells were grown on agar plates containing Trp, Leu, and His dropout nutritional selection media in the presence of 12 mM 3-amino-1,2,4-triazole. To reduce the false positives after selection, the selected yeast colonies were transferred to Trp, Leu, His, and Ade dropout nutritional selection media and assayed for lacZ activity. Prey plasmids encoding human bone marrow proteins that interacted with ATP6v0d2 in the lacZ-positive yeast clones were isolated by transforming them into Escherichia coli and selecting the bacteria on carbenicillin-containing culture plates. Isolated plasmids were then subjected to DNA sequence analysis.
Coimmunoprecipitation and Western blotting
Western blotting and immunoprecipitation analyses were performed as described previously [20]. Briefly, cells were incubated in buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 10% glycerol, and protease and phosphatase inhibitors for 30 min on ice and scraped into microcentrifuge tubes. The whole cell extracts were vigorously vortexed and then microcentrifuged for 20 min at 10,000 × g. Protein concentrations in the supernatants were determined using a DC Protein Assay Kit (Bio-Rad), and 10–20 µg of the cellular proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was probed with a 1:1000 dilution of the antibodies described in the figure legends. For immunoprecipitation, 300–500 µg of the cellular proteins were mixed with 5 µg of the antibodies described in the figure legends for 4–6 hours at 4°C. This mixture was incubated with 50 µl of protein G-Sepharose beads for 2 hours at 4°C. The beads were washed five times in lysis buffer and subjected to Western blotting.
RNAi expression vectors, retroviral infection, and analysis of osteoclasts
RNAi oligonucleotides (Adrm1, 5’-GAA AGA CGA AGA AGA AGA TAT-3’; GFP, 5’-CAT GGA TGA ACT ATA CAA A-3’) were synthesized by Integrated DNA Technologies and cloned into the retroviral small interference RNA (siRNA) vector pSuper-retro-Puro (OligoEngine). BMMs were infected with retroviruses and cultured with M-CSF (50 ng/ml) and sRANKL (150 ng/ml) for 3–5 days to generate osteoclasts as described previously [20]. For osteoclast analyses, osteoclasts were fixed with 10% formalin and stained for TRAP activity, whereas bone slices were stained with 0.5% toluidine blue as described previously [9].
Cell adhesion and migration assays
Cell migration assays were performed as previously described [21]. Briefly, preosteoclasts were cultured with 50 ng/ml M-CSF and 150 ng/ml RANKL for 60–72 hours. Cells were starved in 0.5% FBS for 3 hours, detached with enzyme-free dissociation solution, and resuspended in low-serum α-MEM containing 1% FBS and 2% BSA. Cells were plated in the upper chamber of a Boyden chamber with polycarbonate filters containing 8- µm pore membranes and incubated for 2 hours. The medium was then exchanged to low-serum α-MEM containing 50 ng/ml MCSF and 150 ng/ml RANKL. After 12 hours of incubation, the cells were fixed in 3.7% formaldehyde for 10 minutes, stained for TRAP (Sigma), and counted. Cell adhesion assays were performed as previously described [21]. Briefly, 96-well culture plates were coated with 20 µg/ml fibrinogen, 20 µg/ml fibronectin, or 10 µg/ml vitronectin in PBS for 12 hours at 37°C. The plates were washed with PBS and incubated with PBS containing 0.2% BSA for 1 hour at 37°C. Preosteoclasts were cultured in the presence of 50 ng/mL M-CSF plus 150 ng/mL RANKL for 60 to 72 hours. Cells were detached using cell dissociation solution and suspended in serum-free α-MEM. Cells were seeded in each fibrinogen-, fibronectin-, or vitronectin-coated well of a 96-well plate and incubated for 10 minutes at 37°C. Nonadherent cells were removed by washing with PBS. Adherent cells were fixed in 3.7% formaldehyde for 10 minutes and stained with hematoxylin or TRAP solution. Stained cells were counted.
Results
Adrm1 interacts with Atp6v0d2
To identify possible intracellular interaction partners for Atp6v0d2, we performed yeast two-hybrid screening using a cDNA library prepared from human bone marrow cells. When the human bone marrow cDNA library was screened using full-length Atp6v0d2 as bait, we isolated two independent clones encoding a protein subsequently identified as Adrm1. Previous studies have described Adrm1 as a cell adhesion–promoting protein that is expressed in several tissues, with upregulated expression levels observed in metastatic cancer cells [11, 15]. This protein does not belong to any of the previously known families of cell adhesion molecules.
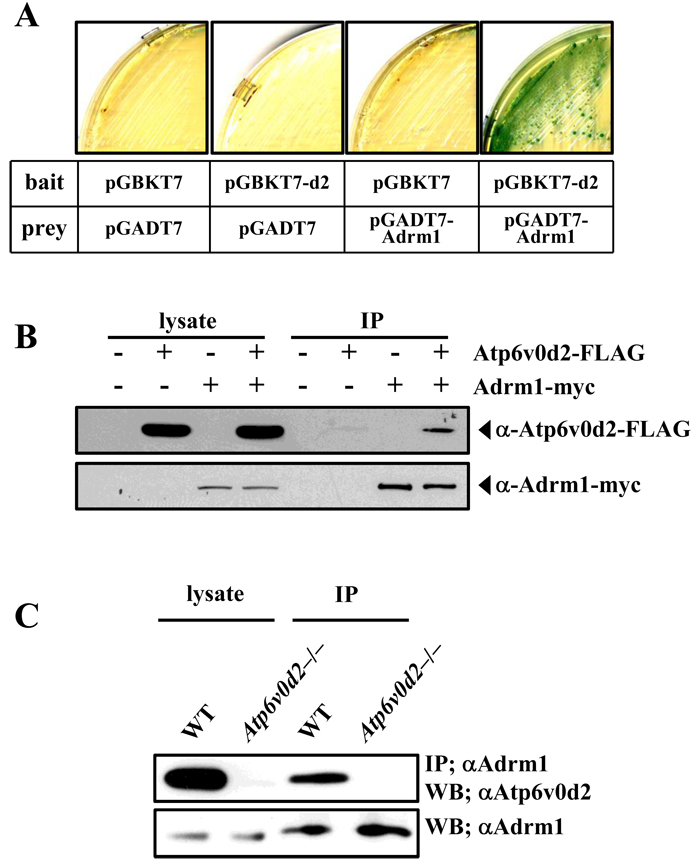
To confirm the interaction between Adrm1 and Atp6v0d2, we cotransfected yeast with pGBKT7 or pGBKT7-d2 along with pGADT7 or pGADT7-Adrm1. The results showed that three reporter genes (lacZ, His, and Ade) were only expressed when Atp6v0d2 and Adrm1 were simultaneously overexpressed, demonstrating that neither of these proteins was capable of activating transcription on its own (Fig. 1A). These results confirm the interaction between Adrm1 and Atp6v0d2 in yeast. To test the Adrm1–Atp6v0d2 interaction in mammalian cells, we expressed FLAG-tagged Atp6v0d2 together with myc-tagged Adrm1, FLAG-tagged Atp6v0d2 alone, or myc-tagged Adrm1 alone in 293T cells. Each cell lysate was subjected to coimmunoprecipitation analysis using anti-myc antibodies and Western blotting using the indicated antibodies. Atp6v0d2 was detected in myc immunoprecipitates from cells coexpressing Adrm1 and Atp6v0d2, but was not observed in cells expressing either Adrm1 or Atp6v0d2 (Fig. 1B). Additionally, the interaction between endogenous Atp6v0d2 and Adrm1 was examined in primary bone marrow–derived osteoclasts using coimmunoprecipitation analysis. To exclude nonspecific interactions between Adrm1 and Atp6v0d2, lysates of Atp6v0d2-deficient bone marrow–derived osteoclasts were used as control samples. This analysis showed that Adrm1 and Atp6v0d2 were detected as a complex in the primary cells (Fig. 1C). Taken together, these results demonstrate that Adrm1 interacts with Atp6v0d2.
Fig. 1.
Adrm1 interacts with ATP6v0d2. (A) Yeast strain AH109 was transformed with pGBKT7 or pGBKT7-d2 (bait) along with pGADT7 or pGADT7-Adrm1 (prey). Transformed yeast cells were then spread on Trp, Leu, His, and Ade dropout nutritional selection media and assayed for lacZ activity. (B) 293T cells were transfected with the indicated plasmids. Cell lysates were prepared and immunoprecipitation was performed with anti-FLAG antibodies. The precipitates were subjected to Western blotting with anti-Myc or anti-FLAG antibodies. (C) Cell lysates were prepared from osteoclasts derived from the bone marrow of wild-type or Atp6v0d2-deficient mice, and immunoprecipitation was performed with anti-Adrm1 antibodies. The precipitates were subjected to Western blotting with anti-ATP6v0d2 or anti-Adrm1 antibodies.
Adrm1 expression levels during osteoclast differentiation
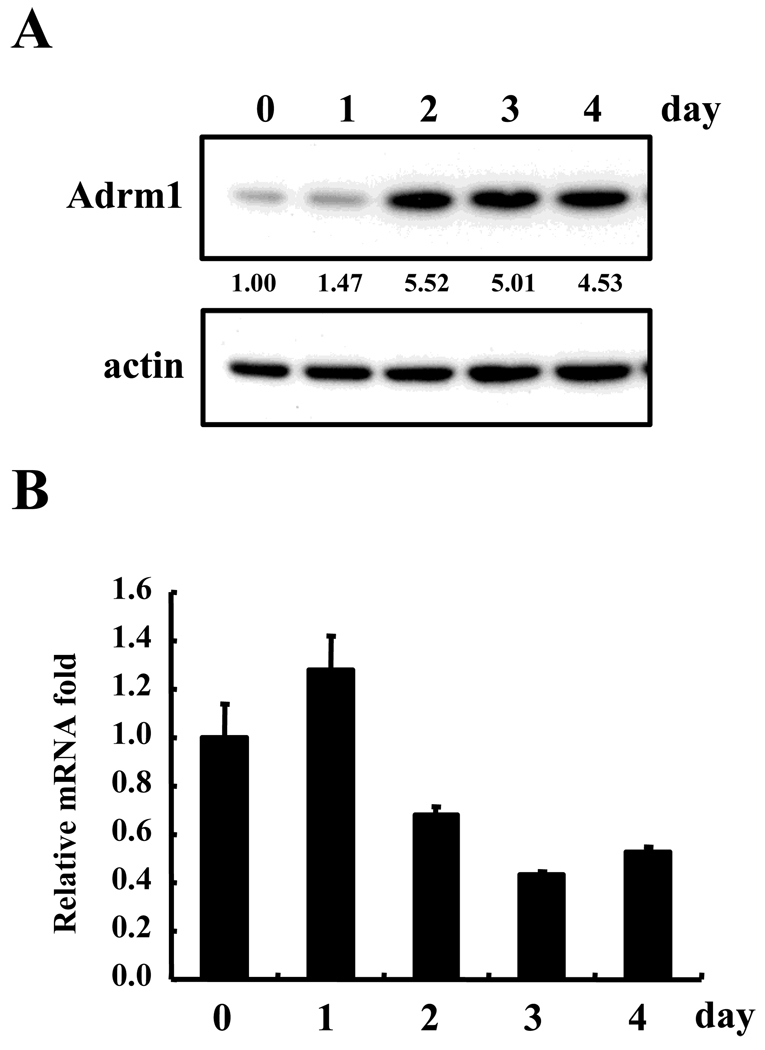
To examine the roles of Adrm1 in osteoclast differentiation, we measured the expression levels of Adrm1 in osteoclast-lineage cells. BMMs from wild-type mice were cultured with RANKL, and Adrm1 expression levels were analyzed using Western blots and real-time PCRs. As seen in Figure 2, the protein levels of Adrm1 increased as the osteoclasts matured, whereas the mRNA levels did not change significantly. This result suggested that the Adrm1 protein became more stable during osteoclast differentiation in the presence of RANKL.
Fig. 2.
Adrm1 expression during osteoclast differentiation. (A) BMMs from wild-type mice were cultured with M-CSF and RANKL for the indicated period of time. Whole-cell extracts were subjected to Western blot analysis to detect Adrm1. Day 0 indicates the day RANKL was added to the BMM cultures. (B) RNA was isolated on the indicated days after incubation with M-CSF and RANKL, and subjected to real-time RT-PCRs to analyze the expression of Adrm1. Data are expressed as the mean ± SD and are representative of at least three independent experiments.
Effect of Adrm1 on osteoclast differentiation
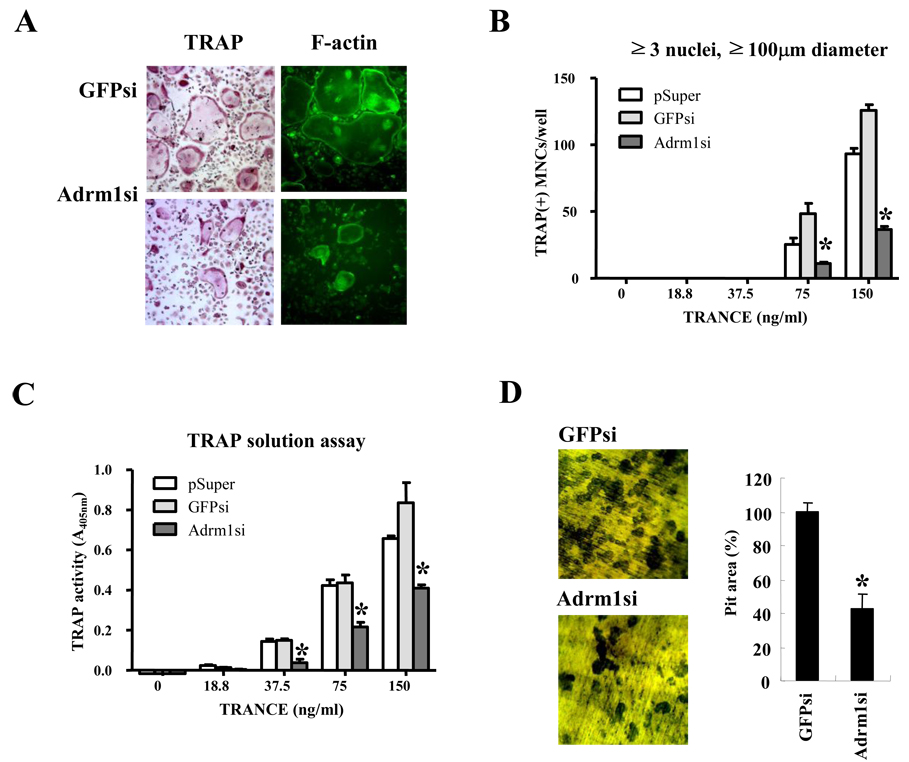
To access whether Adrm1 regulates osteoclast differentiation and bone resorption, we knocked down Adrm1 expression levels using retrovirus harboring Adrm1-specific siRNA (Adrm1 siRNA). In a preliminary knockdown experiment, we cloned siRNA specific for Adrm1 into pSuper-retro-Puro vectors, and carried out Western blot analyses and real-time PCRs to determine the effects of the siRNA. We found that the expression of Adrm1 in osteoclasts significantly decreased in response to the Adrm1 siRNA (Supplemental Fig. 1). We cultured BMMs infected with retrovirus harboring Adrm1 siRNA in the presence of M-CSF and various concentrations of RANKL. The samples were then stained for TRAP and the actin ring, and the osteoclasts that formed after 4 days were counted (Fig. 3A). Interestingly, approximately 3-fold fewer osteoclasts formed among Adrm1 siRNA-infected BMMs compared with BMMs infected with control vector or control siRNA (Fig. 3B). Furthermore, TRAP activity in osteoclasts derived from Adrm1 siRNA-infected BMMs was also lower than that observed in control samples (Fig. 3C). In addition, cells in which Adrm1 expression was knocked down showed decreased pit formation on dentine slices owing to defects in the maturation of the osteoclasts (Fig. 3D). These data suggest that Adrm1 is involved in osteoclastogenesis.
Fig. 3.
Adrm1 expression during osteoclast differentiation. BMMs were infected with retrovirus alone (pSuper), control siRNA (GFPsi), or siRNA specific for Adrm1 (Adrm1si). The infected cells were cultured with 60 ng/ml M-CSF and with the indicated concentrations of RANKL. (A) After 4 days of culture, cells were fixed and stained for TRAP and F-actin. (B) TRAP+ MNCs with more than three nuclei and a diameter larger than 100 µm were counted as osteoclasts. (C) TRAP solution assays were performed as described in the Materials and Methods. (D) Pit formation. BMMs were cultured on dentine slices with M-CSF and RANKL for 4 days. Resorption pits were stained with 0.5% toluidine blue and the pit area was measured using image analysis software. Data are expressed as the mean ± SD and are representative of at least three independent experiments. *, P < 0.05 versus vector controls.
Roles of Adrm1 in osteoclast differentiation
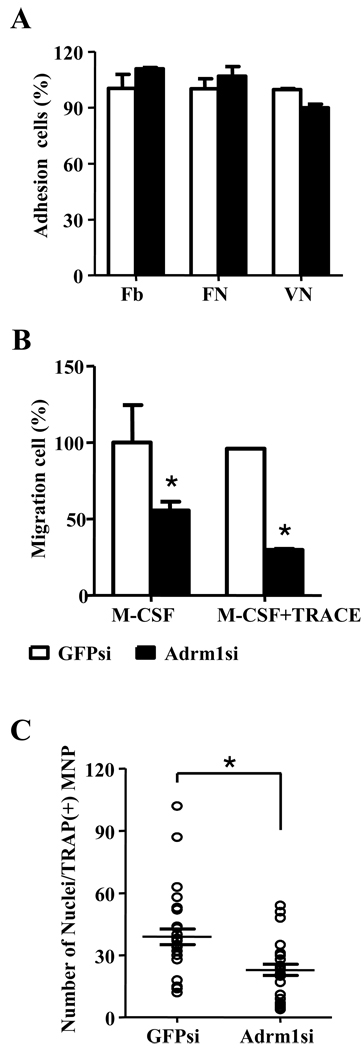
Osteoclasts adhere to the bone matrix, leading to cytoskeletal reorganization, a process that enables these cells to migrate to and between the resorption sites, and that polarizes the cells during resorption. The adhesion and migration of osteoclasts allows them to interact with cells and bone matrix, which leads to the differentiation of multinuclear giant cells and degradation of bone [1, 3]. Previous studies have suggested that Adrm1 regulates cell adhesion in endothelial cells and mediates cell migration in human colon carcinoma RKO cells [12, 22]. We therefore investigated whether Adrm1 contributes to cell adhesion during osteoclast differentiation. Control and Adrm1 knockdown preosteoclasts were attached to wells that were coated with fibrinogen, fibronectin, or vitronectin. As shown in Figure 4A, Adrm1 knockdown did not affect the adhesion of preosteoclasts. We next examined the effects of Adrm1 on cell migration using a transwell culture system. The migration of cells in which Adrm1 expression was knocked down was significantly decreased compared with control samples (Fig. 4B).
Fig. 4.
Effects of Adrm1 on the adhesion, migration, and fusion of osteoclast precursors. (A) BMMs were infected with control siRNA (GFPsi) or siRNA specific for Adrm1 (Adrm1si). The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for 3 days. Osteoclasts precursors were washed with PBS, suspended in low-serum -MEM containing 1% FBS and 2% BSA, and loaded into the upper well of transwell chambers. After 6 to 8 hours, cells that had migrated into the lower well were fixed and stained with hematoxylin. (B) Osteoclasts precursors were attached to fibrinogen (Fb)-, fibronectin (FN)-, or vitronectin (VN)-coated culture plates for 10 minutes. Nonadherent cells were washed away with PBS, and the adherent cells were stained with hematoxylin and counted under a light microscope. Data are expressed as the mean ± SD and are representative of at least three independent experiments. *, P < 0.05 versus vector controls. (C) BMMs were infected with control siRNA (GFPsi) or siRNA specific for Adrm1 (Adrm1si). The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for 4 days. The efficiency of osteoclast fusion was calculated by dividing the total number of nuclei within TRAP+ MNCs that were stained with 4’,6-diamidino-2-phenylindole by the number of TRAP+ MNCs. Data are expressed as the mean ± SD and are representative of at least three independent experiments. *, P < 0.05 versus vector controls.
In a previous study of Atp6v0d2-deficient mice, we reported that Atp6v0d2 regulates osteoclast maturation and bone formation [9]. To investigate Adrm1 as a regulator of Atp6v0d2, we examined the effects of Adrm1 on preosteoclasts cell fusion. After counting the average number of nuclei in each TRAP+ MNC, we observed a significant reduction in the relative fusion efficiency of Adrm1 knockdown preosteoclasts cells compared with control cells (Fig. 4C). Furthermore, we performed real-time PCRs to investigate the effects of Adrm1 knockdown on the expression of genes involved in cell–cell fusion and osteoclast differentiation (supplemental Fig. 2). Adrm1 knockdown and control BMMs were cultured for 3 days in the presence of M-CSF and RANKL, and the expression levels of genes encoding putative fusion and adhesion molecules (Atp6v0d2, DC-STAMP, MFR, CD9, CD47, integrin αν, integrin 3, c-Src, Pyk2, and FAK), and those coding for transcription factors involved in osteoclast differentiation (NFATc1, Mitf, and c-Fos) were examined. Consistent with previous data, RANKL upregulated the expression levels of genes encoding Atp6v0d2, DC-STAMP, integrin β3, c-Src, and NFATc1. Induction of these genes by RANKL was, however, not affected by Adrm1 knockdown. RANKL is a critical osteoclastogenic factor that activates NF-κB, JNK, p38 MAP kinase, and ERK. To examine whether Adrm1 affects immediate signaling pathways, BMMs in which Adrm1 was knocked down were stimulated with RANKL for the indicated periods of time (supplemental Fig. 3). Consistent with previous results, RANKL activated JNK, ERK, p38 MAP kinase, and NF-κB in control BMMs. Adrm1 knockdown did not affect the RANKL-induced activation of these signaling pathways, however.
Discussion
Atp6v0d2, an isoform of V-ATPase subunit d, is highly expressed during osteoclast differentiation. Recently, we reported that targeted disruption of the gene encoding this Atp6v0d2 subunit resulted in a marked increase in bone mass, without affecting V-ATPase-mediated acidification. In addition, osteoclast precursor cells derived from Atp6v0d2-deficient mice showed decreased fusion capacity [6, 9]. To investigate further the biological functions of Atp6v0d2, we search for Atp6v0d2 binding partners using a yeast-two hybrid screen. We identified Adrm1 (previously called GP110 or ARM-1) as an Atp6v0d2-interacting protein and confirmed this interaction in Western blotting and coimmunoprecipitation experiments.
Adrm1 was previously described as a cell adhesion–promoting receptor with a possible role in certain types of cancer. This protein was initially reported to be heavily glycosylated, which increased the mass of the 42-kDa peptide to 110 kDa [13, 23]. Several studies, however, failed to find the 110-kDa form [11, 12, 15]. We also did not detect a 110-kDa form of Adrm1 in osteoclasts or 293T cell lines in which the protein was exogenously overexpressed. Adrm1 is expressed in several tissues, including brain, lung, heart, spleen, kidney, and liver [11, 24]. Adrm1 protein accumulated in murine osteoclasts in response to RANKL stimulation. In contrast, Adrm1 mRNA levels did not change during osteoclast differentiation.
In the present study, we performed in vitro osteoclast differentiation experiments using murine BMMs cultured in the presence of RANKL and M-CSF. To investigate the functional roles of Adrm1, we performed siRNA-mediated Adrm1 knockdown experiments. We observed that siRNA-mediated knockdown of Adrm1 expression significantly inhibited osteoclast differentiation in vitro. Further analysis revealed that knocking down Adrm1 expression decreased the total number of osteoclasts and the average number of nuclei in each osteoclast. These results suggest that Adrm1 regulates the fusion process, although further studies are needed to prove this hypothesis definitively. In addition, several studies have suggested that Adrm1 participates in cell adhesion and cell migration [12, 15, 22]. In this study, we also observed a defect in cell migration by osteoclast precursors in which Adrm1 expression was knocked down. We also observed that knocking down Adrm1 expression during osteoclast differentiation did not markedly affect the expression profiles of a number of genes thought to be involved in cell adhesion, preosteoclast fusion, and/or osteoclast differentiation (Supplemental Fig. 2). RANKL-induced activation of NF-κB and MAP kinase signaling also was not reduced in response to Adrm1 knockdown (Supplemental Fig. 3). These results suggest that Adrm1 may not be involved in the early stages of osteoclast differentiation. In conclusion, our data suggest that Adrm1, a new Atp6v0d2-interacting protein, plays an important role in osteoclast differentiation, and in particular the fusion of preosteoclasts.
Supplementary Material
Supplemental Fig. 1. BMMs were infected with control siRNA (GFPsi) or siRNA specific for Adrm1 (Adrm1si). (A) The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for the indicated period of time. Whole-cell extracts were subjected to Western blot analysis to detect Adrm1. (B) RNA was isolated on Day 4 and subjected to real-time RT-PCRs to analyze the expression of Adrm1. Data are expressed as the mean ± SD and are representative of at least three independent experiments. *, P < 0.05 versus vector controls.
Supplemental Fig. 2. Real-time quantitative PCR analysis of genes encoding cell adhesion and fusion-related proteins, and genes encoding transcription factors that are involved in osteoclastogenesis. BMMs were infected with control siRNA (GFPsi) or siRNA for Adrm1 (Adrm1si). The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for the indicated period of time. RNA was collected at the indicated times after stimulation and was subjected to real-time PCRs to analyze the expression of genes encoding putative fusion and adhesion molecules. Data are expressed as the mean ± SD and are representative of at least three independent experiments.
Supplemental Fig. 3. BMMs were stimulated with 500 ng/ml RANKL for the indicated period of time. Whole-cell extracts were subjected to Western blot analysis with the indicated antibodies. p-JNK, phospho-JNK; p-ERK, phospho-ERK; p-p38, phospho-p38; p-IkB, phospho-IκB.
Acknowledgments
This work was supported by National Research Foundation of Korea Grant funded by the Korean Government (2009-0076101, to S.H.L) and by NIH grant AR55903 (to Y. C.).
Abbreviations
- Adrm1
adhesion-regulating molecule 1 protein
- Atp6v0d2
d2 subunit of the vacuolar (H+) ATPase V0 domain
- RANKL
Receptor activator of nuclear factor-κB ligand
- MCSF
Macrophage-colony stimulating factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 2.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 3.Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J, Choi Y. Osteoimmunology: interplay between the immune system and bone metabolism. Annu Rev Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]
- 4.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 5.Cipriano DJ, Wang Y, Bond S, Hinton A, Jefferies KC, Qi J, Forgac M. Structure and regulation of the vacuolar ATPases. Biochim Biophys Acta. 2008;1777:599–604. doi: 10.1016/j.bbabio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishi T, Forgac M. The vacuolar (H+)-ATPases--nature' s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 7.Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Xu G, Li YP. Atp6v0d2 is an essential component of the osteoclast-specific proton pump that mediates extracellular acidification in bone resorption. J Bone Miner Res. 2009;24:871–885. doi: 10.1359/JBMR.081239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 10.Fejzo MS, Dering J, Ginther C, Anderson L, Ramos L, Walsh C, Karlan B, Slamon DJ. Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosomes Cancer. 2008;47:873–883. doi: 10.1002/gcc.20592. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen JP, Lauridsen AM, Kristensen P, Dissing K, Johnsen AH, Hendil KB, Hartmann-Petersen R. Adrm1, a putative cell adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol. 2006;360:1043–1052. doi: 10.1016/j.jmb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Lamerant N, Kieda C. Adhesion properties of adhesion-regulating molecule 1 protein on endothelial cells. FEBS J. 2005;272:1833–1844. doi: 10.1111/j.1742-4658.2005.04613.x. [DOI] [PubMed] [Google Scholar]
- 13.Shimada S, Ogawa M, Takahashi M, Schlom J, Greiner JW. Molecular cloning and characterization of the complementary DNA of an M(r) 110,000 antigen expressed by human gastric carcinoma cells and upregulated by gamma-interferon. Cancer Res. 1994;54:3831–3836. [PubMed] [Google Scholar]
- 14.Cherix N, Froquet R, Charette SJ, Blanc C, Letourneur F, Cosson P. A Phg2-Adrm1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol Biol Cell. 2006;17:4982–4987. doi: 10.1091/mbc.E06-07-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simins AB, Weighardt H, Weidner KM, Weidle UH, Holzmann B. Functional cloning of ARM-1, an adhesion-regulating molecule upregulated in metastatic tumor cells. Clin Exp Metastasis. 1999;17:641–648. doi: 10.1023/a:1006790912877. [DOI] [PubMed] [Google Scholar]
- 16.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 20.Lee SH, Kim T, Jeong D, Kim N, Choi Y. The tec family tyrosine kinase Btk Regulates RANKL-induced osteoclast maturation. J Biol Chem. 2008;283:11526–11534. doi: 10.1074/jbc.M708935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak HB, Lee SW, Jin HM, Ha H, Lee SH, Takeshita S, Tanaka S, Kim HM, Kim HH, Lee ZH. Monokine induced by interferon-gamma is induced by receptor activator of nuclear factor kappa B ligand and is involved in osteoclast adhesion and migration. Blood. 2005;105:2963–2969. doi: 10.1182/blood-2004-07-2534. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Hu XT, Shi QL, Zhang FB, He C. Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncol Rep. 2009;21:531–537. [PubMed] [Google Scholar]
- 23.Ratovitski EA, Bao C, Quick RA, McMillan A, Kozlovsky C, Lowenstein CJ. An inducible nitric-oxide synthase (NOS)-associated protein inhibits NOS dimerization and activity. J Biol Chem. 1999;274:30250–30257. doi: 10.1074/jbc.274.42.30250. [DOI] [PubMed] [Google Scholar]
- 24.Nakane T, Inada Y, Itoh F, Chiba S. Rat homologue of the human M(r) 110000 antigen is the protein that expresses widely in various tissues. Biochim Biophys Acta. 2000;1493:378–382. doi: 10.1016/s0167-4781(00)00204-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. BMMs were infected with control siRNA (GFPsi) or siRNA specific for Adrm1 (Adrm1si). (A) The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for the indicated period of time. Whole-cell extracts were subjected to Western blot analysis to detect Adrm1. (B) RNA was isolated on Day 4 and subjected to real-time RT-PCRs to analyze the expression of Adrm1. Data are expressed as the mean ± SD and are representative of at least three independent experiments. *, P < 0.05 versus vector controls.
Supplemental Fig. 2. Real-time quantitative PCR analysis of genes encoding cell adhesion and fusion-related proteins, and genes encoding transcription factors that are involved in osteoclastogenesis. BMMs were infected with control siRNA (GFPsi) or siRNA for Adrm1 (Adrm1si). The infected cells were cultured with 60 ng/ml M-CSF and 150 ng/ml RANKL for the indicated period of time. RNA was collected at the indicated times after stimulation and was subjected to real-time PCRs to analyze the expression of genes encoding putative fusion and adhesion molecules. Data are expressed as the mean ± SD and are representative of at least three independent experiments.
Supplemental Fig. 3. BMMs were stimulated with 500 ng/ml RANKL for the indicated period of time. Whole-cell extracts were subjected to Western blot analysis with the indicated antibodies. p-JNK, phospho-JNK; p-ERK, phospho-ERK; p-p38, phospho-p38; p-IkB, phospho-IκB.