Summary of Recent Advances
Glycomics is the comprehensive study of all glycans expressed in biological systems. The biosynthesis of glycan relies on a number of highly competitive processes involving glycosyl transferase. Glycosylation is therefore highly sensitive to the biochemical environment and has been implicated in many diseases including cancer. Recently, interest in profiling the glycome has increased due to the potential of glycans for disease markers. In this regard, mass spectrometry is emerging as a powerful technique for profiling the glycome. Global glycan profiling of human serum based on mass spectrometry has already led to several potentially promising markers for several types of cancer and diseases.
Introduction
The glycome, which is the glycan analog to the proteome and the genome, is loosely defined as the glycan components of a biological source. With the large diversity of glycans, vis á vis proteomics, it is not technically possible to obtain the complete representation of the glycome. Classifying the glycome relative to the genome, the proteome, and the metabolome is difficult because it is often bound to proteins directly correlated to the genome. However, glycans are produced by proteins making them more like metabolic products. In this regard, glycans are unique in that they link the three major areas of genomic, proteomic, and metabolomics since saccharides are found in all three groups. As disease markers glycans have major advantages: (1) Changes in glycosylation in disease states are supported by 50 years of glycobiology, particularly in the study of cancer. (2) N- and O-linked glycans are specifically small molecules and are therefore easy to quantitate like metabolites. Glycomics, therefore, holds considerable promise specifically as disease markers (1–3).
Proteins are often modified by the attachment of glycans during the normal synthesis of protein production. It is estimated that over 70% of all human proteins are glycosylated (4). Glycosylation is often found on cell surfaces and in extracellular matrices making it the first point of contact in cellular interactions (5, 6). Therefore, glycan biosynthesis is more significantly affected by disease states than by protein production. It is now well established that altered glycosylation varies significantly for cancer cells compared to normal cells (7, 8).
The large diversity of glycans makes a general discussion of all types of glycans and their glycoconjugates difficult. In human serum, the abundant glycoconjugates are glycoproteins. Therefore, only N- and O-linked glycans attached to glycoproteins are discussed in this review. Other glycoconjugates such as glycolipids, peptidoglycans, and glycosaminoglycans are not covered because they require different analytical technique. Glycomics represents a new paradigm for cancer biomarker discovery. This short review focuses on mass spectrometry methods for glycan-based disease markers.
Glycomics and Mass Spectrometry
In the glycomics approach, glycans are harvested and used to determine whether the glycosylation has changed in disease-state samples compared to healthy controls (9–13) without prior knowledge of the associated proteome. Advances in glycomics analysis rely on the analytical tools for glycomic profiling. Mass spectrometry provides a rapid and sensitive tool for component analysis. Mass spectrometry has therefore emerged as a central tool for glycomics analysis. It is also a precise tool for structural elucidation resulting in significant progress towards understanding the role of the glycome in many biological systems (14, 15). As the methods for profiling oligosaccharide composition and structures are becoming refined, the search for biomarkers is intensifying (16, 17).
Mass spectrometers capable of glycan analysis typically employ either matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI) with or without chromatographic separation. High mass accuracy instruments such as modern time-of-flight (TOF), ion cyclotron resonance (ICR), and Orbitrap mass analyzers are useful for glycan detection because low or even sub ppm mass errors are often required for exact mass annotation. Lower performance instruments can make glycan analysis challenging by causing false assignments due to the high uncertainties associated with their measurements (18). Tandem MS can be used to remedy false assignments by providing structural information and glycan compositions. Structural elucidation can be obtained using glycan fragments, exact mass, and exoglycosidase digestion.
Glycomics Methods
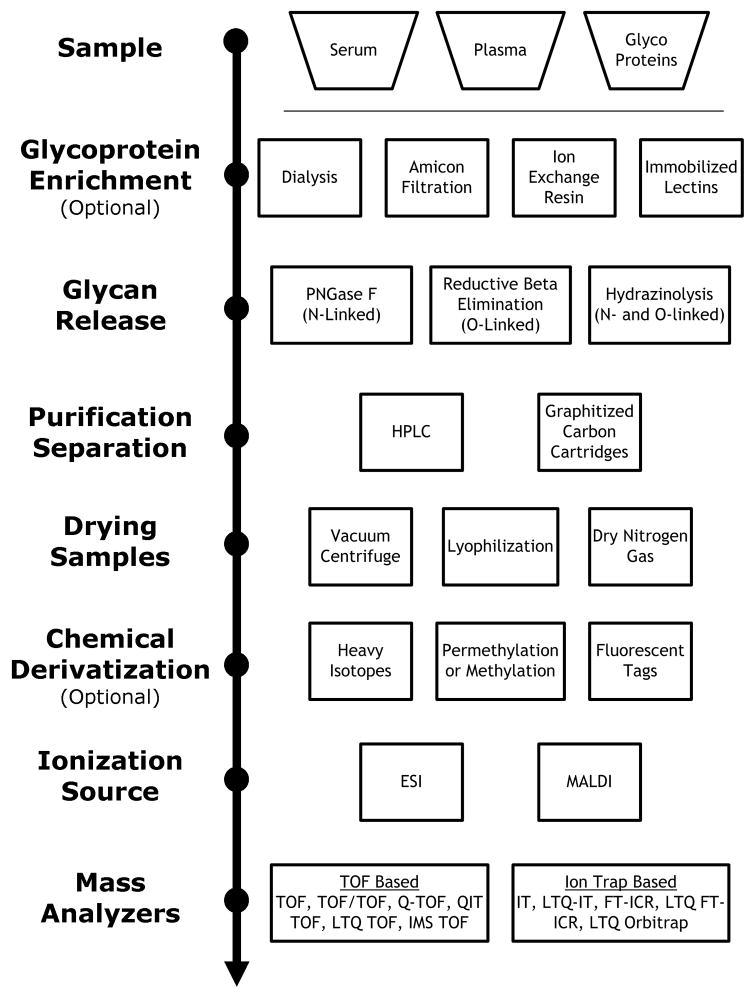
Although there are many groups studying glycans with mass spectrometry, there are some common features to their chemical processes. The workflows for glycan analysis are illustrated in Figure 1. The scheme is broken into seven tasks (listed in left side) that are almost universally implemented in some form in all of the methods.
Figure 1.
The common features of glycan processing are illustrated here. The common categories are listed on the left and the different methods implemented are on the right. Depending on the nature of the samples used, some of the steps were not always needed. The order of procedures had some variability as well.
Aside from proper study design and sample selection, the chemical work up of the samples and mass spectrometric analyses also have a large impact on the discovery and quality of glycan disease markers. Proper sample preparation improves sensitivity and reproducibility of the analysis. Glycans are generally isolated from glycoprotein containing samples by first cleaving off the glycans and then purifying them with solid phase extraction or high performance liquid chromatography (HPLC). Samples are often concentrated with vacuum centrifuges or lyophilizers prior to mass spectrometry analysis. The glycans may be derivatized (e.g., methylation) to improve ionization and stability of ions in the mass spectrometer.
Many of the biological sources of glycoproteins contain levels of salts that are too high for mass spectrometric analysis because the excess salt decreases the ionization of the desired analyte. Desalting is often accomplished by filtration (19, 20), dialysis (21, 22), cation exchange resin (23, 24), or carbon-based column adsorption (11, 13, 16, 18).
Methods for releasing the glycans from glycoproteins depend on the attachment of the glycans. N-linked glycans (N-glycans) attach via nitrogen on an asparagines, while O-linked glycans (O-glycans) attach via oxygen on a serine or threonine. Reductive beta elimination (10, 13) is most common for releasing O-glycans, while Peptide N-Glycosidase F (PNGase F) enzymatic cleavage (11, 18) is the most common for releasing N-glycans. One problem with the beta elimination reaction is the “peeling” reaction where the strong base cleaves off a monosaccharide from the glycan producing degraded structures. One alternative to the beta elimination reaction is the ammonia-based beta elimination reaction (22), which uses milder conditions. Alternatively, hydrazinolysis (21) can be used to release N-linked and O-linked glycans simultaneously but is rarely used due to the relative difficulty and hazards of the process. Hydrazine monohydrate substitutions have been used to decrease explosion danger of anhydrous hydrazine but were not as effective in the release of the glycans (25).
Purification of the glycan mixture brings its own challenges. Isolating the glycans from peptides or proteins present in the solution is necessary because these compounds ionize more readily than glycans and their presence in the mixture will severely suppress glycan signals. Purification methods take advantage of the differences in polarity between peptides and glycans. Large amounts of proteins can be precipitated from the solution using methanol (22, 24), ethanol (11, 13, 16, 18) or acetone (24). Lectin affinity enrichment has also been used to concentrate the glycoproteins prior to glycan release (20). C18 stationary phases (20, 26–30) are used to retain residual amounts of proteins. Amine (30), amide (30–32), and graphitized carbon (11, 13, 16, 18, 24, 26–29, 33, 34) media can be used to retain glycans for further elution into solution. Solvent removal is critical for sample concentration since the quantity of glycans is often too low for sensitive detection. Since increasing the temperature to remove the solution will degrade or destroy the glycans, vacuum centrifugation, lyophilization (21, 23, 29, 34) or bubbling dry nitrogen gas (22) through the sample, is implemented instead.
Mass spectrometric analysis of oligosaccharides can be performed in the native or derivatized state. The question of derivatization versus native prior to MS analysis is a matter of preference. Derivatization can increase sensitivity and decrease fragmentation. The latter is particularly important with fucosylated and sialylated glycans. Even with the “soft” ionization sources of electrospray and less “soft” MALDI, there is still impetus to modify the sialic acid for stability. However, derivatization appears to miss modifications on oligosaccharides such as sulfation. Furthermore, derivatization prevents the prefractionation by solid phase extraction (SPE). As sialylated glycans make up the most abundant species in human serum, they can suppress other less abundant components such as the high mannose glycans. Elution of the glycans from SPE with varying ratios of acetonitrile and water increases the number of glycans observed by mass spectrometry and allows better observation of less abundant yet important components such as the high mannose glycans. Nonetheless, a wide variety of chemical procedures can be employed to improve glycan ionization and ion stability in the mass spectrometers. Methylation (35), permethylation (23, 24, 26–28), and pyridylamination (25, 26) are examples of methods used to stabilize the sialic acid by forming esters. These methods have the additional benefit of positive ion detection of sialylated glycans concurrently with the neutral glycans.
Other chemical modifications can be useful for quantitating oligosaccharides. The reducing end of the glycans is the target for incorporating stable isotopes (23) that enable relative quantitation or fluorescent tags (22, 30–32) that are readily monitored during liquid chromatography.
Glycan Profiling
Glycan Mass Profiling using Mass Spectrometry
Glycan mass profiling (GMP) is a method for analyzing the complete mixture by obtaining masses with little or no separation. This method yields the compositional profile with regard to the number of sialic acids, fucose, hexose, and N-acetylhexosamine. GMP provides rapid analysis based on mass and, therefore, composition; however, it cannot distinguish between isomeric species (compounds with identical mass and composition). Glycan mass profiling is usually performed with MALDI or ESI as the ionization source with only a crude method of glycan purification.
In our laboratory, glycan mass profiling is done using a matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance (MALDI-FTICR) mass spectrometer equipped with 7.0 T magnet and infrared multiphoton dissociation (IRMPD) for tandem mass spectrometry. The FTICR mass analyzer, with its high resolution of 105–106 at full width half maximum and high mass accuracy of less than 10 parts per million (ppm) on internal calibration, allows us to profile the glycans using exact masses. IRMPD and collision-induced dissociation (CID) allows us to confirm the monosaccharide compositions based on the mass losses. The reproducibility of MALDI has been established in several studies, including one from our laboratory(11).
The extent of glycosylation and the types of O- and N-glycans have been shown to change in cancer (5, 7). The selection of one group for biomarker discovery over the other is, however, not a trivial choice. The release procedures and the analyses are distinct. Previous studies from this laboratory were focused on profiling O-glycans because the research shows more profound changes in these compounds. Indeed, a limited study on ovarian (13, 36) and breast (12) cancer show differentiation between disease and control, however it was found that the O-glycans were contaminated with a large number of more abundant N-glycans even when the release procedure was reported to produce predominantly O-glycans. The composition and the structures were confirmed by tandem MS including CID and IRMPD. Given that the changes were possibly N-glycans, we repeated these mass profiling studies specifically with procedures to release only N-glycans (11). N-linked glycans are readily identified by trimannosyl core (Man3GlcNAc2) and complex type N-glycans containing sialic acid were observed as major glycans in human serum (16, 18).
For N-glycans, putative structures can be obtained from the composition. We recently developed a theoretical library for the human serum N-glycome, which is highly effective for automatically annotating mass spectrum with a low false positive rate (18).
Glycan Chromatographic Profiling (GCP)
Until now, the focus on glycan marker discovery has involved glycan mass profiling. Individual oligosaccharides (specific isomeric structures) may provide more robust glycan markers with higher specificity than composition alone. Changes in specific linkages have been attributed to some diseases (37, 38). The number of isomers greatly surpasses the number of compositions, providing a significantly larger set number of potential glycan markers (16, 28). To gain access to individual glycans, separation of the isomers is required. Analytical methods for separating components to isomers, such as nanoflow liquid chromatography (16, 28, 39) and capillary electrophoresis (40, 41), coupled with mass spectrometry have proved highly useful. A method to assess the diversity of the N-glycans in human serum without derivatization has been developed by our laboratory employing porous graphitized carbon as stationary phase (16). Glycans of individual serum samples were profiled and compared. Complex type N-linked glycans were the most abundant glycans in human serum accounting for ~96% of the total glycans indentified, while hybrid and high mannose type glycans comprise the remaining ~4% [16]. The ability to separate and simultaneously analyze neutral and anionic N-glycans from human serum without derivatization demonstrates a rapid yet highly sensitive and highly specific tool for disease biomarker discovery.
Potential Glycan Markers based on Disease Type
There are now several recent publications that focus on N- and O-glycans as disease markers (11–13, 27, 28, 34, 42–53). Alterations in the degree of branching and levels of sialylation and fucosylation in N-glycans have been reported as a consequence of diseases (1, 54). Similarly, changes in O-glycans in mucins have also been associated with diseases (50, 54). In cancer, changes in the branching of N-glycans, truncation of O-glycans, changes in the amount, linkage, acetylation and expression of sialic acids have all been suggested to signify the diseases(54).
Table 1 shows a summary of some of the potential N- and O-glycan markers for cancer gathered from literature. The table is by no means complete, but it provides a limited overview and illustrates the potential of glycans as markers for cancers. The biological sources were mostly sera and cell lines, but many other tissue samples have been examined. In prostate cancer, for example, there are differences in mannosylation and fucosylation, and specifically complex N-glycans were found to vary with the disease (11, 44, 51). In breast cancer, a fucosylated N-glycan was found as a potential marker but there was also an increase in intensity in the high-mannose N-glycans and fucosylation in O-glycans (27, 31, 50, 53). Fucosylation and sialylation levels show conflicting results in studies (27, 53). In liver cancer, fucosylation, whether in the core or outer arm, tends to be elevated (28, 48). The same increase in levels of fucosylation can be seen in ovarian and pancreatic cancer (46, 47, 49). Although there are some conflicting results in some of the markers found in cancer, e.g. some studies show an increase and others a decrease in mannosylation, there are clearly distinct differences in the levels of N- and O-glycans.
Table 1.
Changes in glycosylation and potential N- and O-glycan biomarkers for several types of cancer. The arrows indicate either an increase (↑) or a decrease (↓) in intensity levels of the biomarker or glycosylation change.
| Cancer | Biological Sample | Glycosylation Change/Potential Glycan Biomarker | ↑ or ↓ |
|---|---|---|---|
| General | ▪ Branched N-glycans [54], specifically β1–6 GlcNAc branching | ↑ | |
| ▪ Truncated O-glycans [54] | ↑ | ||
| ▪ Sialic acids – amount, linkage, acetylation and expression[54] | ↑ or ↓ | ||
| Prostate | Sera[11,45,52] | ▪ Complex N-glycans[11] | ↑ or ↓ |
| ▪ High-mannose glycans/mannosylation[44] | ↑ and ↓ | ||
| ▪ Total fucosylation[44] | ↑ | ||
| ▪ Fucose and α(2,3)-linked sialic acid content of PSA[50] | ↓ | ||
| Breast | Cell Lines[54] Sera[32,51,28] |
▪ An α(1,3)-fucosylated monogalactosylated triantennary glycan[31] | ↑ |
| ▪ High mannose N-glycans[53] | ↑ or ↓ | ||
| ▪ N-glycan branching | ↑ | ||
| ▪ Hybrid structures | ↓ | ||
| ▪ Fucosylation in O-glycans | ↑ | ||
| ▪ N-glycan sialylation and fucosylation[27, 53] | ↓ and ↑ | ||
| ▪ Core 1 type O-glycans[50] | ↑ or ↓ | ||
| ▪ O-glycan sialylation[50] | ↑ | ||
| Liver | Cell Lines[49] Sera[43,46,53] |
▪ Core fucosylated structures[48] | ↑ |
| ▪ Outer-arm fucosylation in highly branched glycans[52] | ↑ | ||
| ▪ A branched α(1,3)-fucosylated triantennary N-glycan[45] | ↑ | ||
| Ovarian | Sera[50] | ▪ Core fucosylated, agalactosyl biantennary glycans and sialyl Lewis x[49] | ↑ |
| Pancreatic | Cell Lines[47] Sera[48] |
▪ Fucosylation[46,47] | ↑ |
| Gastric | Cell Lines[44] | ▪ Free complex-type N-glycans[43] | ↑ |
Conclusions
Glycan markers hold considerable opportunities and challenges for disease diagnosis because the glycosylation machinery is highly sensitive to the biochemical environment. Mass spectrometry-based glycomics has the potential to provide a single platform to monitor several diseases simultaneously. The main glycomics method currently involves mass mapping strategies where composition profiles are obtained. However, structure and protein-specific glycomics has yet to be explored. These strategies will require further analytical development. The serum N-glycome will need to be annotated, and glycoproteomic approaches need to be developed that describe both protein and glycan simultaneously. Furthermore, as the strategies become more refined and the sample sets become larger the glycomics approach for biomarker discovery will not only impact diagnosis of disease, but will also provide a new paradigm for understanding the role of the glycome in many biological areas.
Acknowledgments
We are grateful for funds provided by the National Institute of Health (RO1GM049077), the Ovarian Cancer Research Fund, and the Ann Garat Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1•.Lebrilla CB, An HJ. The prospects of glycan biomarkers for the diagnosis of diseases. Mol Biosyst. 2009;5:17–20. doi: 10.1039/b811781k. The opinion article on glycan biomarkers. The author argues the intrinsic advantages of using glycans as disease markers over other biomolecules and the potential of glycan profiling for diagnosing and determining the progression of disease. [DOI] [PubMed] [Google Scholar]
- 2.Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: bioinformatics and biomarkers in disease. An NIH white paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11–13, 2006) Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- 3.Turnbull JE, Field RA. Emerging glycomics technologies. Nat Chem Biol. 2007;3:74–77. doi: 10.1038/nchembio0207-74. [DOI] [PubMed] [Google Scholar]
- 4.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6•.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. A thorough review of the regulation of cellular mechanism by glycans. [DOI] [PubMed] [Google Scholar]
- 7.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 8•.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. A comprehensive review of pathogenic mechanisms in tumor progression that glycan facilitate It also covers glycan-targeted therapy for cancer treatment. [DOI] [PubMed] [Google Scholar]
- 9.An HJ, Ninonuevo M, Aguilan J, Liu H, Lebrilla CB, Alvarenga LS, Mannis MJ. Glycomics analyses of tear fluid for the diagnostic detection of ocular rosacea. J Proteome Res. 2005;4:1981–1987. doi: 10.1021/pr0501620. [DOI] [PubMed] [Google Scholar]
- 10.Cooke CL, An HJ, Kim J, Canfield DR, Torres J, Lebrilla CB, Solnick JV. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 11.de Leoz MLA, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, Miyamoto S, White RD, Lam KS, Lebrilla C. Glycomic approach for potential biomarkers on prostate cancer: Profiling of N-linked glycans in human sera and pRNS cell lines. Disease Markers. 2008;25:243–258. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S, Lebrilla CB, Miyamoto S. A Serum Glycomics Approach to Breast Cancer Biomarkers. Molecular and Cellular Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 13•.An HJ, Miyamoto S, Lancester KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of Glycans in Serum for the Discovery of Potential Biomarkers for Ovarian Cancer. Journal of Proteome Research. 2006;5:1626–1635. doi: 10.1021/pr060010k. The first glycan mass profiling based on mass spectrometry for biomarker study. Without protein separation and identification glycans were globally released from human serum and profiled for biomarker discovery. [DOI] [PubMed] [Google Scholar]
- 14.Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2003–2004. Mass Spectrom Rev. 2009;28:273–361. doi: 10.1002/mas.20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park Y, Lebrilla CB. Application of Fourier transform ion cyclotron resonance mass spectrometry to oligosaccharides. Mass Spectrom Rev. 2005;24:232–264. doi: 10.1002/mas.20010. [DOI] [PubMed] [Google Scholar]
- 16.Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster KS, An HJ, Li B, Lebrilla CB. Interrogation of N-Linked oligosaccharides using infrared multiphoton dissociation in FT-ICR mass spectrometry. Anal Chem. 2006;78:4990–4997. doi: 10.1021/ac0600656. [DOI] [PubMed] [Google Scholar]
- 18•.Kronewitter SR, An HJ, de Leoz MLA, Lebrilla CB, Miyamoto S, Leiserowitz GS. The developmet of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. A systematic approach for high throughput glycomics with the use of N-linked glycan library models. The theoretical models are used to rapidly predict glycan profiles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woosley BD, Kim YH, Kolli VSK, Wells L, King D, Poe R, Orlando R, Bergmann C. Glycan analysis of recombinant Aspergillus niger endo-polygalacturonase A. Carbohydrate Research. 2006;341:2370–2378. doi: 10.1016/j.carres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Qiu YH, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, Jin G, Ruffin MT, Turgeon DK, Synal S, Bresalier R, Marcon N, Brenner DE, Lubman DM. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. Journal of Proteome Research. 2008;7:1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Merry AH, Neville DCA, Royle L, Matthews B, Harvey DJ, Dwek RA, Rudd PM. Recovery of intact 2-aminobenzamide-labeled O-glycans released from glycoproteins by hydrazinolysis. Analytical Biochemistry. 2002;304:91–99. doi: 10.1006/abio.2002.5620. The first report of the released O-glycans with anhydrous hydrazine and produced intact glycans free from the “peeling” reaction. [DOI] [PubMed] [Google Scholar]
- 22•.Huang YP, Mechref Y, Novotny MV. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Analytical Chemistry. 2001;73:6063–6069. doi: 10.1021/ac015534c. This paper was the first to perform O-linked oligosaccharide release with an ammonia based beta elimination method. This method is softer than the sodium hydroxide based beta elimination reactions. [DOI] [PubMed] [Google Scholar]
- 23•.Botelho JC, Atwood JA, Cheng L, Alvarez-Manilla G, York WS, Orlando R. Quantification by isobaric labeling (QUIBL) for the comparative glycomic study of O-linked glycans. International Journal of Mass Spectrometry. 2008;278:137–142. This paper provides the method for the quantitation of glycans via heavy isotope labeling similar to methods used for the quantitation of proteins. [Google Scholar]
- 24.Costello CE, Contado-Miller JM, Cipollo JF. A glycomics platform for the analysis of permethylated oligosaccharide alditols. Journal of the American Society for Mass Spectrometry. 2007;18:1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakakita SI, Sumiyoshi W, Miyanishi N, Hirabayashi J. A practical approach to N-glycan production by hydrazinolysis using hydrazine monohydrate. Biochemical and Biophysical Research Communications. 2007;362:639–645. doi: 10.1016/j.bbrc.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 26.Bleckmann C, Geyer H, Reinhold V, Lieberoth A, Schachner M, Kleene R, Geyer R. Glycomic Analysis of N-Linked Carbohydrate Epitopes from CD24 of Mouse Brain. Journal of Proteome Research. 2009;8:567–582. doi: 10.1021/pr800729r. [DOI] [PubMed] [Google Scholar]
- 27.Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clinical Chemistry. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 28•.Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. Journal of Proteome Research. 2008;7:1109–1117. doi: 10.1021/pr700702r. Used a capillary reactor approach for permethylating N-glycans. This was also applied to disease marker discovery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YG, Jang KS, Joo HS, Kim HK, Lee CS, Kim BG. Simultaneous profiling of N-glycans and proteins from human serum using a parallel-column system directly coupled to mass spectrometry. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2007;850:109–119. doi: 10.1016/j.jchromb.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Kamoda S, Nakano M, Ishikawa R, Suzuki S, Kakehi K. Rapid and sensitive screening of N-glycans as 9-fluorenylmethyl derivatives by high-performance liquid chromatography: A method which can recover free oligosaccharides after analysis. Journal of Proteome Research. 2005;4:146–152. doi: 10.1021/pr049825o. [DOI] [PubMed] [Google Scholar]
- 31.Hamid UMA, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Pardo M, Antrobus R, Chapman CJ, Zitzmann N, Robertson JF, Dwek RA, Rudd PM. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18:1105–1118. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- 32.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM, Dwek RA. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Analytical Biochemistry. 2008;376:1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JH, Xie YM, Hedrick JL, Lebrilla CB. Profiling the morphological distribution of O-linked oligosaccharides. Analytical Biochemistry. 2004;334:20–35. doi: 10.1016/j.ab.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Bereman MS, Williams TI, Muddiman DC. Development of a nanoLC LTQ Orbitrap Mass Spectrometric Method for Profiling Glycans Derived from Plasma from Healthy, Benign Tumor Control, and Epithelial Ovarian Cancer Patients. Analytical Chemistry. 2009;81:1130–1136. doi: 10.1021/ac802262w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An HJ, Lebrilla CB. Suppression of sialylated by sulfated oligosaccharides in negative MALDI-FTMS. Israel Journal of Chemistry. 2001;41:117–127. [Google Scholar]
- 36.Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, Duong H, Kirmiz C, Li B, Liu H, Lam KS. Glycomics analysis of serum: a potential new biomarker for ovarian cancer? Int J Gynecol Cancer. 2008;18:470–475. doi: 10.1111/j.1525-1438.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. Journal of Proteome Research. 2006;5:1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 38.Dereggi M, Capon C, Gharib B, Wieruszeski JM, Michel R, Fournet B. The Glycan Moiety of Human Pancreatic Lithostathine - Structure Characterization and Possible Pathophysiological Implications. European Journal of Biochemistry. 1995;230:503–510. doi: 10.1111/j.1432-1033.1995.tb20589.x. [DOI] [PubMed] [Google Scholar]
- 39.Ninonuevo MR, Park Y, Yin HF, Zhang JH, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, Lebrilla CB. A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 40.Nakano M, Higo D, Arai E, Nakagawa T, Kakehi K, Taniguchi N, Kondo A. Capillary electrophoresis-electrospray ionization mass spectrometry for rapid and sensitive N-glycan analysis of glycoproteins as 9-fluorenylmethyl derivatives. Glycobiology. 2009;19:135–143. doi: 10.1093/glycob/cwn115. [DOI] [PubMed] [Google Scholar]
- 41.An HJ, Franz AH, Lebrilla CB. Improved capillary electrophoretic separation and mass spectrometric detection of oligosaccharides. J Chromatogr A. 2003;1004:121–129. doi: 10.1016/s0021-9673(03)00718-0. [DOI] [PubMed] [Google Scholar]
- 42.Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z, Mechref Y, Novotny MV. Detection of Hepatocellular Carcinoma Using Glycomic Analysis. Clinical Cancer Research. 2009;15:1808–1813. doi: 10.1158/1078-0432.CCR-07-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishizuka A, Hashimto Y, Naka R, Kinoshita M, Kakehi K, Seino J, Funakoshi Y, Suzuki T, Kameyama A, Narimatsu H. Accumulation of free complex-type N-glycans in MKN7 and MKN45 stomach cancer cells. Biochem J. 2008;413:227–237. doi: 10.1042/BJ20071562. [DOI] [PubMed] [Google Scholar]
- 44.Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, Hickey RJ, Vinson J, Sweeney CJ, Novotny MV. Alterations in the serum glycome due to metastatic prostate cancer. J Proteome Res. 2007;6:1822–1832. doi: 10.1021/pr060664t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, Wang L, Zhuang H, Callewaert N, Libert C, Contreras R, Chen C. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology. 2007;46:1426–1435. doi: 10.1002/hep.21855. [DOI] [PubMed] [Google Scholar]
- 46.Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: Detailed analyses of oligosaccharide structures. Proteomics. 2008;8:3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 47.Nakano M, Nakagawa T, Ito T, Kitada T, Hijioka T, Kasahara A, Tajiri M, Wada Y, Taniguchi N, Miyoshi E. Site-specific analysis of N-glycans on haptoglobin in sera of patients with pancreatic cancer: a novel approach for the development of tumor markers. Int J Cancer. 2008;122:2301–2309. doi: 10.1002/ijc.23364. [DOI] [PubMed] [Google Scholar]
- 48.Norton PA, Comunale MA, Krakover J, Rodemich L, Pirog N, D’Amelio A, Philip R, Mehta AS, Block TM. N-linked glycosylation of the liver cancer biomarker GP73. J Cell Biochem. 2008;104:136–149. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saldova R, Royle L, Radcliffe CM, Hamid UMA, Evans R, Arnold JN, Banks RE, Hutson R, Harvey DJ, Antrobus R, Petrescu SM, Dwek RA, Rudd PM. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 50.Storr SJ, Royle L, Chapman CJ, Hamid UMA, Robertson JF, Murray A, Dwek RA, Rudd PM. The O-linked glycosylation of secretory/shed MUC1 from an advanced breast cancer patient’s serum. Glycobiology. 2008;18:456–462. doi: 10.1093/glycob/cwn022. [DOI] [PubMed] [Google Scholar]
- 51.Tabares G, Radcliffe CM, Barrabes S, Ramirez M, Aleixandre RN, Hoesel W, Dwek RA, Rudd PM, Peracaula R, Llorens Rd. Different Glycan Structures in Prostate-specific Antigen from Prostate Cancer Sera in Relation to Seminal Plasma PSA. Glycobiology. 2006;16:132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 52.Tanabe K, Deguchi A, Higashi M, Usuki H, Suzuki Y, Uchimura Y, Kuriyama S, Ikenaka K. Outer arm fucosylation of N-glycans increases in sera of hepatocellular carcinoma patients. Biochem Biophys Res Commun. 2008;374:219–225. doi: 10.1016/j.bbrc.2008.06.124. [DOI] [PubMed] [Google Scholar]
- 53.Goetz JA, Mechref Y, Kang P, Jeng MH, Novotny MV. Glycomic profiling of invasive and non-invasive breast cancer cells. Glycoconj J. 2009;26:117–131. doi: 10.1007/s10719-008-9170-4. [DOI] [PubMed] [Google Scholar]
- 54•.Varki ACR, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Cold Spring Harbor Laboratory Press 2009. 2. 2009. Essentials of Glycobiology. Comprehensive book of principles and applications of glycobiology. It discusses the different classes of glycans and the glycosylation changes observed in diseases. [PubMed] [Google Scholar]