Abstract
The human Ab repertoire exhibits restrictions during fetal life characterized by biases of variable gene segment usage and lack of junctional diversity. We tested the hypotheses that Ab repertoire restriction persists in the early postnatal period and contributes to the observed poor quality of specific Ab responses made by neonates to viruses and vaccines. We analyzed the molecular determinants of B cell responses in humans to respiratory syncytial virus (RSV). Analysis of the variable gene segment usage of adult RSV-specific B cells revealed a repertoire profile in these cells similar to that seen in randomly-selected B cells, which was VH3-dominant, Four gene segments (VH3–23, VH3–30, VH3–33 and VH4–04) accounted for almost half of the VH genes used. In contrast, very young infant RSV-specific antibodies exhibited a biased repertoire characterized by comparable use of the VH1, VH3, and VH4 families, and less common use of the four immunodominant gene segments. Infants and children older than three months used an antibody repertoire similar to that of adults. Mutational analysis revealed that the antibody variable genes of infants under three months of age also possessed significantly fewer somatic mutations in both framework and CDR regions than those of adults, even in a child with recurrent RSV infection. These data suggest that neonates use a biased antibody gene repertoire that is less VH3-focused and that possesses a dramatically lower frequency of somatic mutations. These biased features of the RSV-specific repertoire likely contribute to the poor functional Ab response in very young infants.
Keywords: Human, B cells, repertoire development, antibodies, viral
Introduction
Young infants are predisposed to a variety of serious infections based on lack of prior antigenic exposure and reduced functional quality of their primary immune responses. Numerous studies have documented the poor humoral immune response of infants to respiratory syncytial virus (RSV) and other pathogens that commonly cause infection early in life (Brandenburg et al., 1997; Murphy et al., 1986a; Murphy et al., 1986b; Roca et al., 2003). Passively-acquired maternal antibodies suppress the primary antibody response in infants. Careful analysis of infant immune responses reveals, however, that the quantitatively and qualitatively poor nature of infant antibody responses also occurs independent of the presence of maternally-derived antibodies. Furthermore, the magnitude of the immune response in early life is age-dependent, with older infants able to mount higher titers of high-affinity neutralizing antibodies than the youngest infants (Murphy et al., 1986a; Murphy et al., 1986b). Although the first few months of life are brief in terms of immunological development, the clinical burden of disease during this period is enormous. The leading causes of death in the first year of life are infectious diseases, and the majority of morbidity and hospitalization from RSV, for example, occurs in the first few months of life (Glezen et al., 1981; Hall, 2001; Parrott et al., 1973). Thus, the apparently delayed maturation of the infant antibody response is highly significant for human disease.
Another issue affected by the immaturity of the neonatal antibody response is the difficulty of inducing effective immune responses in this population by vaccination. Currently licensed vaccines require multiple-dose schedules to achieve protective efficacy in infancy. For some of these diseases, such as pertussis, the most severe disease and highest mortality occur before three months of life, when infants have not completed a primary vaccine series and are incompletely protected (Halasa et al., 2003; Healy and Baker, 2006). The development of live, attenuated vaccines for RSV has been hindered by the poor responses of neonates to vaccine candidates that are sufficiently immunogenic in older infants, for whom the need for an effective vaccine is less important (Karron et al., 2005; Polack and Karron, 2004; Wright et al., 2000). Thus, the immature neonatal antibody response has implications both for infectious diseases and the preventive strategies for those diseases.
Multiple mechanisms contribute to the production of diversity needed for high-affinity, neutralizing antibodies: preferential utilization of particular germline gene segments, recombination of those segments, and somatic hypermutation in antigen-stimulated B cells. Some studies have suggested that the random peripheral B cell repertoire of young infants was biased in gene segment usage or complementarity-determining region 3 (CDR3) length compared to adults (Mortari et al., 1992; Mortari et al., 1993; Raaphorst et al., 1992; Schroeder et al., 1987), while others have suggested that infants are capable of mounting adult-like Ab responses (Bona et al., 2004; Kolar et al., 2004). There have been few studies investigating antigen-specific Ab repertoires in infants due to technical limitations. Our group previously developed a novel method for the generation of human antigen-specific mAbs that incorporated several novel features, including physical selection of Ag-specific B cells from human subjects by flow cytometry and single B cell expansion in vitro (Weitkamp et al., 2003b). We previously used these techniques to show that rotavirus-specific B cell responses in infants and adults shared a striking VH1 and VH4 segment usage bias, with three VH segments dominating the rotavirus-specific repertoire (Weitkamp et al., 2003a). We also showed that somatic mutations were much less frequent in B cells isolated from infants infected with rotavirus (Weitkamp et al., 2005). However, clinical disease due to rotavirus occurs later during the first year of life than that due to RSV. Severe RSV disease typically occurs within the first few months of life, allowing the opportunity to determine the molecular basis of virus-specific Ab responses in more immunologically immature infants.
In the current study we sought to investigate the molecular basis for RSV-specific antibody responses in young RSV-infected infants compared with RSV-infected older children and adults. RSV fusion (F) protein is the dominant protective antigen and the target of neutralizing Abs. RSV F-specific B cells were isolated from acutely infected infants, children or adults, or from previously-infected healthy adults. Single F-specific B cells were expanded in culture and heavy and light chain genes from the clones were sequenced. We found that infants less than 3 months expressed a less focused variable gene repertoire, and they less commonly used dominant VH segments. The expressed antibody genes of infants exhibited remarkably fewer somatic mutations in the antibody genes of RSV-specific B cells than did those of older individuals. These data suggest that the ability of human neonates to generate highly functional antigen-specific Ab responses is significantly limited at a molecular level.
Materials and Methods
Generation of RSV F-specific human B cell clones
Whole blood was obtained by venipuncture from healthy adult blood donors, RSV-infected adults or RSV-infected infants or children. Infants with RSV infection (documented by rapid antigen assay on nasopharyngeal secretions) from both outpatient and inpatient populations were recruited. Blood from healthy adult blood donors was obtained from fresh leukofilters as previously described (Weitkamp and Crowe, 2001). The Vanderbilt University Institutional Review Board approved the study. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation over Histopaque (Sigma) and washed twice with phosphate-buffered saline (PBS). PBMCs then were enriched for B cells by positive selection using CD19-Dynabeads and DetachaBEAD (Dynal, Sweden) according to the manufacturer’s instructions. Purified B cells were washed with PBS and resuspended in PBS with 1% fetal bovine serum (PBS/FBS) for immunostaining. An aliquot was stained with trypan blue to determine viability and counted in a hemacytometer.
We developed a novel method to sort RSV F-specific B cells. B cells first were incubated with 1 µg of immunoaffinity-purified RSV F protein (PFP, kindly provided by Wyeth-Lederle Vaccines and Pediatrics) at 4 °C for one hour. B cells then were washed twice with PBS/FBS and incubated with a mixture of three mouse monoclonal antibodies (MAbs) specific for different epitopes on RSV F protein. MAb 1214 is an IgG1 isotype specific to antigenic site A, mAb 1243 is an IgG2b specific for antigenic site C and mAb 1331H is an IgG2a specific for antigenic site C that only partially competes with mAb 1243. B cells were incubated with the mAbs at 4 °C for one hour, washed twice with PBS/FBS, and re-suspended in PBS/FBS. The B cell suspension then was incubated with goat anti-mouse IgG1-FITC (Southern Biotech, Birmingham, AL), goat anti-mouse IgG2a-PE (Southern Biotech, Birmingham, AL), goat anti-mouse IgG2b-PE (Southern Biotech) and mouse anti-human CD19-CyChrome (Becton Dickinson) at 4 °C for one hour. The cells then were washed twice with PBS/FBS and re-suspended in PBS/FBS at a concentration of 5 × 105 cells/mL for FACS. Samples were analyzed on a FACStar Plus equipped with a 96-well deposition device (Becton Dickinson). Acquisition was gated first on viable lymphocytes, based on forward and side scatter parameters. Samples then were gated on CD19-positive B cells. Cells contained within this gate that showed high fluorescent signal with both FITC and PE were considered doubly positive for RSV F protein, and thus RSV-F specific. Preliminary studies (not shown) suggested that single-color detection of RSV F-specific B cell was less specific than dual-color detection. B cells specific for RSV F protein were sorted singly into 96-well sterile culture plates (Costar, Corning, NY) containing 100 µl of B cell culture media.
Expansion of single RSV F-specific B cells into clones
For the expansion of single B cells, we used a B cell culture system that we previously described to amplify and identify antigen-specific or randomly-selected B cell clones (Weitkamp et al., 2003b). Briefly, 50,000 irradiated EL-4-B5 mouse thymoma cells (kindly provided by Dr. R. H. Zubler, Geneva University Hospital, Geneva, Switzerland) per well of 96-well culture plates were used as feeder cells immediately following single cell isolation. A combination of 100 U of recombinant human IL-2, 5 ng/ml PMA, and 10% (v/v) of supernatant from pokeweed mitogen-activated human T cells (T cell replacing factor) was added. The culture plates were incubated for 7 days, then we removed 100 µl of supernatant, and added 10,000 irradiated fibroblastic L cells stably transfected with human CD154 to each well (Garrone et al., 1995). This cell line was kindly provided by DNAX via the American Type Culture Collection (CRL 12095; Manassas, VA). We also added 5 ng/ml recombinant human IL-4 in addition to the B cell culture media described above. The cultures were kept for another 2 wk with a second addition of CD154 fibroblasts, cytokines, PMA, and T cell replacing factor on day 14.
Secreted human Ig was detected by capture ELISA on day 14 as described (Weitkamp et al., 2003b). To maximize the identification of human Ig-producing wells secreting RSV-specific Abs, we used anti-Ig reagents that detected Abs of any isotype. Therefore, the isotype of the Abs secreted by cell lines derived from single B cells was not determined. Wells that tested positive for human Ig production at day 14 were rescreened for RSV F-specific antibodies on day 21, using an enhanced ELISA method. Plates were pre-coated with immunoaffinity-purified RSV F protein (15 ng/well) overnight at 4 °C in carbonate buffer, pH 9.0. Plates were blocked as described (Weitkamp et al., 2003b). Supernatant from B cell cultures (100 µL) was added to wells and incubated at room temperature for two hours. Plates were washed as before and goat anti-human Ig-alkaline phosphatase (Southern Biotech) at 1 µg/mL in blocking buffer was added to the wells and incubated at room temperature for one hour. The plates then were washed with buffer provided in the AmpliQ kit (DAKO, Sweden) and reagents A and B added at 50 µL each. Plates were read after 15 minutes at 490 nm, and also at 650 nm to determine background for subtraction. Supernatant from wells with all components except B cells was used as negative control and human serum with a high anti-RSV titer was included as positive control. Samples were considered positive at twice the mean absorbance of negative control wells.
RT-PCR amplification of antibody variable genes
Wells containing B cells that produced antibodies to RSV F were harvested and the cells pelleted. For RT-PCR amplification of heavy and light chain variable gene regions we used a commercial oligo-dT based isolation method (mRNA Capture Kit; Roche, Basel, Switzerland), a single-tube RT-PCR method (Titan One Tube system) and pooled primer mixes. The heavy and light chain primer sequences used were designed to amplify all heavy and light chain genes in the VBASE collection of genomic variable gene sequences (Ruiz et al., 2000; Sblattero and Bradbury, 1998). We further amplified the first-round PCR products using heavy and light chain nested primers hybridizing to framework regions (FR) 1 and FR4 (forward and reverse, respectively) under the same conditions as above. The primers used and the amplification conditions were described previously (Weitkamp et al., 2003b). Combinations of different primer mixtures and reaction conditions were extensively optimized; heavy chain variable gene amplification was generally more efficient (not shown).
Variable gene sequence analysis
We ligated gel-extracted PCR products into a TA cloning vector (Promega, Madison, WI), generated bacterial clones, and purified plasmid DNA from overnight bacterial cultures. Plasmid DNA was digested with restriction enzymes to identify clones with proper ligation of variable gene regions. The nucleotide sequences of both strands of plasmids that contained a heavy or light chain insert were determined in the Vanderbilt DNA Sequencing Core on an ABI3700 instrument. Sequences were aligned using ClustalW alignment in the MacVector package (Accelrys) and primer sequences were removed. We analyzed heavy or light chain variable gene region sequences using DNAPLOT to search the international ImMunoGeneTics database (http://imgt.cines.fr/) using the junction analysis program, reporting results with an updated nomenclature of the human Ig genes, as previously summarized (Giudicelli et al., 2006; Ruiz et al., 2000). All VH and VL lengths, and VH, D and JH segment assignments were reviewed and confirmed by manual inspection. Mutations in the junctional region were confirmed manually, and mutations in the remaining regions were scored manually and tabulated. D gene segment assignments were performed by database alignment, and confirmed by manual inspection, using the genomic sequences of all D gene segments (Corbett et al., 1997). The assignment of D segments was defined using similarity with D segment germline sequences on the basis of 100% amino acid identity over a stretch of at least three consecutive amino acids. For a definite assignment of a D or J segment, we required at least three consecutive amino acids to be identical to germline sequences. Unassignable D segments were considered to be mutated, though these were not formally counted in the analysis of numbers of mutations (Corbett et al., 1997; Weitkamp et al., 2003a). The number of altered amino acid residues in the N or C terminus of D segments within the HCDR3 was determined only in HCDR3 sequences in which a definite D segment assignment was possible. All VH, D and JH segment assignments and mutation scores were entered into a database constructed by the authors, using Access software (Microsoft). The sequences were deposited into GenBank; accession numbers and details of each antibody sequence are provided in tabular form in supplementary data online.
Statistical analysis of genetic data
Comparisons between the use of variable gene segments of particular gene families were made by using a permutation test statistic (an extension of the Fisher’s exact test for 2 × C contingency tables) and were performed using the StatExact software package (Cytel Software Corporation). Pair-wise comparisons for gene segment differences were made between adult randomly-selected and adult RSV-specific cells, or cells from infants < 3 or ≥3 months of age, as well as for adult RSV-specific and adult random cells compared to the infant RSV-specific cells (both infant age groups combined). The counts of total framework and CDR mutations should follow a Poisson distribution. However, there were higher numbers of zero counts (i.e., complete lack of change) for CDR mutations for both heavy and light chain sequences than can be explained using a simple Poisson distribution. Therefore, we used a zero-inflated Poisson (ZIP) model (Lambert, 1992) to provide point estimates and test for differences in CDR mutations between donor groups. The ZIP model allowed us to test for both differences in the number of zero counts and differences in the mean number of mutations that was adjusted for the number of excess zeros. This analysis was implemented using the R version 2.01 package for Windows (http://www.R-project.org). The distribution of total framework mutations did not exhibit the zero-inflation problem. Instead, these data were modeled using Poisson regression, adjusting for over-dispersion.
Results
Number of subjects and B cells analyzed
We isolated B cells from 12 hospitalized RSV-infected infants younger than 3 months of age (mean age 8 weeks, range 3–12 weeks), 9 older RSV-infected children from 7 to 60 months of age (mean age 23 months), and 12 RSV-infected adults. One of the RSV-infected infants had been born prematurely at 29 weeks’ gestation and had a documented previous infection with RSV at 3 weeks of age. This infant was recruited for this study during a second documented RSV infection at 11 weeks of age. Two other infants had been born prematurely at 28 and 33 weeks. Cells isolated from nine leukofilters from healthy adult blood donors (RSV-immune by virtue of age and exposure) also were sorted for RSV-specific B cells. The number of individual B cells sorted and yield of Ig-producing and F-specific Ig-producing clones is shown in Table I. B cells from young infants proliferated in culture and produced Ig at rates equal to B cells from adults, suggesting similar ability to respond to CD40L-driven stimulation in the setting of the appropriate cytokine milieu. The overall percentage of sorted B cells that produced human Ig after 14 days of culture was 21% (7,775/36,572 wells sorted) and did not differ between infants and adults. Of those wells secreting Ig, 451/7,775 (6%) were secreting RSV F-specific antibodies after 21 days in culture.
Table I.
Total number of B cells sorted by donor group and yield of single B cell-derived cultures secreting human immunoglobulin (Ig) or RSV F-specific antibodies.
| Donor group | Number of subjects |
Number of wells sorted |
Number secreting human Ig (%) |
Number secreting RSV F- specific antibodies (%) |
|---|---|---|---|---|
| Adult blood donors | 9 | 11,927 | 2,732 (21) | 193 (7) |
| RSV-infected adults | 12 | 3,691 | 401 (26) | 21 (5) |
| RSV-infected children (≥3 months) |
9 | 6,290 | 1,347 (21) | 91 (7) |
| RSV-infected infants (< 3 months) |
12 | 14,664 | 3,295 (22) | 146 (5) |
| Total | 42 | 36,572 | 7,775 (21) | 451 (6) |
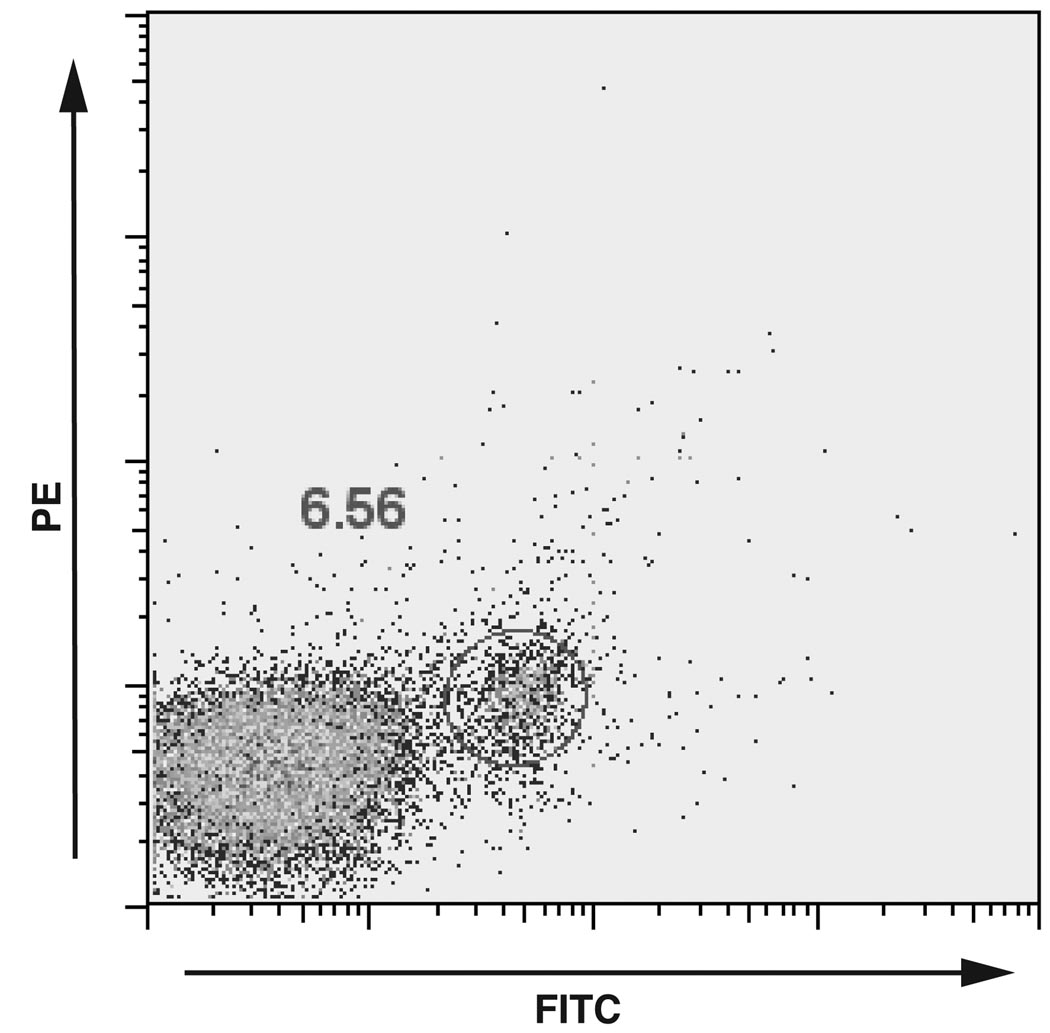
The percentage of RSV-specific B cells ranged from <0.05% (<1 in 2000) in randomly-selected adult blood donors to 6.6% (1 in 14) in RSV-infected infants (Figure 1). The recovery of RSV-specific B cells in culture overall represented a yield of 1% of total circulating B cells.
Figure 1.
FACS scatter plot of RSV F-specific B cells isolated from a 2-month-old RSV-infected infant. Cells were gated on small lymphocytes using forward and side scatter and B cells using CD-19 CyChrome. Shown are RSV-F-specific PE and FITC labeling.
The Ab gene repertoire of RSV-specific B cells in adults but not the youngest infants was VH3-dominated
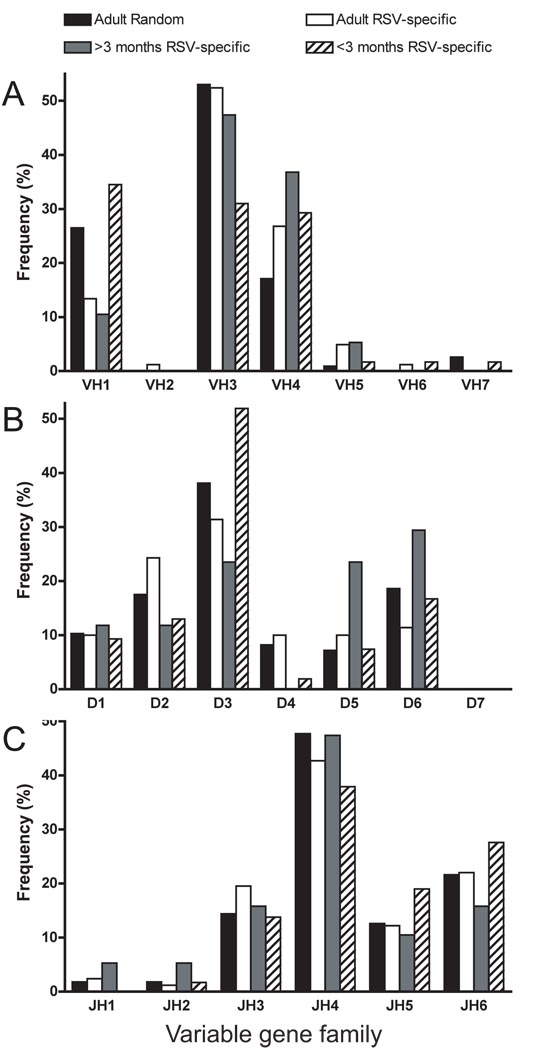
We analyzed 117 randomly-selected B cells from adult donors, 82 RSV-specific B cells from adults (healthy or RSV-infected), 19 RSV-specific B cells from RSV-infected children ≥ 3 months and 58 RSV-specific B cells from RSV-infected infants <3 months old (Figure 2A). All VH families were represented, showing that the primer sets used were capable of amplifying a broad repertoire (Weitkamp et al., 2003a; Weitkamp et al., 2003b). The randomly-selected adult B cell antibody gene repertoire was VH3-dominant, with the VH3 family accounting for 53% of the repertoire, as has been described previously (Brezinschek et al., 1995; Corbett et al., 1997; Weitkamp et al., 2003b). The RSV-specific antibody gene repertoire in adults used the VH4 family more prominently (p=0.04) than did the randomly-selected repertoire, although the VH3 family still accounted for 52% of all B cells analyzed. The RSV-specific repertoire in children ≥3 months did not differ statistically from the randomly-selected repertoire, and VH3 accounted for 47.4% of the clones. In contrast, the repertoire in infants <3 months was not VH3-dominant, but rather used the VH1, VH3 and VH4 families almost equally, accounting for 35%, 31% and 27% of the clones analyzed, respectively. RSV-specific B cells from infants and adults utilized segments from the VH5, VH6 and VH7 families, but much less frequently than VH1, VH3 and VH4. Thus, the distinguishing feature of the VH gene segment repertoire analysis was a repertoire bias in the youngest infants, who used a much broader, less focused, repertoire to respond to RSV F protein than did older children and adults.
Figure 2.
VH, D and JH gene family utilization in circulating human random or RSV-specific B cells. Single circulating random or RSV-specific B cells from healthy blood donors, RSV-infected adults, RSV-infected children >3 months old and RSV-infected infants <3 months old were sorted, amplified in culture, and tested for RSV F-specific antibody production by ELISA, followed by sequence analysis of the expressed VH gene segment. Frequencies of VH, D and JH gene family utilization in all four subsets are presented in (A), (B) and (C), respectively.
D and JH segment usage
The D2, D3 and D6 families predominated in the repertoire of all the groups analyzed (Figure 2B). The D3 family accounted for 52% of all D segments used in RSV-specific B cells from infants <3 months old, but this did not reach statistical significance compared to the frequency of D3 usage by other groups. The D5 and D6 families were used more frequently in RSV F-specific B cells from children ≥3 months old, but this did not differ significantly from other groups. In 17 (15%) of the randomly-selected adult B cells, D segment assignment could not be made, suggesting a high frequency of somatic mutations. The percent of D segments that could not be recognized was 14% in RSV-specific adult B cells, 10% in RSV-specific B cells from children ≥3 months old and 7% in RSV-specific B cells from infants <3 months old. Thus, the number of unassignable D segments was reduced in infants. JH4 was the most frequently used segment in all subgroups, and the distribution of JH segment usage between groups did not differ (Figure 2C). RSV-specific B cells from infants <3 months old more frequently utilized JH5 and JH6, but this finding was not statistically significant.
VH segment usage
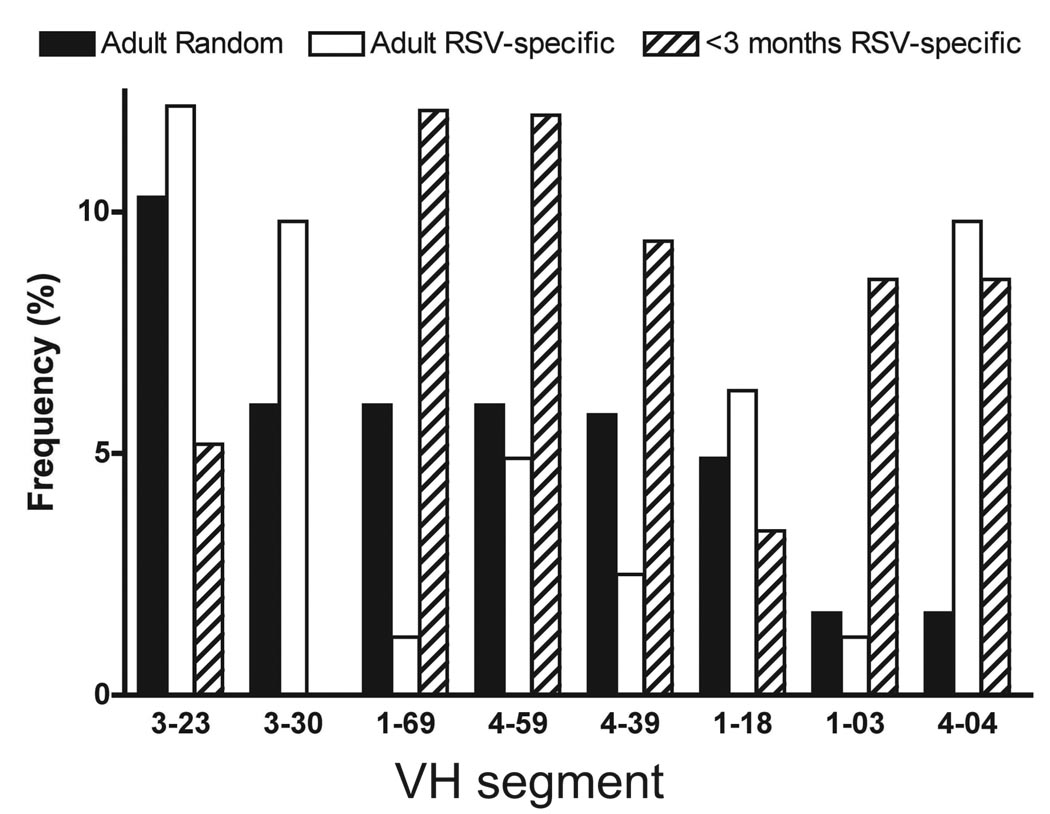
We compared the most common VH gene segments used in the randomly selected B cell repertoire with those of the RSV-specific adult and young infant repertoires (Figure 3). The most common VH segment in randomly-selected or RSV-specific adult B cells was VH3–23, while this segment accounted for only 5% of the repertoire in RSV-specific B cells from infants <3 months old. The next most frequently used segment in random and RSV-specific adult B cells was VH3–30; this segment was not used in any of the 58 RSV-specific B cells from infants <3 months old. VH1–69, VH4–59 and VH4–39 segments were used with similar frequency to VH3–30 in adult random B cells, each accounting for ~5% of total VH repertoire. In contrast, VH1–69 and VH4–59 segments were the most frequent segments in RSV-specific B cells from infants <3 months old, accounting for 24% of the total repertoire in this population. VH1–03 also was used commonly in RSV-specific B cells from infants <3 months old, but rarely observed in random or RSV-specific adult B cells. VH4–04 was commonly used in RSV-specific cells from both adults and infants, accounting for 10% and 9% of VH segments, respectively, but this segment was found in less than 2% of the randomly selected B cells. Thus, the repertoire of particular VH gene segments used in the youngest infants exhibited a clear bias.
Figure 3.
VH segment utilization in circulating human random or RSV-specific B cells. Single circulating random or RSV-specific B cells from healthy blood donors, RSV-infected adults and RSV-infected infants <3 months old were sorted, amplified n culture, and tested for RSV F-specific antibody production by ELISA, followed by sequence analysis of the expressed VH gene segment. Frequencies of VH segment utilization in all three subsets are presented.
VL and JL family usage
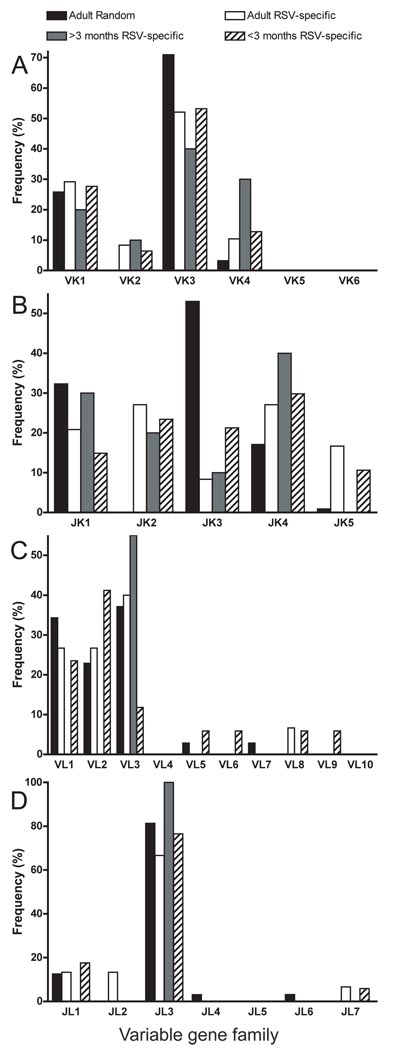
Fewer light chain genes than heavy chains were recovered by RT-PCR for most groups despite extensive optimization of the light chain RT-PCR assay. Thus, the number of sequences analyzed for the light chain repertoire was smaller: 66 randomly-selected B cells from adult donors, 63 RSV-specific B cells from adults (healthy or RSV-infected), 13 RSV-specific B cells from RSV-infected children ≥3 months and 64 RSV-specific B cells from RSV-infected infants <3 months old. The proportions of kappa and lambda light chain gene usage in randomly-selected adult B cells were 47% and 53%. However, kappa chains were expressed more frequently by RSV-specific B cells in all age groups: 76% of adult, 77% of RSV-infected children ≥3 months and 73% of RSV-infected infants <3 months old. VK3 was the most commonly expressed light chain gene segment in all groups (Figure 4A). However, JK usage was distributed more broadly in RSV-specific B cells, compared to the randomly-selected repertoire that was dominated by JK3 (Figure 4B). VL1, VL2 and VL3 were the dominant VL gene segments expressed in all groups. Interestingly, RSV-specific B cells from children ≥3 months all used the VL3 gene segment, while RSV-specific B cells from infants < 3 months utilized a broader repertoire of VL gene segments (Figure 4C). A majority of B cells in all groups expressed the JL3 segment (Figure 4D). Thus, although the repertoire for RSV-specific cells was less tightly focused on VK3 and JK3 compared to random B cells, there was little evidence of repertoire bias in the light chain sequences when infants and older subjects were compared.
Figure 4.
VK, JK, VL and JL gene family utilization in circulating human random or RSV-specific B cells. Single circulating random or RSV-specific B cells from healthy blood donors, RSV-infected adults, RSV-infected children >3 months old and RSV-infected infants <3 months old were sorted, amplified in culture, and tested for RSV F-specific antibody production by ELISA, followed by sequence analysis of the expressed gene segment. Frequencies of VK, JK, VL and JL gene family utilization in all four subsets are presented in (A), (B), (C) and (D), respectively.
N-nucleotide insertions
The adjusted mean number of heavy chain N sequence nucleotides was significantly fewer for infants <3 months old compared to infants ≥3 months old (p=0.0013, Table II), but not for infants <3 months old vs. adults (p=0.12). RSV F-specific Abs from infants ≥3 months old exhibited a higher number of N nucleotide insertions than adult Ab sequences (p=0.025). The adjusted mean number of light chain N sequence nucleotides was significantly fewer for infants <3 months old compared to adults (p=0.0398) and approached significance for infants ≥3 months old (p=0.0586). The number of N-nucleotides in light chain sequences from infants ≥3 months old did not differ from adult Ab sequences.
Table II.
Pair-wise comparisons of N nucleotides of RSV F-specific antibodies derived from subjects of varying ages
| Age group comparisons |
p-value for the difference in adjusted mean number of N nucleotides |
|
|---|---|---|
| Heavy chain | ||
| < 3 mon vs ≥3 mon | 0.0013 | |
| < 3 mon vs adult | 0.1201 | |
| ≥3 mon vs adult | 0.0250 | |
| Light chain | ||
| < 3 mon vs ≥3 mon | 0.0586 | |
| < 3 mon vs adult | 0.0398 | |
| ≥3 mon vs adult | 0.9999 | |
The table shows the p-values for the pair-wise differences in the number of N sequence additions. P values <0.05 are shown in bold type.
Somatic mutations in variable regions
CDR3 lengths did not differ between groups analyzed (data not shown). We compared the frequency and number of mutations in the CDRs of variable regions between groups using a zero-inflated Poisson analysis, since many of the sequences had zero mutations. Table III shows the total number of mutations in the CDR regions, and Table IV shows the pair-wise comparisons between groups. Ab sequences from infants <3 months old had a significantly higher proportion of heavy chain CDR regions with zero mutations (88%) compared to infants ≥3 months old (53%) and adults (55%); this comparison approached significance for the adjusted mean number of heavy chain CDR mutations (p=0.0545). The proportion of light chain CDR regions with zero mutations did not differ significantly between infants <3 months old, infants ≥3 months old and adults. The adjusted mean number of light chain CDR region mutations was significantly fewer for infants <3 months old compared to infants ≥3 months old and adults (p=0.0250 and 0.0139, respectively). The number of mutations in the heavy chain framework regions was significantly fewer in RSV F-specific Ab sequences from infants <3 months old compared to infants ≥3 months old (p=0.0018) and adults (p=0.0003)(Table V). Likewise, the number of light chain framework region mutations was significantly fewer in sequences from infants <3 months old compared to infants ≥3 months old (p=0.0002) and adults (p<0.0001). Thus, the youngest infants exhibited reduced numbers of somatic mutations in both heavy and light chains when compared with adults.
Table III.
Frequency of mutations in the heavy and light chain CDR sequences of RSV F-specific antibodies derived from subjects of indicated ages
| Antibody Region | Number of clones with zero mutations (%) |
Estimated adjusted mean number of mutations (SEM) |
||||
|---|---|---|---|---|---|---|
| < 3 mon | ≥ 3 mon | Adult | < 3 mon | ≥ 3 mon | Adult | |
| Heavy chain CDR | 50 (88) | 10 (53) | 46 (55) | 3.0 (1.3) | 5.1 (1.2) | 3.7 (1.1) |
| Light chain CDR | 43 (83) | 10 (78) | 35 (57) | 0.96 (1.5) | 2.7 (1.2) | 2.8 (1.1) |
| All | 46 (88) | 19 (90) | 57 (93) | 0.61 (2.0) | 2.8 (1.6) | 2.8 (1.0) |
The adjustment was based on using a zero-inflated Poisson model, as described in Methods.
Table IV.
Pair-wise comparisons of mutations in the heavy and light chain CDR sequences of RSV F-specific antibodies derived from subjects of varying ages
| Age group comparisons |
p-value for the difference in proportion of zero mutations |
p-value for the difference in adjusted mean number of mutations |
|
|---|---|---|---|
| Heavy chain | |||
| < 3 mon vs ≥ 3 mon | 0.0033 | 0.0545 | |
| < 3 mon vs adult | 0.0002 | 0.3763 | |
| ≥ 3 mon vs adult | 0.8772 | 0.0730 | |
| Light chain | |||
| < 3 mon vs ≥ 3 mon | 0.1032 | 0.0250 | |
| < 3 mon vs adult | 0.2174 | 0.0139 | |
| ≥ 3 mon vs adult | 0.4354 | 0.8712 | |
The table shows the p-values for the pair-wise differences in the number of zero mutations, and differences in the adjusted mean number of mutations. P values <0.05 are shown in bold type.
Table V.
Pair-wise comparisons of mutations in the heavy and light chain framework regions of RSV F-specific antibodies derived from subjects of varying ages
| Donor Group | Age | Total number framework mutations in indicated chain (mean +/− SEM) |
|
|---|---|---|---|
| Heavy chain* | Light chain** | ||
| A | < 3 month | 0.86 +/− 1.3 | 0.17 +/− 1.5 |
| B | ≥3 month | 4.37 +/− 1.5 | 1.57 +/− 1.5 |
| C | Adult | 3.13 +/− 1.2 | 1.36 +/− 1.3 |
Group A vs B, p=0.0018; A vs C, p=0.0003; B vs C, p=0.4941.
Group A vs B, p=0.0002; A vs C, p<0.0001; B vs C, p=0.7656.
One infant had been born at 29 weeks of gestation and hospitalized previously for laboratory-confirmed RSV infection at 3 weeks of age. We recruited this subject for this study at 11 weeks of age during a second laboratory-confirmed RSV infection, which did not require hospitalization. We isolated nine distinct B cell clones from this child, with differing heavy and light chain gene segments. Remarkably, only one nucleotide per clone was mutated in three of the nine clones, and the remaining sequences were identical to the predicted germline sequences.
Discussion
Very young infants <3 months old are highly susceptible to bacterial and viral infections, and this is a target age group for many licensed and investigational vaccines. Thus, it is of major interest to determine the quality of infant Ab responses during the first few months of life. The majority of studies of human infant Ab repertoire and diversity have analyzed unselected immune repertoires. This study defined a viral antigen-specific Ab repertoire in very young infants at a detailed molecular level with sufficient numbers of RSV F-selected Ab genes to perform rigorous statistical analysis. The central finding is that RSV F-specific Ig repertoires from infants <3 months of age exhibit a biased use of antibody variable genes and a lack of somatic mutations.
We used novel flow cytometry and culture methods to isolate a large set of RSV F-specific B cell clones isolated from infants or adults with RSV infection. Previous studies have shown the frequency of antigen-specific B cells in the peripheral blood to be between 1 in 2,000 to 1 in 10,000 (Lagerkvist et al., 1995; Zubler et al., 1992); thus, this yield represents an enhanced recovery of 20–1,000 fold. These techniques have potential to identify other rare populations of antigen-specific peripheral B cells. B cells from infants proliferated in culture and expressed Ig at frequencies similar to that of B cells isolated from adults, making it unlikely our findings were biased by differential recovery from infants.
We found that RSV F-specific Ab gene repertoires expressed by B cells from infants <3 months old differed from those of older children and adults. Expression of VH1 and VH4 families was more prominent in infants <3 months old. Previous studies have suggested that fetal-like constrained repertoires may persist into early infancy (Lucas et al., 1993; Mortari et al., 1992; Mortari et al., 1993; Schroeder et al., 1987; Schroeder et al., 1995; Shiokawa et al., 1999). Our data support the hypothesis that infants in the first few months of life exhibit a less focused repertoire in an antigen-specific response. Similar trends were observed for D and JH gene segment family usage but significant differences between groups were not detected. In random adult B cells or RSV-specific B cells from adults or infants, we observed a uniform bias toward the DH3 and JH4 families (Fig. 2, B and C). These data are consistent with previously identified usage frequencies of randomly-selected B cells in other studies.
VH segment usage in RSV F-specific B cells from infants <3 months old differed from the most frequently used segments in adult unselected or adult RSV-specific repertoires. Several segments that are uncommon in the random B cell expressed VH gene repertoire represented close to 10% or greater of the RSV F-specific repertoire in infants <3 months old (VH1–69, VH4–59, VH4–39, VH1–03 and VH4–04). These data suggest that the youngest infants differ in their expressed repertoire from adults. The RSV-specific light chain repertoire was similar between young infants, older infants and adults, and was similar to the random light chain repertoire. However, the number of light chain genes recovered for analysis was smaller than for heavy chain genes.
The most striking molecular feature in these studies was the finding that RSV F-specific antibodies from infants <3 months old exhibited many fewer somatic mutations than RSV F-specific antibodies from older children or adults. This finding was true for both VH and VL gene segments, and in CDR and framework regions. The majority of heavy and light chain CDR regions from infants <3 months old had no mutations. It is possible that this lack of mutations was due to a lack of prior antigenic stimulation. However, several aspects of the study suggest that developmental limitations play a role in addition to lack of prior exposure. First, the majority of the older infants studied here likely had only experienced one prior RSV infection based on their age and the seasonality of RSV, yet had a greater number of mutations. One infant in this study who was experiencing a second documented RSV infection within the first few months of life still lacked mutations in RSV-specific gene segments. These data strongly suggest that inefficiencies in the molecular and cellular mechanisms controlling somatic hypermutation may underlie poor infant antibody responses to vaccines and viral infections (Brandenburg et al., 1997; Murphy et al., 1986a; Murphy et al., 1986b; Roca et al., 2003). Zemlin et al analyzed global Ig repertoire and discovered that preterm infants exhibited reduced somatic mutations compared with term infants (Zemlin et al., 2007). Other studies have described differences between infant and adult B cell repertoire and somatic hypermutation despite repeated antigenic stimulation by vaccines (Weller et al., 2008). Our study shows that this phenomenon extends to antigen-specific B cells.
It is not clear why very young infants do not introduce somatic mutations into virus-specific antibody genes during primary infection. We previously showed that transcriptional control of activation-induced cytidine deaminase and error-prone DNA polymerases is functionally mature in cord blood (Bowen et al., 2006). However, neonatal B cells clearly differ functionally from those of older subjects. For example, neonatal B cells exhibit reduced expression of the interleukin (IL)-4 receptor alpha chain and reduced IL-4-induced signaling (Tian et al., 2006), which has important implications for antigen-specific responses early in life. Immaturity of control of expression of factors extrinsic to B cells, such as cell-surface and soluble molecules from T cells, antigen presenting cells and bone marrow stromal cells also likely contributes to the lack of somatic mutations. Further studies are necessary to elucidate the mechanisms underlying the limited extent of somatic hypermutation in the antibody genes of very young infants.
Supplementary Material
Acknowledgements
The authors thank Kathy Allen for assistance with flow cytometry and Mina Baisch, Kitty Miller, and Alice O’Shea for assistance with patient recruitment, and Koichi Kusuhara and Elizabeth Bures for technical assistance and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bona C, Radu D, Kodera T. Molecular studies on the diversification of hemagglutinin-specific human neonatal repertoire subsequent to immunization with naked DNA. Vaccine. 2004;22:1624–1630. doi: 10.1016/j.vaccine.2003.10.045. [DOI] [PubMed] [Google Scholar]
- Bowen AL, Tian C, LaFleur BJ, Crowe JE., Jr Transcriptional control of activation-induced cytidine deaminase and error-prone DNA polymerases is functionally mature in the B cells of infants at birth. Hum Immunol. 2006;67:43–46. doi: 10.1016/j.humimm.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Brandenburg AH, Groen J, van Steensel-Moll HA, Claas EC, Rothbarth PH, Neijens HJ, Osterhaus AD. Respiratory syncytial virus specific serum antibodies in infants under six months of age: limited serological response upon infection. J Med Virol. 1997;52:97–104. doi: 10.1002/(sici)1096-9071(199705)52:1<97::aid-jmv16>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, "minor" D segments or D-D recombination. J Mol Biol. 1997;270:587–597. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- Garrone P, Neidhardt EM, Garcia E, Galibert L, van Kooten C, Banchereau J. Fas ligation induces apoptosis of CD40-activated human B lymphocytes. J Exp Med. 1995;182:1265–1273. doi: 10.1084/jem.182.5.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc MP. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezen WP, Paredes A, Allison JE, Taber LH, Frank AL. Risk of respiratory syncytial virus infection for infants from low-income families in relationship to age, sex, ethnic group, and maternal antibody level. J Pediatr. 1981;98:708–715. doi: 10.1016/s0022-3476(81)80829-3. [DOI] [PubMed] [Google Scholar]
- Halasa NB, Barr FE, Johnson JE, Edwards KM. Fatal pulmonary hypertension associated with pertussis in infants: does extracorporeal membrane oxygenation have a role? Pediatrics. 2003;112:1274–1278. doi: 10.1542/peds.112.6.1274. [DOI] [PubMed] [Google Scholar]
- Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001;344:1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- Healy CM, Baker CJ. Prospects for prevention of childhood infections by maternal immunization. Curr Opin Infect Dis. 2006;19:271–276. doi: 10.1097/01.qco.0000224822.65599.5b. [DOI] [PubMed] [Google Scholar]
- Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis. 2005;191:1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- Kolar GR, Yokota T, Rossi MI, Nath SK, Capra JD. Human fetal, cord blood, and adult lymphocyte progenitors have similar potential for generating B cells with a diverse immunoglobulin repertoire. Blood. 2004;104:2981–2987. doi: 10.1182/blood-2003-11-3961. [DOI] [PubMed] [Google Scholar]
- Lagerkvist AC, Furebring C, Borrebaeck CA. Single, antigen-specific B cells used to generate Fab fragments using CD40-mediated amplification or direct PCR cloning. Biotechniques. 1995;18:862–869. [PubMed] [Google Scholar]
- Lambert D. Zero-Inflated Poisson Regression, With an Application to Defects in Manufacturing. Technometrics. 1992;34:1–14. [Google Scholar]
- Lucas AH, Azmi FH, Mink CM, Granoff DM. Age-dependent V region expression in the human antibody response to the Haemophilus influenzae type b polysaccharide. J Immunol. 1993;150:2056–2061. [PubMed] [Google Scholar]
- Mortari F, Newton JA, Wang JY, Schroeder HW., Jr The human cord blood antibody repertoire. Frequent usage of the VH7 gene family. Eur J Immunol. 1992;22:241–245. doi: 10.1002/eji.1830220135. [DOI] [PubMed] [Google Scholar]
- Mortari F, Wang JY, Schroeder HW., Jr Human cord blood antibody repertoire. Mixed population of VH gene segments and CDR3 distribution in the expressed C alpha and C gamma repertoires. J Immunol. 1993;150:1348–1357. [PubMed] [Google Scholar]
- Murphy BR, Alling DW, Snyder MH, Walsh EE, Prince GA, Chanock RM, Hemming VG, Rodriguez WJ, Kim HW, Graham BS, et al. Effect of age and preexisting antibody on serum antibody response of infants and children to the F and G glycoproteins during respiratory syncytial virus infection. J Clin Microbiol. 1986a;24:894–898. doi: 10.1128/jcm.24.5.894-898.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Graham BS, Prince GA, Walsh EE, Chanock RM, Karzon DT, Wright PF. Serum and nasal-wash immunoglobulin G and A antibody response of infants and children to respiratory syncytial virus F and G glycoproteins following primary infection. J Clin Microbiol. 1986b;23:1009–1014. doi: 10.1128/jcm.23.6.1009-1014.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott RH, Kim HW, Arrobio JO, Hodes DS, Murphy BR, Brandt CD, Camargo E, Chanock RM. Epidemiology of respiratory syncytial virus infection in Washington, D.C. II. Infection and disease with respect to age, immunologic status, race and sex. Am J Epidemiol. 1973;98:289–300. doi: 10.1093/oxfordjournals.aje.a121558. [DOI] [PubMed] [Google Scholar]
- Polack FP, Karron RA. The future of respiratory syncytial virus vaccine development. Pediatr Infect Dis J. 2004;23:S65–S73. doi: 10.1097/01.inf.0000108194.71892.95. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, Timmers E, Kenter MJ, Van Tol MJ, Vossen JM, Schuurman RK. Restricted utilization of germ-line VH3 genes and short diverse third complementarity-determining regions (CDR3) in human fetal B lymphocyte immunoglobulin heavy chain rearrangements. Eur J Immunol. 1992;22:247–251. doi: 10.1002/eji.1830220136. [DOI] [PubMed] [Google Scholar]
- Roca A, Quinto L, Abacassamo F, Loscertales MP, Gomez-Olive FX, Fenwick F, Cane PA, Saiz JC, Toms G, Alonso PL. Antibody response after RSV infection in children younger than 1 year of age living in a rural area of Mozambique. J Med Virol. 2003;69:579–587. doi: 10.1002/jmv.10348. [DOI] [PubMed] [Google Scholar]
- Ruiz M, Giudicelli V, Ginestoux C, Stoehr P, Robinson J, Bodmer J, Marsh SG, Bontrop R, Lemaitre M, Lefranc G, Chaume D, Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000;28:219–221. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sblattero D, Bradbury A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology. 1998;3:271–278. doi: 10.1016/s1380-2933(97)10004-5. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Hillson JL, Perlmutter RM. Early restriction of the human antibody repertoire. Science. 1987;238:791–793. doi: 10.1126/science.3118465. [DOI] [PubMed] [Google Scholar]
- Schroeder HW, Jr, Mortari F, Shiokawa S, Kirkham PM, Elgavish RA, Bertrand FE., 3rd Developmental regulation of the human antibody repertoire. Ann N Y Acad Sci. 1995;764:242–260. doi: 10.1111/j.1749-6632.1995.tb55834.x. [DOI] [PubMed] [Google Scholar]
- Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- Tian C, Kron GK, Dischert KM, Higginbotham JN, Crowe JE., Jr Low expression of the interleukin (IL)-4 receptor alpha chain and reduced signalling via the IL-4 receptor complex in human neonatal B cells. Immunology. 2006;119:54–62. doi: 10.1111/j.1365-2567.2006.02405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkamp JH, Crowe JE., Jr Blood donor leukocyte reduction filters as a source of human B lymphocytes. Biotechniques. 2001;31:464–466. doi: 10.2144/01313bm01. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Bures E, Williams JV, LaFleur B, Greenberg HB, Crowe JE., Jr Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J Immunol. 2003a;171:4680–4688. doi: 10.4049/jimmunol.171.9.4680. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, Crowe JE., Jr Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003b;275:223–237. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Lafleur BJ, Greenberg HB, Crowe JE., Jr Natural evolution of a human virus-specific antibody gene repertoire by somatic hypermutation requires both hotspot-directed and randomly-directed processes. Hum Immunol. 2005;66:666–676. doi: 10.1016/j.humimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, Weill JC, Reynaud CA. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J Exp Med. 2008;205:1331–1342. doi: 10.1084/jem.20071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE, Jr, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, Wittek AE, Casey R, Eichelberger M, Thumar B, Randolph VB, Udem SA, Chanock RM, Murphy BR. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis. 2000;182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- Zemlin M, Hoersch G, Zemlin C, Pohl-Schickinger A, Hummel M, Berek C, Maier RF, Bauer K. The postnatal maturation of the immunoglobulin heavy chain IgG repertoire in human preterm neonates is slower than in term neonates. J Immunol. 2007;178:1180–1188. doi: 10.4049/jimmunol.178.2.1180. [DOI] [PubMed] [Google Scholar]
- Zubler RH, Perrin LH, Doucet A, Zhang X, Huang YP, Miescher PA. Frequencies of HIV-reactive B cells in seropositive and seronegative individuals. Clin Exp Immunol. 1992;87:31–36. doi: 10.1111/j.1365-2249.1992.tb06409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.