Abstract
Proper regulation of the Wingless/Wnt signaling pathway is essential for normal development. The scaffolding protein Axin plays a key role in this process through interactions with Drosophila Shaggy and Armadillo. In the current studies, we used a yeast two-hybrid assay to identify ten amino acids in Axin that are critical for in vitro interaction with Shaggy and two for interaction with Armadillo. We then generated five Axin variants in which individual putative contact amino acids were mutated and compared their activity, as assayed by rescue of axin null mutant flies, to that of Axin lacking the entire Shaggy (AxinΔSgg) or Armadillo (AxinΔArm) binding domain. Although we expected these mutants to function identically to Axin in which the entire binding domain was deleted, we instead observed a spectrum of phenotypic rescue. Specifically, two point mutants within the Shaggy binding domain showed loss of activity similar to that of AxinΔSgg and dominantly interfered with complex function, whereas a third mutant allele, AxinK446E retained most function. Two Axin point mutants within the Armadillo binding domain were weak alleles, and retained most function. These findings demonstrate the importance of in vivo verification of the role of specific amino acids within a protein.
Keywords: Axin mutations, Wnt/β-catenin signaling, in vivo analysis, cancer, Drosophila
Introduction
Cell-cell communication is essential to development and homeostasis in the adult organism. A pathway critical to intercellular communication is the Wnt/β-catenin pathway, whose dysregulation causes developmental defects and numerous diseases including colorectal cancer, liver cancer, cardiovascular disease, and osteoporosis (Baron et al., 2006; de Lau et al., 2007; Giles et al., 2003; Grigoryan et al., 2008; Klaus and Birchmeier, 2008; Mani et al., 2007). The central component of the Wnt/β-catenin signaling pathway is a protein complex assembled around the scaffold protein Axin, the ‘destruction complex’; pathway activity is controlled by destruction complex regulation and modulation. The key components of the active complex include APC (Adenomatous Polyposis Coli), the kinase GSK3β (Glycogen synthase kinase3β; Drosophila Shaggy/Zw3, Sgg), and β-catenin (Drosophila Armadillo, Arm). GSK3β phosphorylates β-catenin, providing the signal for its ubiquitination and degradation by the proteasome, thereby preventing β-catenin from completing the signaling cascade by entering the nucleus to regulate transcription of Wnt target genes (Polakis, 2007).
Wnt/β-catenin signaling is induced when secreted glycoproteins of the Wnt family bind to a receptor complex consisting of Frizzled family serpentine receptors and members of the LDL-receptor related protein (LRP) family (LRP5 & LRP6 in vertebrates; Arrow in Drosophila (Wehrli et al., 2000)). Physical proximity of Frizzled and Arr/LRP cytoplasmic domains initiates a signal (Cong et al., 2004; Tolwinski et al., 2003) which is thought to activate Dishevelled (see Malbon and Wang, (2006) for a discussion of additional implicated proteins). Dishevelled plays distinct roles at multiple points in the pathway, including promotion of Axin translocation from the cytoplasm to the membrane and direct inhibition of destruction complex activity through a Dishevelled-Axin interaction (Cliffe et al., 2003; Julius et al., 2000; Mao et al., 2001). The latter interaction, which is thought to occur through aggregated DIX domains present in Axin as well as Dishevelled (Schwarz-Romond et al., 2007), represents only a minor part of Axin complex inhibition (Peterson-Nedry et al., 2008). Following Axin s translocation to the cell membrane, it is bound by Arr/LRP5/6, which appears to result in direct inhibition of GSK3β kinase activity and subsequent degradation of Axin (Davidson et al., 2005; Mao et al., 2001; Piao et al., 2008; Tolwinski et al., 2003; Zeng et al., 2005). The combined direct inhibition of the destruction complex by Dishevelled and Arr/LRP, in combination with other regulatory interactions in the pathway, determines the precise level of signaling mediated by β-catenin s transcriptional activity. Many examples, including graded signaling in the developing fly wing (Zecca et al., 1996; Strigini and Cohen, 2000) and deviation from optimal activity resulting in either osteoporosis or excess bone mass in humans (Glass and Karsenty, 2006), indicate β-catenin signaling is subject to finely graded regulation.
The precise level of Wnt/β-catenin signaling determines distinct cell fates during normal development and homeostasis and, similarly, it is increasingly apparent that disease-causing forms of pathway components exert their effects by only slightly changing signaling levels. For example, mutant forms of APC that retain much of their ability to regulate β-catenin signaling are observed in, and are assumed to promote the development of colorectal cancer. By contrast, complete loss of APC function, which precludes destruction complex activity resulting in maximal signaling levels, is not observed in these tumors. This observation has led to the ‘just-right’ hypothesis that tumors require moderately raised, controlled signaling levels for optimal development (Albuquerque et al., 2002; Smits et al., 2000); a premise that has been tested and confirmed in Drosophila (McCartney et al., 2006). Thus, in vivo experiments led to this recent break-through in our understanding of destruction complex modulation.
A central problem in genetics is predicting consequences of specific mutations. For example, did a mutation identified in a tumor cell contribute to tumorigenesis or is it irrelevant? For example, at least 35 missense mutations in Axin are associated with cancer (Salahshor and Woodgett, 2005), yet their culpability is unclear. One useful criterion that may be applied is whether the mutation resides in a defined motif or domain. In Axin, the binding domains for its partners have been mapped and partial co-crystal structures have been solved for some vertebrate components (Xing et al., 2003; Dajani et al., 2003). The importance of these interaction domains in vivo remained uncertain due to conflicting data in experiments where Axin mutant proteins lacking such binding sites were overexpressed, leaving open the question of whether the composition and regulation of the Axin complex was context dependent or whether the level of over-expression affected the outcome. Expression of mutant Axin proteins at near-physiological levels in the absence of endogenous protein resolved these issues and, surprisingly, demonstrated that deletion of single binding sites in Axin nevertheless allows assembly of a largely functional complex in vivo (Peterson-Nedry et al., 2008). Rather than discounting biochemical data, these experiments revealed the significance in vivo of secondary interactions between APC, β-catenin, and GSK3β, which allow recruitment of components into the complex even in the absence of the binding site on Axin. While embryos appeared largely or completely rescued, viability was not restored by any of the mutant proteins. Therefore loss of a binding partner (e.g. APC) completely disrupted destruction complex function whereas loss of one protein-protein interaction site only moderately affected its function.
Shaggy/GSK3β and Armadillo/β-catenin bind to the central region of Axin (Hart et al., 1998; Sakanaka et al., 1998; Fagotto et al., 1999; Hedgepeth et al., 1999; Hamada et al., 1999; Willert et al., 1999; Yanagawa et al., 2000). However, the binding site for Shaggy on Drosophila Axin is poorly defined, due to limited sequence identity between Drosophila and vertebrate protein sequences (Fig. 1). We previously examined the consequence of loss of the entire Shaggy binding site from Axin as part of a series of Axin deletion mutants all expressed near physiological levels and in the absence of wild type Axin; we demonstrated that each of these mutant proteins retained significant function (Peterson-Nedry et al., 2008). This finding was best explained by highly cooperative assembly, in which components could be recruited indirectly if their binding site on Axin was missing. For example Armadillo would be brought into an AxinΔArm complex (which lacks the entire Armadillo binding site) by interacting with pre-bound APC and Shaggy. These findings raised questions such as (a) where are the precise binding sites for Armadillo and Shaggy on Axin and (b) what is the effect of point mutations in the binding sites compared to deletion of the entire site, a question of particular interest to tumorigenesis since, as mentioned above, many missense mutations in Axin are associated with cancer (Salahshor and Woodgett, 2005).
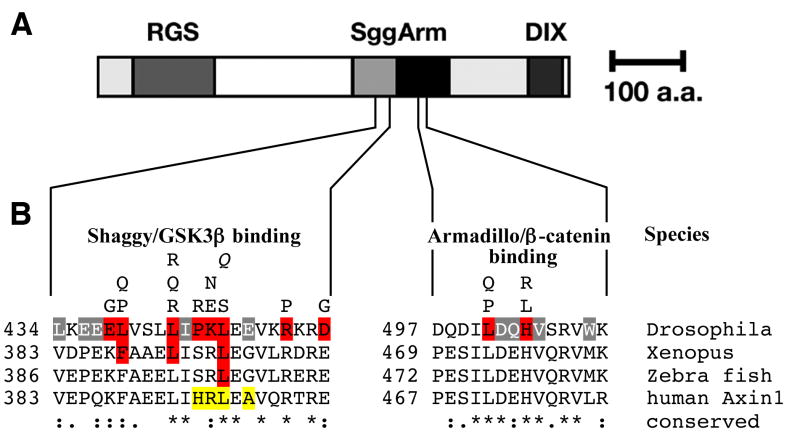
Fig. 1. Identification in vitro of contact residues on Axin critical for binding by Shaggy and Armadillo, respectively.
(A) A schematic Axin structure is shown. Key binding partners and their binding domains are APC (RGS), Shaggy/GSK3β (Sgg), Armadillo/β-catenin (Arm), and the DIX domain required for Axin homodimerization and binding of Dishevelled. In a saturation mutagenesis, we identified the critical residues for Shaggy and Armadillo binding, respectively, using the yeast two hybrid system (Material and methods). (B) An alignment with sequences from the fruit fly (Drosophila melanogaster), frog (Xenopus laevis), zebrafish (Danio rerio) and human is shown. Mutant isolates are shown above the Drosophila sequence and the critical residues are indicated in red. Amino acids highlighted in gray reproducibly disrupted the interaction but depended on the synergy with secondary mutations outside the binding site (Material and methods). L447Q (Q) was not identified as a mutant in the screen but modeled after the zebrafish allele (Heisenberg et al., 2001) and it too disrupts the interaction with Shaggy in yeast. Mutations shown to disrupt the interaction with Xenopus Axin (Hedgepeth et al., 1999) and zebrafish Axin (Heisenberg et al., 2001) are also highlighted in red. Mutations identified in cancers cells but not shown to disrupt the Axin-GSK3β interaction are highlighted in yellow (Salahshor and Woodgett, 2005). The alignment was generated using ClustalW2 and degree of sequence conservation is illustrated (“*”, identity, “:” conserved substitution, “.” semi-conserved substitution).
In this study, we address how single point mutations that change contact amino acids for the Shaggy (Sgg; Drosophila GSK3β) -Axin interaction and the Armadillo (Arm; Drosophila β-catenin) -Axin interaction affect complex function in vivo. We used the yeast two-hybrid system to identify footprints on Axin where Shaggy and Armadillo bind. Several residues identified here are in conserved positions with the vertebrate components where they function as contact amino acids in the respective co-crystal structures (Xing et al., 2003; Dajani et al., 2003). We tested five Axin variants in which these putative contact amino acids were mutated for function in transgenic flies. Although we expected these mutants to function identically to Axin in which the entire binding domain was deleted, we instead observed a spectrum of phenotypic rescue ranging from that seen with the domain deletion mutant to complete rescue to viability. These findings demonstrate that structural information is not sufficient to predict a requirement for specific amino acids and underscores the need to analyze mutant proteins in their normal context in vivo.
Results
Mutational analysis of Drosophila Axin identifies critical amino acids for the interaction with Shaggy/GSK3β and Armadillo/β-catenin
First, we aimed to generate a footprint for the interaction of Armadillo and Shaggy on Axin. We subjected cDNA encoding the C-terminal half of Axin to PCR-mutagenesis (Material and methods) and identified Axin mutations that disrupt the interaction between mutant Axin prey and the respective baits (Arm, Sgg) in the yeast two-hybrid system. Twenty-two different Axin mutant clones disrupting the interaction with Armadillo (Axin[Arm*]) were isolated; these changed four amino acids closely clustered within regions previously identified through immunoprecipitation to contain the Armadillo binding site (Hamada et al., 1999; Willert et al., 1999; Yanagawa et al., 2000; Fig. 1, and not shown). Several clones contained multiple mutations and, in these cases, we introduced single mutations into Axin by site-directed mutagenesis to test whether the loss of interaction was due to single mutations. These experiments demonstrated that L501 and H504 were required for the Axin-Armadillo interaction in yeast. A partial requirement for D502Q and W509R was identified, since loss of interaction was observed in the double mutant D502Q W509R but not if either mutation was present singly, suggesting that cumulative changes in less critical amino acids can have additive or synergistic effects. The independent isolation of 13 cDNAs with mutations in L501 and eight cDNAs with substitutions at H504 suggests the screen was near saturation (see Material and methods). Figure 1 shows that the residues we identified are at the core of the conserved Arm/β-catenin binding domain. L501, D502, H504 are identical between Drosophila and vertebrate orthologs of Axin whereas W509 is substituted by the hydrophobic residues methionine and leucine in vertebrates. Taken together, these data validate screening for loss of interaction mutations in the yeast two-hybrid system as a stringent approach to identify key residues involved in protein-protein interaction.
Next, we mutagenized Axin to identify Axin mutations that disrupt Shaggy-binding, again using the yeast two-hybrid system. We identified 21 Axin mutant clones encoding proteins that failed to interact with Shaggy. The mutations led to substitutions at 8 different amino acids (Fig. 1B), all contained within the 65 amino acid fragment previously shown to contain the binding site for Shaggy (Willert et al., 1999; Yanagawa et al., 2000). A sequence alignment reveals that L439, L443 and L447 show either complete identity with, or are conservative changes relative to residues identified as being essential for the corresponding Axin-GSK3β interaction in Xenopus and zebrafish (Fig. 1; (Hedgepeth et al., 1999; Heisenberg et al., 2001). Three additional residues (E478, P445, D455) are also essential for the Shaggy-Axin interaction in yeast but show a lesser degree of conservation (Fig. 1). Taken together, several amino acids essential for the interaction of Shaggy and Axin in yeast also align well with the vertebrate GSK3β binding site on Axin (Fig. 1). Although automated algorithms previously failed to properly align the binding site (e.g. in Willert et al., 1999), we conclude that we have functionally identified the Shaggy binding site on Drosophila Axin (Fig. 1).
Axin point mutant proteins display a spectrum of activity when assayed for ability to rescue hatching in axinnull mutants
Next, we generated point mutant forms of Axin that are predicted to disrupt Armadillo or Shaggy binding in order to test their ability to support development and survival of transgenic flies. Our previous work demonstrated that Axin lacking the entire Armadillo (AxinΔArm) or Shaggy (AxinΔSgg) binding site failed to rescue axinnull mutant flies to viability (Peterson-Nedry et al., 2008). However, AxinΔArm rescued several phenotypic aspects of embryonic patterning including formation of a nearly wild type cuticle pattern. AxinΔSgg also generated segmented cuticle but displayed partial loss of denticle belts indicative of compromised catalytic function, producing an intermediate phenotype between wild type and the axinnull mutant. In both instances, deletion of the entire binding site would be expected to reveal the maximal phenotype for the loss of this particular interaction. This raised the question of what effect the loss of single contact amino acids might have in vivo.
Two criteria guided our choice of amino acids for mutagenesis. First, we chose amino acids that, when mutated, completely disrupted the interaction of Axin with Arm (L501, H504) or Sgg (L447) in our yeast assay, and that are conserved across species (Fig. 1). Second, we chose residues that are good candidates for contact amino acids (L447, K446) based on the co-crystal structure of vertebrate components. Based on these criteria, we introduced separate mutations into the Armadillo binding domain (L501P and H504L) and the Shaggy binding domain (L447S, L447Q, and K446E) of Axin that would be expected to disrupt the interaction with Armadillo or Shaggy, respectively. Notably, the residue corresponding to Drosophila L447 has been shown to be required for the interaction of Xenopus GSK3β and Axin in vitro (Hedgepeth et al., 1999) and when mutated to glutamine, renders zebrafish Axin temperature-sensitive, generating the masterblind allele (Heisenberg et al., 2001). We therefore sought to determine whether L447Q would similarly affect Drosophila Axin. Furthermore, Drosophila K446 is adjacent to L447 and analogy with the vertebrate co-crystal structure suggests it may similarly form a salt bridge between Shaggy and Axin (see Discussion; Xing et al., 2003). The K446E mutation would cause a charge reversal in this putative salt bridge that might disrupt protein-protein interaction (see Discussion; Dajani et al., 2003; Xing et al., 2003). We introduced each of these five mutations individually into FLAG-tagged Axin, inducibly driven by the tubulin promoter, and generated transgenic flies (Material and methods; see also Peterson-Nedry et al., 2008). Expression was induced in females and levels of each protein were assayed by Western blot analysis of embryo extracts. We previously showed that wild type FLAG-tagged Axin (FLAxin) is expressed at levels ~4.3 fold higher than endogenous Axin (Peterson-Nedry et al., 2008) and the current results show that all of the mutant proteins are expressed at relatively comparable levels, within a two-fold range of each other (Suppl. Fig. 1).
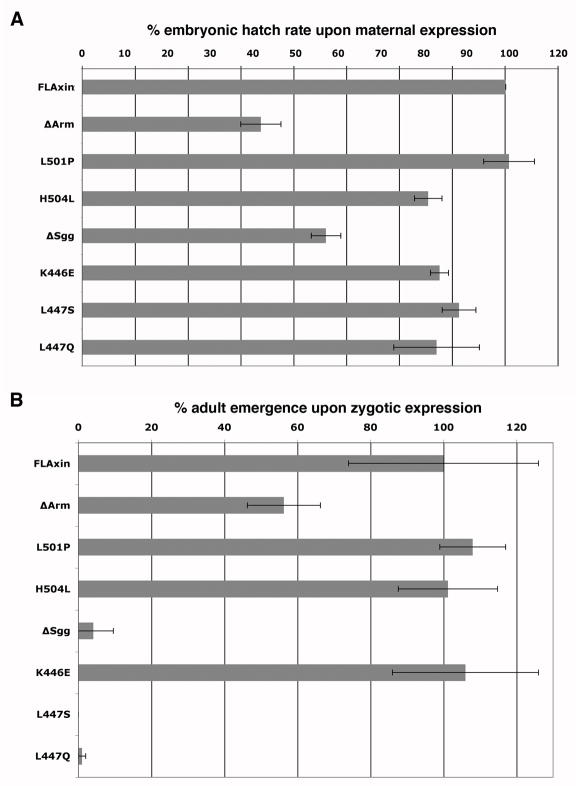
Proper assessment of mutant Axin protein function requires expression at near-physiological levels without interference by endogenous Axin. The onset of embryonic development relies on maternally deposited Axin, which is required to maintain the OFF state of signaling until Wg de-repression induces the signal some three hours later; compromised function of maternally deposited Axin results in embryonic lethality. We expressed wild type or mutant Axin during oogenesis, which results in maternal deposition into eggs, and at the same time removed endogenous Axin by inducing axinnull germ line clones (Material and methods) thereby generating embryos in which the only source of Axin is the wild type or mutant protein that we introduce. Subsequently, fertilized eggs will develop and hatch as larvae unless the mutant Axin is not sufficiently functional to support embryogenesis. The embryonic hatch rate thus provides a semi-quantitative assessment of the function retained by mutant forms of Axin relative to the wild type Axin control (ubiquitously expressed full-length Axin, FLAxin; Peterson-Nedry et al., 2008). Survival rates for embryos expressing Axin containing point mutations in Shaggy or Armadillo binding sites can thus be compared to those for embryos expressing Axin in which the entire Shaggy or Armadillo binding site is deleted, to determine the relative severity of the effect of the point mutations. We previously showed, that FLAxin rescues hatching in 80.0% of embryos, whereas AxinΔArm rescues hatching in 33.8% of embryos (Fig. 2; Peterson-Nedry et al., 2008). When we analyzed embryos expressing the Axin point mutant alleles affecting the Armadillo binding site (AxinL501P and AxinH504L) we did not detect a significant difference in hatch rates compared to embryos expressing Axin lacking the entire Armadillo binding site (AxinΔArm; Fig. 2). Therefore, in this assay, mutation of the contact amino acids L501 or H504 of Axin appears to result in the complete loss of direct Armadillo binding; the partial retention of function is presumably due to binding of Armadillo by other components in the Axin complex (Peterson-Nedry et al., 2008; see also Fig. 6E,F).
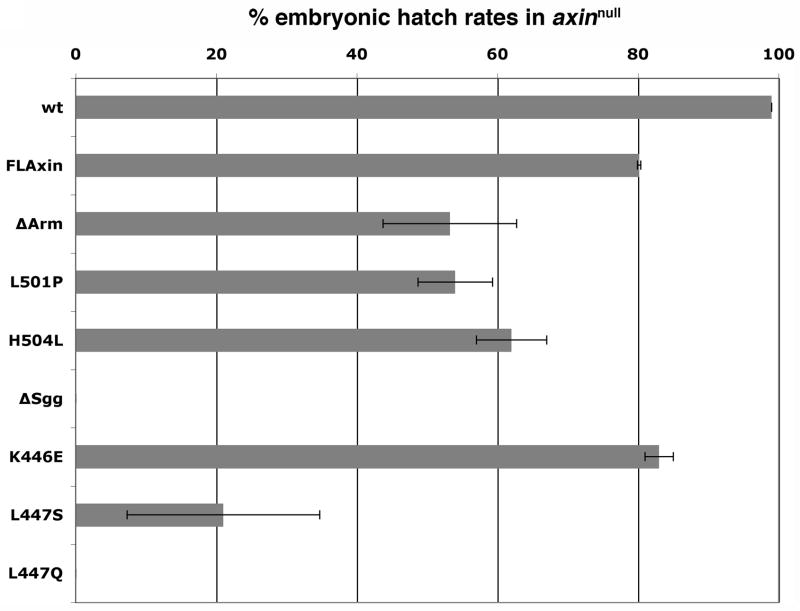
Fig. 2.
The hatch rate of embryos provides a measure for retained function of Axin mutant proteins. Constitutive expression of Axin mutant proteins using the tubulin promoter (tub>Axin*) in axinnull embryos revealed that the point mutations in the Armadillo binding domain result in complete loss of function, equivalent to AxinΔArm. All or nearly all embryos expressing the point mutations in the Shaggy binding domain, AxinL447Q or AxinL447S, die prior to hatching and therefore behave similarly to deletion of the entire site (AxinΔSgg). In contrast, AxinK446E allows hatch rates at similar levels to expressed FLAxin, though animals subsequently die (in K446E). Hatch rates are FLAxin 80% ±0.3, n=554; AxinΔArm, 33.4% ±3.7; AxinL501P, 54% ±5.3, n=599; AxinH504L, 62% ±5.0, n=781; AxinΔSgg, 0% ±0, n=630; AxinK446E, 83% ±2, n=738; AxinL447S, 21.0%±13.7, n=857; AxinL447Q, 0%±0; n=520.
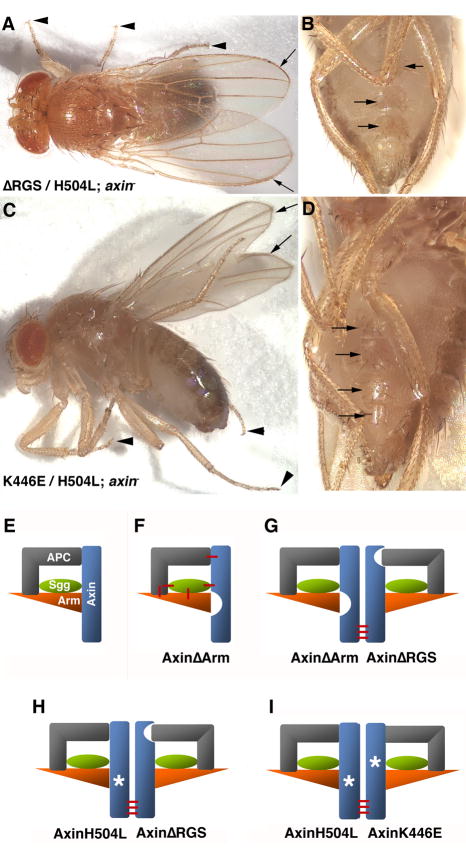
Fig. 6. Complementation between Axin mutant proteins restores viability.
(A,B) Homozygous axin mutant flies are fully rescued by co-expression of AxinΔRGS and AxinH504L. (A) A male fly displaying no defects; the wild type wings with intact wing margins (arrows) and fully formed legs with tarsal claws at their distal tip (arrowheads) are indicated. (B) The abdomen of a female shows the wild type pattern of sternites and their bristles (arrows).
(C,D) Homozygous axin mutant flies rescued by co-expression of the point mutants AxinK446E and AxinH504L. (C) Wings (arrows) and legs with visible tarsal claws (arrowheads) are fully formed, as are sternites in the ventral abdomen shown in (D)(arrows).
(E–I) Schematic representation of wild type and mutant Axin complexes. (E) Wild type Axin recruits APC, Shaggy and Armadillo, which also interact with each other. (F) These additional interactions (red bars) allow the indirect recruitment of Armadillo into an AxinΔArm complex, and by extension into other deletion or point mutant complexes. (G) Dimerization (red bars) between two Axin mutants AxinΔArm and AxinΔRGS restores a functional complex (Peterson-Nedry et al., 2008). (H) Similarly, complementation between AxinH504L and AxinΔRGS rescues, as shown in (A,B), and co-expression of AxinH504L with the Shaggy binding domain mutant AxinK446E also rescues (see panels C,D). Models are modified from Peterson-Nedry et al. (2008).
Next, we compared the function of Axin in which the entire Shaggy binding site is deleted to that of Axin containing single point mutations in residues that are predicted, by crystal structure, to contact Shaggy. Axin lacking the Shaggy-binding domain, or Axin containing the L447Q mutation does not support survival to hatching of any embryos. Axin L447S retains sufficient function to enable a modest number of embryos to hatch (21%) while hatching rates for embryos expressing Axin K446E are in the range of those expressing FLAxin (Fig. 2), indicating substantially retained function. These results show that single point mutations in the Shaggy domain (L447Q) can be as detrimental as the deletion of the entire domain and we note that the same is true for point mutations (L501P and H504L) within the Armadillo binding region.
The effect of Axin point mutants on molecular targets of Wg signaling conforms with binding site deletions, except that AxinK446E that appears to retain wild type function
A refined understanding of Axin function can be obtained from the analysis of Wg signaling events in the early embryo. At embryonic stages 5–10, Wg and Engrailed are expressed in adjacent rows of cells and Engrailed expression becomes dependent on Wg. Wg diffuses away from its source and induces expression of Engrailed in stripes that are approximately 2.2 cells wide in a normal embryo. In axinnull mutants, Wg signaling is fully upregulated and the Engrailed expression domain expands to a maximum width of 5–6 cells, with an average width of 3.6 cells; (Peterson-Nedry et al., 2008; Fig. 3). When Axin function is partially reduced, the Engrailed stripes widen to an intermediate value. When we examined Engrailed stripe width in axinnull mutant embryos expressing AxinL501P, AxinH504L or AxinΔArm, we found that it was indistinguishable from wild type (Fig. 3A–D, I). This finding indicates that these point mutant forms of Axin retain significant function, similar to that of AxinΔArm, as has been shown previously (Peterson-Nedry et al., 2008). In contrast, AxinΔSgg, AxinL447S, and AxinL447Q displayed an average Engrailed stripe width of 3.5 cells, which is not significantly different from the axinnull mutant (Fig. 3, and not shown; (Peterson-Nedry et al., 2008) and is thus indicative of substantial loss of Axin function. Finally, Engrailed stripes in AxinK446E are not significantly different from wild type indicating that this mutant protein retains wild type function, a conclusion that is also supported by analysis of hatch rates (Fig. 2).
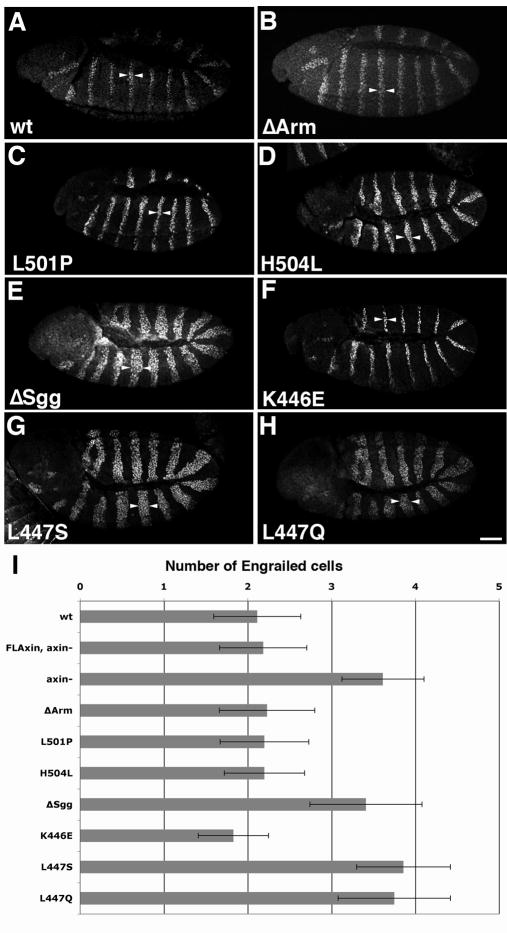
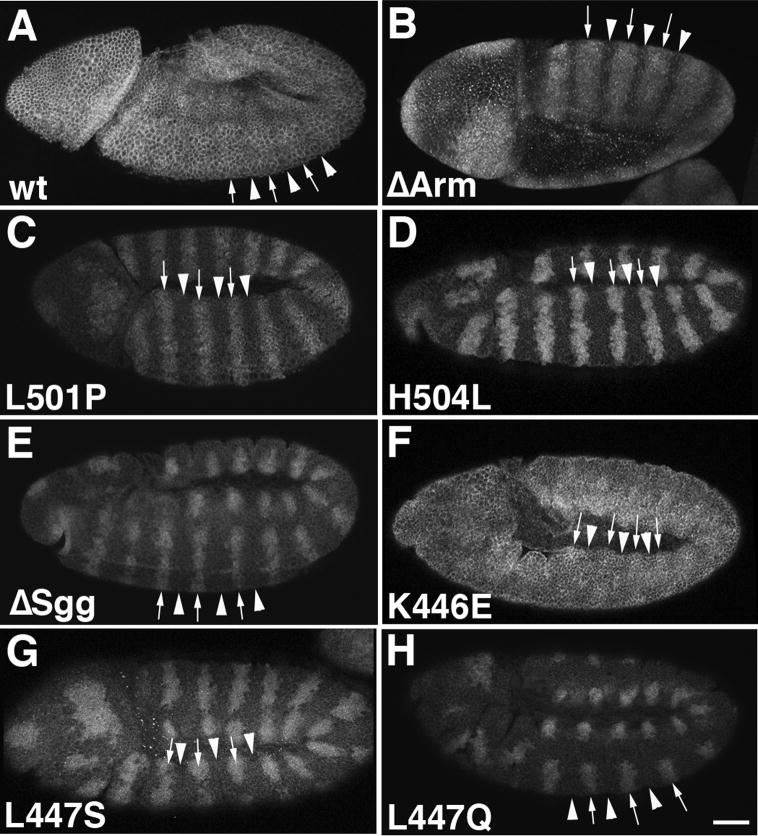
Fig. 3.
The width of Engrailed (En) stripes provides a measure for catalytic Axin activity. ~3.5 cells are competent to express Engrailed (e.g. in the axin mutant) but the wild type Axin complex limits Engrailed stripes to ~2 cells wide. Lateral or ventrolateral view of embryos stained using anti-Engrailed (En) antibodies. Arrowheads indicate the widths of En stripes. (A) Wild type (wt) embryo displaying normal En stripes. (B) An axinnull embryo ubiquitously expressing AxinΔArm (tub>AxinΔArm) shows normal width of En stripes. (C, D) Embryos expressing AxinL501P (C) or AxinH504L, respectively, also display the wild type En pattern. (E) An AxinΔSgg embryo shows widened En stripes (~3.5 cells wide). (F) An embryo expressing AxinK446E has a normal En pattern, whereas AxinL447S (G) and L447Q (H) have wide En stripes, as in AxinΔSgg (E) and the axinnull (not shown; Peterson-Nedry et al., 2008). (I) Quantification of the width of Engrailed stripes as a measure of Axin activity. The number of Engrailed-positive cells was quantified for the point mutants. We determined the data previously for wild type, axinnull, FLAxin and the domain deletions (Peterson-Nedry et al., 2008). Values for stripe width are: wt, 2.1, n=281; FLAxin rescue, 2.2, n=454; AxinΔArm, 2.2, n=566; AxinL501P, 2.2, n=516; AxinH504L 2.2, n= 514; AxinΔSgg 3.4, n=1013;AxinK446E, 1.83, n=451; AxinL447S, 3.7, n=486; AxinL447Q, 3.6, n=650.
Bar=40 μm.
Axin complex activity serves to maintain a low cytoplasmic pool of Armadillo (in contrast to Cadherin-associated Armadillo functioning in adhesion) by causing its degradation. Wg signal blocks Axin complex activity, which becomes apparent near stage 10 as segmentally repeated stripes of cytoplasmic Armadillo that is stabilized in cells that receive the Wg signal, but is degraded by axin in cells outside of this domain (Fig. 4A; Peifer et al., 1994). Loss of Axin s ability to assemble a functional destruction complex, as in axinnull mutants, results in stabilization, and uniformly high accumulation of Armadillo in all cells, and thus loss of the segmentally repeated striped pattern (Peterson-Nedry et al., 2008; Tolwinski and Wieschaus, 2001). We previously showed that AxinΔArm is able to rescue the ubiquitous stabilization of Armadillo seen in axinnull mutant embryos, causing degradation of Armadillo outside of the Wnt signaling domain and thus restoring the striped pattern observed in wild type embryos (Fig. 4B, E; Peterson-Nedry et al., 2008). In this study, we found that AxinL501P and AxinH504Lretained a similar ability to regulate Armadillo degradation (Fig. 4C,D), indicating these Axin point mutant proteins remain catalytically active and are also subject to regulation by Wg. AxinΔSgg, AxinK446E, AxinL447S, and AxinL447Q show a similar ability to restore Armadillo degradation outside of the Wg signaling domain (Fig. 4E–H), We note, however, that embryos expressing AxinΔSgg, AxinL447S, and AxinL447Q have lost the subtle differences between Armadillo levels in cells that do or do not receive the Wg signal and instead display very sharp boundaries between these domains (Fig. 4E,G,H). Notably, the same mutants show a reduced ability to negatively regulate engrailed expression (Fig. 3E, G,H). These observations might be explained if Axin mutants have reduced catalytic activity, which may allow even low levels of Wg to completely block destruction complex activity, thus leading to enhanced Armadillo stabilization and broadened expression of Engrailed, a notion requiring further quantitative analysis.
Fig. 4.
Axin function modulates Armadillo stability, which is essential for proper patterning. Stage 9–10 embryos were immunostained with anti-Armadillo (Arm) antibodies; each preparation is shown in lateral or ventrolateral view. (A) A wild type (wt) embryo, exhibiting the normal pattern of alternating stripes of Armadillo accumulation (arrows) and reduced levels of Armadillo (arrowheads). (B–H) axinnull embryos expressing maternally deposited tub>Axin* mutant protein. In all panels Armadillo stabilization in response to Wnt signaling is apparent indicating both retained catalytic activity (in areas of lowered Armadillo) as well as retained inhibition of Axin complex function by Wg signaling (domains of higher Armadillo levels). (B) Embryo rescued with AxinΔArm. (C) Embryo rescued with AxinL501P. (D) Embryo rescued with AxinH504L. (E) Embryo rescued with AxinΔSgg. (F) Embryo rescued with AxinK446E. (G) Embryo rescued with AxinL447S. (H) Embryo rescued with AxinL447Q. The disruption of Armadillo stripes in (E,G,H) is not evident in wild type (A) but correlates with fading of Wg expression in this domain (van den Heuvel et al., 1989) Bar=40 μm.
The ability of Axin* mutant proteins to pattern the cuticle mirrors their ability to regulate earlier molecular markers of Wg signaling
The interaction between the Wg signal and the Axin destruction complex, resulting in Armadillo stabilization at embryonic stage 10, determines whether the cells produce smooth cuticle or adopt the denticle fate. Formation of smooth cuticle depends on the ability of the Wg signal to block Axin complex activity; thus, encroachment of denticles into the smooth cuticle domain indicates loss of Axin s ability to be regulated by Wg. Conversely, denticle fate relies on the ability of Axin to assemble a catalytically active destruction complex, with diminished activity resulting in the formation of smooth cuticle in place of denticles. We previously observed that mutant forms of Axin lacking the entire binding site for Shaggy or Armadillo retained significant catalytic activity and remained subject to Wg regulation, as evidenced by their ability to rescue cuticle pattern in axinnull embryos (Fig. 5; Peterson-Nedry et al., 2008). Embryos expressing AxinΔArm remained clearly segmented but displayed infrequent loss of parts of denticle belts or individual denticles (Fig. 5B,B′; arrow and brackets). We scored the cuticles on a 1–10 scale where 1 reflects a total absence of denticles, 4 indicates loss of less than half a denticle belt, and 5 is wild type. On this scale, AxinΔArm scored 4.3, indicating about two thirds of embryos had some missing denticles (see Material and Material and methods for scoring criteria; Peterson-Nedry et al., 2008). When we examined axinnull embryos expressing AxinL501P or AxinH504L, they nearly always appeared wild type (scores of 5.0 and 4.9, respectively; Fig. 5C,D), suggesting that these mutants retain nearly full function (further explored below).
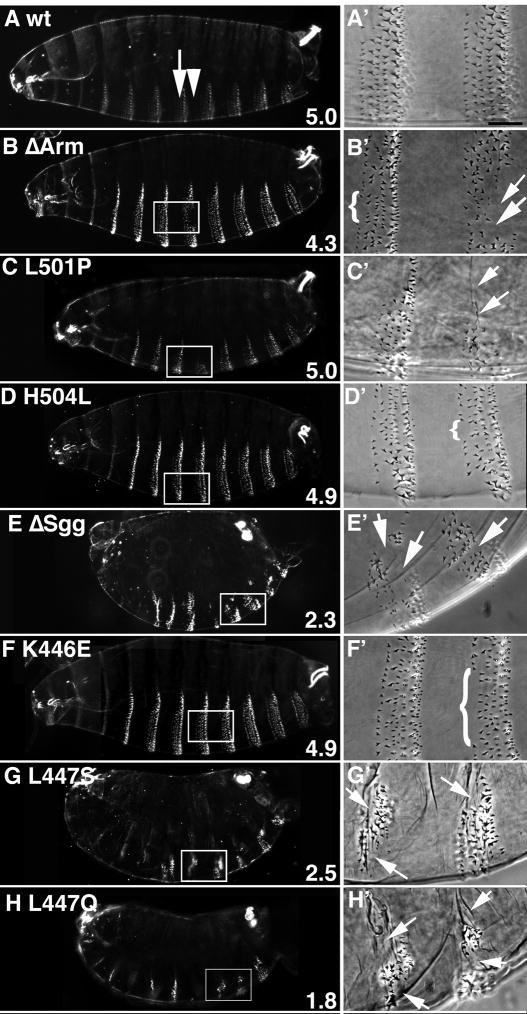
Fig. 5.
Axin mutant proteins with mutated contact residues nevertheless retain significant functional activity in vivo. The mutations in the Armadillo binding site, L501P and H504L resemble in phenotype the loss for the binding site (AxinΔArm). Similarly, L447S and L447Q produce a similar phenotype as AxinΔSgg, whereas AxinK446E resembles wild type. axinnull embryos expressing maternally deposited tub>Axin* mutant protein were scored for their ability to restore the segmentally repeated cuticle pattern of abdominal denticle belts, consisting of six rows of denticles (A,A′, arrow) alternating with smooth cuticle (arrowhead). Cuticle scoring criteria: The abdominal region, consisting of 8 denticle belts and 7 bands of smooth cuticle, was scored on 1–10 scale. Embryos were scored as follows: 1=loss of 7–8 denticle belts (=axin phenotype); 2=loss of 4–7 denticle belts; 3=loss of 1–3 denticle belts; 4=no loss of denticle belts, but minor loss of denticle within the belts; 5=wild type; 6=the presence of some ectopic denticles in smooth cuticle bands; 7=loss of 1–2 smooth cuticle bands; 8=loss of 3–4 smooth cuticle bands; 9=loss of 5–6 smooth bands; 10=no smooth cuticle (wingless phenotype). Bands were scored as missing if more than 50% of the denticle band was deleted. Phenotypic scores are indicated in the lower right corner of the dark field image.
(A) Wild type (wt). (B, B′) Embryos expressing AxinΔArm showed modest loss of denticles, including a frequent loss of row 1 denticles (brackets in panel B′), loss of posterior denticles (arrowhead) and replacement of part of the belt by smooth cuticle. (C) Embryo expressing AxinL501P displays loss of part of a denticle belt (arrows in C′). (H) An AxinH504L expressing embryo is lacking some row1 denticles (bracket in D′). (E) Embryos expressing AxinΔSgg cuticles showed a marked loss of denticles. (E′) A magnified ventral view of another embryo, showing significant portions of the abdominal belts did not form (arrows). (F) An embryo expressing AxinK446E appears nearly wild type except for the occasional loss of row 1 denticles (bracket in F′). (G) AxinL447S expressing embryo, which displays a marked loss of denticles (arrows in G′). (H) Similar to AxinΔSgg and AxinL447S, embryos expressing AxinL447Q also lack significant parts of denticle bands (arrows in H′). Bar=100 μm in panels A H; 25 μm in panels A′–H′.
In contrast to the wild type phenotype of embryos expressing Armadillo-domain binding mutants, embryos expressing AxinΔSgg displayed severe defects in cuticle pattern. Although segmentation remained clearly apparent, several denticle bands showed a loss of most denticles (Fig. 5E–E′; Peterson-Nedry et al., 2008). These defects indicated that the catalytic activity of AxinΔSgg was compromised, although regulation by Wg was retained. Analysis of embryos expressing AxinL447S and AxinL447Q revealed cuticle defects that were not significantly different from those observed in embryos expressing AxinΔSgg (Fig. 5G,H), indicating that substitution of this single amino acid, which we expect to be a contact amino acid in hydrophobic interaction with Shaggy, disrupted function to a similar extent as deletion of the entire Shaggy-binding domain. Thus, both mutations display the ‘null’ phenotype for the interaction domain. Surprisingly, a mutant form of Axin containing a charge reversal in another presumed contact amino acid, K446, generated embryos with infrequent loss of few denticles (bracket in Fig. 5F′) and therefore functioned nearly as wild type.
L447Q fails to render Drosophila Axin temperature-sensitive
The zebrafish Axin mutant masterblind contains an L→Q substitution at the equivalent position to Drosophila L447 that specifically inactivates zebrafish Axin at high temperatures (Heisenberg et al., 2001). This raised the possibility that the L447Q mutation, or mutation of other putative contact amino acids would render Axin temperature sensitive. We tested our five mutant Drosophila Axin proteins (K446E, L447S, L447Q, L501P, and H504L; Fig. 1) for the ability to rescue cuticle pattern of axinnull embryos when raised across the physiological temperature range for flies (18°C and 30°C) relative to their ability to rescue when raised at 25°C. We did not observe a significant temperature dependence of Axin function for any mutant (Suppl. Fig. 2). We conclude that none of these mutations render the Axin protein complex or the Axin protein itself temperature-sensitive.
Mutated contact amino acids in Axin can produce weak alleles in vivo
Our analysis so far revealed that three of the Axin mutants tested (L501P, H504L and K446E) generated the normal width of Engrailed stripes (Fig. 3C,D,F), regulated Armadillo accumulation (Fig. 4C,D,F), rescued normal cuticle pattern (Fig. 5C,D,F), and allowed a significant number of axin germ line clone embryos to hatch (Fig. 2). We next wished to know whether any of our mutants could replace wild type Axin during larval stages, metamorphosis, and in the adult organism. To do so, we asked whether expression of the Axin point mutant constructs could rescue axinnull animals if they had received maternally deposited wild type Axin. Here, maternal Axin would allow animals to complete embryogenesis and hatch; our Axin mutant proteins would replace wild type Axin during later stages of development and in adults. Flies expressing AxinH504L survived to adulthood without defects, indicating near-wild type function of this protein (Suppl. Fig. 3). Subsequently, axinnull animals expressing AxinH504L were mated with each other, producing a few larvae and misshapen pupae that subsequently died (not shown). This indicates that both sexes were fertile but AxinH504L fell short of fully rescuing axinnull animals. By contrast, no survivors were found for AxinL501P, AxinK446E, AxinL447S, or Axin447Q, indicating compromised function or dominant lethality (see below).
Heteroallelic complementation by weak Axin alleles
Above we examined the function of Axin mutant proteins in the axinnull mutant, which reveals retained function. Our previous work revealed an additional aspect of retained function: two mutant forms of Axin can complement each other by forming a heterodimeric complex. One such combination, AxinΔRGS + AxinΔArm, restored viability, demonstrating Axin dimerization can be functionally significant, at least in particular circumstances (Peterson-Nedry et al., 2008). We expected point mutations in the Armadillo binding domain would similarly restore viability in combination with AxinΔRGS unless the point mutation dominantly interfered with complex activity. Indeed, AxinH504L combined with AxinΔRGS could rescue axinnull mutants to adult viability and normal morphology (Fig. 6). However, unlike the domain deletion AxinΔArm, the point mutant AxinL501P failed to fully rescue in concert with AxinΔRGS. For some animals survival extended into the pupal stage, an improvement over expression of AxinΔRGS or AxinL501P in the axinnull mutant, which results in larval lethality. We interpret this somewhat unexpected behavior of AxinL501P as a combination of loss of a contact amino acid and a structural effect of proline in this particular position.
In our previous experiments with Axin deletion mutants we found no complementation between AxinΔSgg and AxinΔArm. We were therefore interested to determine whether point mutations in either domain complemented each other. We tested the mutations K446E, L447S and L447Q for complementation with L501P and H504L. Flies carrying AxinL447S or AxinL447Q were unable to rescue in combination with AxinL501P or AxinH504L (see Material and methods, not shown). Only the combination of the weakest alleles, AxinK446E and AxinH504L, was able to fully rescue flies (not shown). As noted above, AxinH504L comes close to rescuing the axin mutant and the finding that it synergizes with AxinK446E to permit full rescue underscores that both are weak alleles, that neither functions as a dominant negative (see also below), and that complementation between weak alleles can restore viability.
Stage-specific dominant interference with wild type signaling by certain Axin point mutant proteins
Above, we examined the function of Axin mutant proteins in the absence of wild type Axin, which is important because it reveals their intrinsic function. Another important question is whether Axin mutant proteins can dominantly interfere with the function of wild type Axin. In Drosophila, one wild type copy of Axin is sufficient to support normal development (Hamada et al., 1999) and we previously demonstrated that some Axin mutant proteins, when expressed at near-physiological levels, dominantly interfere with wild type Axin and disrupt development (Peterson-Nedry et al., 2008). Specifically, we previously observed that AxinΔArm and AxinΔSgg expression in wild type flies dominantly reduced survival rates (Fig. 7; Peterson-Nedry et al., 2008), which provided us with an approach to assess the dominant effect of mutated contact amino acids in vivo. We expressed the Axin point mutant proteins in wild type animals and compared survival rates to those of flies expressing AxinΔArm and AxinΔSgg. Two different expression approaches highlight context-specific effects of dominant negative Axin. Maternal co-expression of wild type and mutant Axin reveals interference with normal Axin function. The Axin complex needs to maintain the OFF state prior to the onset of Wg signaling at the cellular blastoderm stage and this activity is quantified by determining embryonic hatch rates (Fig. 7A). In contrast, zygotic expression of Axin mutant proteins reveals interference with the precise regulation of Wnt signaling during the remainder of development and becomes apparent as diminished survival to adulthood (Fig. 7B).
Fig. 7.
Dominant interference with developmental processes by Axin mutant proteins causes death. Mutant Axin proteins expressed at near-physiological levels can disrupt development if they interfere with destruction complex function and result in death (Peterson-Nedry et al., 2008). Conversely, weak Axin alleles do not interfere with the function of endogenous Axin.
(A) The effect of maternally expressed Axin point mutant proteins (tub>Axin*) alongside endogenous wild type Axin is assayed on the ability of embryos to complete development and hatch as larvae and are compared to rates for wild type (wt), full length Axin (FLAxin), as well as the deletion mutants AxinΔArm and AxinΔSgg (Peterson-Nedry et al., 2008). Values are standardize to embryos expressing wild type Axin () and for our reference points (FLAxin, AxinΔArm, AxinΔSgg) were published previously (Peterson-Nedry et al., 2008)). Values are tub>FLAxin, 100% ±1.8, n=554; AxinΔArm, 42.3% ±4.75, n=529; AxinL501P, 101% ±6.0, n=581; AxinH504L 81.9 ±3.3, n= 513; AxinΔSgg 57.8% ±3.5, n=529; AxinK446E, 84.6% ±2.1, n=568; AxinL447S, 89.3% ±4, n=578; AxinL447Q, 83.9% ±10.1, n=569.
(B) Zygotic expression of mutant proteins allows us to assay predominantly late embryonic and post-embryonic development, as the maternal contribution of wild type Axin is depleted. Rates of adult emergence are determined. Again, the mutant proteins are constitutively expressed in the presence of wild type Axin. Here, the point mutants AxinL501P, AxinH504L and AxinK446E do not interfere, while the Shaggy domain point mutants dominantly kill animals. An escaper of Axin L447Q expression is shown in Suppl. Fig. 4. Survival rates are determined by comparing tub>Axin* flies to siblings marked by the slightly detrimental dominant mutation Sp, which results in survival rates greater than 100% if dominant mutations in the control siblings reduce viability; (Materials and methods). Error bars represent standard deviation. Values are compared to FLAxin, AxinΔArm, AxinΔSgg from (Peterson-Nedry et al., 2008) and are derived to the number of surviving non-expressing control sibling (nc): FLAxin, 100% ±26, nc=442; AxinΔArm, AxinL501P, 108% ±9, nc=345; Axin504H, 101.2% ± 13.5, nc=494; 56.3% ±10, nc=417; AxinΔSgg, 4.1% ±5, nc=620; AxinK446E, 106% ±20, nc=362; AxinL447S, 0% ±0, nc=467; AxinL447Q, 1% ±1, nc=366.
Embryos maternally loaded with AxinΔArm hatch at a rate of only 42.3% (Peterson-Nedry et al., 2008) whereas maternal expression of AxinL501P (101% ±6.0) has little effect on survival, similar to FLAxin (Fig. 7A). By contrast, maternal expression of AxinH504L has a moderately dominant negative effect (hatch rate of 81.9%) intermediate between that of AxinΔArm and FLAxin. These survival rates indicate that AxinH504L interferes with regulation by wild type Axin present in these animals but that AxinL501P does not.
Embryos maternally loaded with AxinΔSgg hatch at a rate of only 57.8%, compared to wild type. By contrast, each of the Axin mutants with amino acid substitutions in the Sgg binding domain displayed only mild dominant negative effects (survival rates: AxinK446E, 84.6% ±2.1, n=568; AxinL447S, 89.3% ±4, n=578; AxinL447Q, 83.9% ±10.1, n=569). Thus, mutation of individual putative contact residues in the Armadillo or Shaggy binding domains of Axin generates proteins with no (L501P) or mild to moderate abilities to disrupt endogenous Axin complex function whereas deletion of the entire binding site generates proteins with much stronger dominant interfering effects.
Next, we examined how continued expression of these mutant proteins, starting with the onset of zygotic transcription, would affect subsequent development as reflected in rates of emergence as adult flies, about two weeks later (Fig. 7B). As we previously demonstrated (Peterson-Nedry et al., 2008), expression of AxinΔArm reduces survival to 56.3%. By contrast, zygotically expressed Axin containing point mutants in the Arm binding domain, AxinL501P and Axin504H did not interfere with development and behaved like wild type Axin. Zygotic expression of Axin lacking the entire Sgg binding domain, AxinΔSgg, dominantly interfered with endogenous Axin function and killed nearly all animals (4.1%). We saw an identical effect with two Axin point mutants in the Sgg binding domain, AxinL447S (0%) and AxinL447Q (1%), whereas a third mutant (AxinK446E) had no effect on survival (106%, relative to controls). Surviving animals had no visible defects, with exception of three L447Q escapers, which displayed partial or complete loss of sternites (Suppl. Fig. 4, and not shown), which differentiate in the ventral abdomen in response to Wg signaling. In this instance, AxinL447Q dominantly interferes with Wg signaling in the abdomen, even though other Wg-dependent tissues differentiate normally (Suppl. Fig. 4).
Taken together, both Axin[Arm*] point mutants L501P and H504L, as well as the Axin[Sgg*] point mutant K446E, do not dominantly interfere in signaling during development, whereas mutants L447S and L447Q both dominantly interfere with wild type signal regulation by Axin. Survival to adulthood appears to be more sensitive than to dominant interference by Axin mutants than is embryonic survival, possibly because of more extensive requirements for Wnt signaling during larval development and metamorphosis. The pronounced dominant lethal effect of AxinΔSgg is recapitulated by AxinL447S and AxinL447Q, in contrast to AxinK446E that appears to have no discernable dominant negative effect. These effects may be due to complex interactions with wild type Axin, which is present in these animals.
Discussion
Axin mutations that significantly disrupt destruction complex function
A major obstacle to predicting cancer risks is uncertainty in how point mutations affect protein function in vivo. Several dozen mutations identified in human Axin are associated with cancer (Salahshor and Woodgett, 2005), yet their causality remains unclear. Here, we identified point mutations in Drosophila Axin that disrupted the interaction with either Shaggy or Armadillo using yeast as an in vitro system, and then tested their function in an in vivo, developmental system. Because Wnt signal regulation is conserved in development and disease, our findings, which are summarized in Table 1, are likely to have relevance to understanding Wnt function in both contexts
Table 1.
| In vitro (Y2H) | Expression in wild type (dominant interference) | Expression in the axin null mutant (indicates retained function) | ||||||
|---|---|---|---|---|---|---|---|---|
| Interaction | Maternally: Embryonic survival | Zygotically: Survival to adult | Rescue of embryonic survival | Control of Engrailed expression | Control of Armadillo levels# | Cuticle rescue# | Rescue to viability | |
| Full length | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| ΔArm | - | ++ | ++ | ++ | ++++ | +++ | +++ | - |
| L501P | - | ++++ | ++++ | ++ | ++++ | ++(+) | ++++ | - |
| H504L | - | +++ | ++++ | ++ | ++++ | +++ | ++++ | ++ |
| ΔSgg | - | ++ | -* | - | - | ++ | ++ | - |
| K446E | - | +++ | ++++ | ++++ | ++++ | +++ | ++++ | - |
| L447S | - | +++ | - | + | - | ++ | ++ | - |
| L447Q | - | +++ | -* | - | - | ++ | ++ | - |
Semi-quantitative representation: ‘++++’, 100%; ‘+++’, >75%; ‘++, >50%; ‘+’, >25%;
escapers observed;
semi-quantitative
The finding that Axin mutant proteins retained significant function in vivo was not unexpected; it had been previously observed by us and is likely due to secondary interactions that promote complex assembly (Peterson-Nedry et al., 2008). Surprisingly, however, different mutations that disrupted interaction of Axin with Arm or Sgg in yeast displayed a spectrum of retained function relative to domain deletion mutants. Specifically, change of one amino acid (L447) produced the maximal effect, equivalent to loss of the interaction domain, two changes exhibited moderate loss of function (K446, L501), and one even appeared largely dispensable (H504) when compared to deletion of the entire binding site (Table 1). These findings were unanticipated because each of the four residues tested appears to be a contact residue for the interaction in vertebrates, as discussed below.
Importantly, a number of mutations in human Axin are associated with disease. Of 50 defects identified in human Axin, 32 are point mutations and four map to the GSK3β binding site whereas none are found in the β-catenin binding domain (reviewed in Salahshor and Woodgett, 2005). H394N, R395C, L396M and A398V are located in the same motif of Axin as is Drosophila L447; the first three are associated with colorectal cancer and the fourth with endometrial cancer. Are any or all of these mutations likely to have been causal in carcinogenesis? The most critical of these mutations is likely L394M, which disrupts a site of hydrophobic interaction with Shaggy, as does Drosophila L447S/Q. The Axin L447S/Q mutants show a severe reduction in function in Drosophila, equivalent to that of the domain deletion mutant (AxinΔSgg). Human Axin1-L394M would be expected to show a similarly severe loss of function. In contrast, the other three mutations would be expected to be weak alleles, similar to Drosophila K446E.
How can we identify Axin residues important in disease?
What factors determine whether a mutation is likely to contribute to disease? Different types of cancer or diseases are likely to be promoted by different levels of Wnt/β-catenin signaling. Colorectal cancer appears to depend on moderately raised, rather than maximal, levels of signaling; this is the ‘just right’ hypothesis based on the analysis of APC mutants in humans and flies (Albuquerque et al., 2002; McCartney et al., 2006). When equivalent APC mutants were tested in flies, they displayed partial loss of catalytic activity, raising signaling to levels similar to those of AxinΔSgg, AxinL447S and AxinL447Q (McCartney et al., 2006; Peterson-Nedry et al., 2008; this study). All other Axin domain deletions or point mutations tested by us (Peterson-Nedry et al., 2008; this study) retained more function and are therefore not be expected to promote colorectal carcinogenesis. By extension, we consider the human mutations H394N, R395C and A398V unlikely to independently promote colorectal cancer, although they could do so in concert with additional mutations in the Axin complex.
It is quite possible that different types of cancer depend on different levels of Wnt pathway activation. For example, several large deletions in Axin remove multiple domains and are associated with medulloblastoma (Salahshor and Woodgett, 2005); this may indicate an instance where the maximal level of signaling is tumor-promoting. Conversely, chronic diseases, such as increased bone mass or cardiovascular disease may result from subtle increases in Wnt signaling by mutant destruction complex components, similar to our weak alleles (L501P, H504L and K446E).
Our previous and current studies of the Axin complex highlight an important aspect of destruction complex assembly that likely applies to many, but not all protein complexes – as a scaffold protein, Axin brings together several other proteins that interact among themselves. These additional interactions provide for very robust complex assembly and explain why loss of contact residues or entire binding sites can be largely tolerated (Fig. 6E,F; Table 1; Peterson-Nedry et al., 2008; this study). Therefore many mutations, even those in seemingly critical positions, may only mildly interfere with human development and the viability of mutant cells or their potential to develop tumors may not be significantly affected.
Crystal structures may reflect in vitro interactions rather than function in vivo
Our mutational analysis in yeast identified amino acids critical for the interaction of Axin with Shaggy and Armadillo and these amino acids are contained within protein domains that are conserved with those of vertebrate Axin for which partial crystal structures are available. A co-crystal structure of Xenopus Axin and β-catenin peptides shows seven Axin residues that contact the β-catenin peptide with the motif LDXH at its core. Structural analysis suggests that this motif is critical for Axin/β-catenin interactions, and this is confirmed by immunoprecipitation of mutant peptides (Xing et al., 2003). Our saturating yeast two-hybrid screen identified two residues that are important for interaction with Armadillo in the LDXH motif of Drosophila Axin, L501 and H504; all others appeared redundant. However, even mutation of these two residues had only a modest effect when tested in transgenic flies under rescue conditions. AxinH504L even rescued zygotic axin mutants to viability and fertility, although their progeny failed to thrive.
Conservation in sequence and requirements for binding in vitro between vertebrate and Drosophila proteins suggests similar modes of molecular interaction between Axin and Shaggy/GSK3β. The co-crystal structure of human GSK3β with a 19 amino acid Axin peptide revealed that Axin binds to GSK3β through a α-helix hydrophobic interaction, hydrogen bonding, and a salt bridge (Dajani et al., 2003). Eight Axin residues appeared critical for the interaction as had been shown in vitro and, for one, genetically (Hedgepeth et al., 1999; Smalley et al., 1999; Heisenberg et al., 2001). Our in vitro analysis identified eight residues critical for the interaction in yeast; an additional five depended on synergy with secondary interactions. However, only four of our eight critically interacting amino acids (Drosophila L439, L443, K446, L447) matched those proposed by crystal structure analysis (Dajani et al., 2003). Two of those, R452 and D455, extend beyond the Axin peptide structure (Dajani et al., 2003) and thus suggest additional interactions with the kinase. It is possible the interactions of human and fly proteins do not exactly match. For example, the presumed Drosophila Axin α-helix contains a proline (P445) that is not present in the vertebrate structures, which may slightly perturb the helix. This change in structure may be sufficient to account for the apparent difference in interactions between human and Drosophila proteins.
The yeast two-hybrid system allowed us to identify critical interactions in vitro while available co-crystal structures further refined molecular understanding of the interactions. However, both in vitro data and crystal structures had limited value for predicting the activity of point mutants in vivo, For example, the Axin K446E mutation reverses an electrostatic charge that should destroy a predicted salt bridge between K446 of Axin and D262 of Shaggy (based on structural modeling of vertebrate proteins), and yet this had little effect on activity in vivo compared to mutations interrupting the hydrophobic interface between the two proteins in L447S/Q. The most likely explanation for this unexpected finding is that activity is modified by additional interactions between complex components. Although such additional interactions cannot be quantified, they can certainly never be excluded. Since unknown interactions are intrinsic to in vivo experiments, such tests are essential to understanding the activity of a mutant protein in its normal context in vivo. These findings highlight that crystal structures can help to identify critical amino acids for function but experimental verification in vivo is essential.
The analysis of Axin mutant proteins may serve as a model for the understanding of complex protein assemblies. Our current and previous studies demonstrated that (a) secondary interactions within the complex can compensate for defects in Axin, (b) heteroallelic complementation can occur between two mutant components, (c) dominant interference can disrupt wild type function and, (d) several of these different phenomena can occur simultaneously and depending on the particular mutations present. Therefore, disease-causing defects in constituents of multiprotein complexes require confirmation in a model system.
Material and methods
Axin constructs and identification of binding site mutants
FLAG-Axin and Axin deletion constructs were described previously (Peterson-Nedry et al., 2008). The yeast two-hybrid screen was conducted as described (Tolwinski et al., 2003). We followed the mutagenesis and screening protocol of Erdeniz et al. (2005) to generate and identify Axin mutants failing to interact with Armadillo- or Shaggy-bait. A 1 kb Axin fragment was amplified by mutagenic polymerase chain reaction (PCR); the product was directly recombined into a vector in yeast to generate a library. The library cells were mated to two yeast strains containing the bait plasmids, either Arm or Sgg, by replica plating. Differential loss of binding to one bait but not the other, was identified by differential lack of growth under selective conditions revealed by examination of the replica plates. Of ~10,000 clones screened, 401 clones were retested by Western blotting to eliminate lost interaction due to stop codons. In the yeast assay, 26 Axin mutant clones failed to produce the interaction with Arm, while 27 clones display lost interaction with Shaggy. On average, clones contained three mutations and thus we screened about 30,000 mutations in the mutagenized 1 kb region. To identify critical mutations, single point mutations were subsequently introduced into the Axin yeast two-hybrid prey plasmid using the Stratagene QuikChange® Site-Directed Mutagenesis kit. Of these, five mutations were assembled into the Drosophila constructs tub>w+>FLAG-Axin*.
Analysis of Axin point mutants in vivo
Simultaneous removal of endogenous Axin from the female germ line and induction of tub>Axin* constructs is as described previously (Peterson-Nedry et al., 2008; for an illustration, see Suppl. Fig. 4 in Peterson-Nedry et al., 2008), as are quantification of embryonic hatch rates, adult emergence rates, immuno-staining of embryos and cuticle analysis. Cuticle scoring criteria: The abdominal region, consisting of 8 denticle belts and 7 bands of smooth cuticle, was scored on 1–10 scale. Embryos were scored as follows: 1=loss of 7–8 denticle belts (=axin phenotype); 2=loss of 4–7 denticle belts; 3=loss of 1–3 denticle belts; 4=no loss of denticle belts, but minor loss of denticle within the belts; 5=wild type; 6=the presence of some ectopic denticles in smooth cuticle bands; 7=loss of 1–2 smooth cuticle bands; 8=loss of 3–4 smooth cuticle bands; 9=loss of 5–6 smooth bands; 10=no smooth cuticle (wingless phenotype). Bands were scored as missing if more than 50% of the denticle band was deleted.
Immunohistochemistry
Antibodies and procedures were as described Peterson-Nedry et al., (2008), except that some pictures were collected on an Olympus Fluoview FV1000 laser scanning confocal microscope.
Photography
Cuticles were photographed on an Axioplan2 microscope with an AxioCam MRm Zeiss digital camera. Photographs of living flies were collected as Z-stacks using a Leica MZFL-III stereomicroscope and photographed with an Optronics Magna Fire CCD Camera; Z-stack projections were then generated using an Image Pro Plus workstation. Blank areas in pictures are artifacts produced by the projection algorithm. Photomontages were assembled using Adobe Photoshop CS3.
Supplementary Material
Supplemental Fig. 1
Western blot analysis reveals similar levels of construct expression. Expression was induced in females and eggs collected to assess maternally and zygotically expressed protein. Axin constructs were detected by their FLAG tag and α-catenin was used as input control.
Supplemental Fig. 2.
Mutant Axin proteins are not temperature-sensitive. Axin mutant embryos expressing the Axin point mutants AxinL501P, AxinH504L, AxinK446E, AxinL447S or AxinL447Q developed at the indicated temperature. The phenotypic score of cuticle preparations was then determined. Error bars represent standard deviation. Note that the 1–10 scale is not linear but rather magnifies subtle changes (scores 4–6, loss of single denticles rather than denticle belts), which is reflected in larger error bars.
Supplemental Fig. 3.
AxinH504L rescues zygotic axin mutants to adulthood without visible defects (though it fails to rescue maternal/zygotic axin mutants). tub>AxinH504L expression was induced in flies receiving maternal wild type Axin with one quarter of the flies homozygous mutant for axin. Two tub>AxinH504L; axin/axin rescued flies are shown. (A) Male fly displaying fully formed legs with tarsal claws at their distal tip (arrowheads) and wild type wings including an intact wing margin (arrows). (B) The head of the fly shown in (A) has wild type antenna (arrowhead) and eyes (arrow) with the regular array of lenses. (C) The ventral abdomen of a rescued female shows wild type sternites (arrowheads) with regular bristles (arrows). Also visible is the wild type wing margin (large arrowhead). Blank areas, e.g. adjacent to the legs, are artifacts of the projection.
Supplemental Fig. 4
Subtle defects are observed in a rare zygotically expressing tub>AxinL447Q escaper. (A) Ventral view of the fly shows wild type legs (black arrows) and wings with intact margins (white arrows) (the wings are curved up, due to the dominant marker Curly). (B) On the head of the fly, with wild type antenna (arrowhead) and a compound eye (arrow) with regular arrays of lenses is visible, indicating normal differentiation. (C) The only defects observed are in the ventral abdomen where sternites (black arrow) and their bristles (white arrows) require Wg signaling and are absent (large arrowheads) or partially formed (small arrowheads). Z stacks were collected and projected, which results in some unstructured areas with sharp boundaries, for example below the head in (B).
Acknowledgments
We are very grateful to Jackie Parker for editing, Drs. David Morton and Jan Christian for comments on the manuscript. We also acknowledge the Bloomington Stock Center and the Developmental Studies Hybridoma Bank (DSHB) for providing us with fly stocks and antibodies, respectively. We thank Drs. Jan Christian, Philip Copenhaver, Richard Maurer, Caroline Enns, Sarah Smolik, Mike Forte and David Morton for discussions and advice. Kaitlin Leonard and Jean Kitzman provided excellent technical assistance. This work was supported by NIH grant GM67029 to M.W.
Footnotes
Author contributions
M.W. and N.E. conceived and designed the experiments. S.A.K, N.E., W.P.-N., S.B.-L., E.A.S and M.W. performed the experiments. S.A.K, N.E., W.P.-N. and M.W. analyzed the data. M.W. wrote the paper.
Competing interests
The authors have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, Leitao CN, Fodde R, Smits R. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet. 2002;11:1549–60. doi: 10.1093/hmg/11.13.1549. [DOI] [PubMed] [Google Scholar]
- Baron R, Rawadi G, Roman-Roman S. Wnt signaling: a key regulator of bone mass. Curr Top Dev Biol. 2006;76:103–27. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- Cliffe A, Hamada F, Bienz M. A role of Dishevelled in relocating Axin to the plasma membrane during wingless signaling. Curr Biol. 2003;13:960–6. doi: 10.1016/s0960-9822(03)00370-1. [DOI] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development. 2004;131:5103–15. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. EMBO J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–72. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci. 2007;12:471–91. doi: 10.2741/2076. [DOI] [PubMed] [Google Scholar]
- Erdeniz N, Dudley S, Gealy R, Jinks-Robertson S, Liskay RM. Novel PMS1 alleles preferentially affect the repair of primer strand loops during DNA replication. Mol Cell Biol. 2005;25:9221–31. doi: 10.1128/MCB.25.21.9221-9231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Jho E, Zeng L, Kurth T, Joos T, Kaufmann C, Costantini F. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J Cell Biol. 1999;145:741–56. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- Glass DA, 2nd, Karsenty G. Molecular bases of the regulation of bone remodeling by the canonical Wnt signaling pathway. Curr Top Dev Biol. 2006;73:43–84. doi: 10.1016/S0070-2153(05)73002-7. [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–41. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F, Tomoyasu Y, Takatsu Y, Nakamura M, Nagai S, Suzuki A, Fujita F, Shibuya H, Toyoshima K, Ueno N, Akiyama T. Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science. 1999;283:1739–42. doi: 10.1126/science.283.5408.1739. [DOI] [PubMed] [Google Scholar]
- Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–81. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- Hedgepeth CM, Deardorff MA, Rankin K, Klein PS. Regulation of glycogen synthase kinase 3beta and downstream Wnt signaling by axin. Mol Cell Biol. 1999;19:7147–57. doi: 10.1128/mcb.19.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, Dale TC, Wilson SW, Stemple DL. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001;15:1427–34. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius MA, Schelbert B, Hsu W, Fitzpatrick E, Jho E, Fagotto F, Costantini F, Kitajewski J. Domains of axin and disheveled required for interaction and function in wnt signaling. Biochem Biophys Res Commun. 2000;276:1162–9. doi: 10.1006/bbrc.2000.3607. [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Malbon CC, Wang HY. Dishevelled: a mobile scaffold catalyzing development. Curr Top Dev Biol. 2006;72:153–66. doi: 10.1016/S0070-2153(05)72002-0. [DOI] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–82. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–9. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- McCartney BM, Price MH, Webb RL, Hayden MA, Holot LM, Zhou M, Bejsovec A, Peifer M. Testing hypotheses for the functions of APC family proteins using null and truncation alleles in Drosophila. Development. 2006;133:2407–18. doi: 10.1242/dev.02398. [DOI] [PubMed] [Google Scholar]
- Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Peterson-Nedry W, Erdeniz N, Kremer S, Yu J, Baig-Lewis S, Wehrli M. Unexpectedly robust assembly of the Axin destruction complex regulates Wnt/Wg signaling in Drosophila as revealed by analysis in vivo. Dev Biol. 2008;320:226–41. doi: 10.1016/j.ydbio.2008.05.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, Lee SJ, Lee J, Oh S, Han JK, Park BJ, Weis WI, Ha NC. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS ONE. 2008;3:e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Sakanaka C, Weiss JB, Williams LT. Bridging of beta-catenin and glycogen synthase kinase-3beta by axin and inhibition of beta-catenin-mediated transcription. Proc Natl Acad Sci U S A. 1998;95:3020–3. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahshor S, Woodgett JR. The links between axin and carcinogenesis. J Clin Pathol. 2005;58:225–36. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat Struct Mol Biol. 2007;14:484–92. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- Smalley MJ, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, Fryer LG, Hutchinson L, Fry MJ, Dale TC. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J. 1999;18:2823–35. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits R, Hofland N, Edelmann W, Geugien M, Jagmohan-Changur S, Albuquerque C, Breukel C, Kucherlapati R, Kielman MF, Fodde R. Somatic Apc mutations are selected upon their capacity to inactivate the beta-catenin downregulating activity. Genes Chromosomes Cancer. 2000;29:229–39. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1033>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. Wingless gradient formation in the Drosophila wing. Curr Biol. 2000;10:293–300. doi: 10.1016/s0960-9822(00)00378-x. [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3beta activity. Dev Cell. 2003;4:407–18. doi: 10.1016/s1534-5807(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Tolwinski NS, Wieschaus E. Armadillo nuclear import is regulated by cytoplasmic anchor Axin and nuclear anchor dTCF/Pan. Development. 2001;128:2107–17. doi: 10.1242/dev.128.11.2107. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Nusse R, Johnston P, Lawrence P. Distribution of the wingless gene product in Drosophila embryos: A protein involved in cell-cell communication. Cell. 1989;59:739–749. doi: 10.1016/0092-8674(89)90020-2. [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–30. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Willert K, Logan CY, Arora A, Fish M, Nusse R. A Drosophila Axin homolog, Daxin, inhibits Wnt signaling. Development. 1999;126:4165–73. doi: 10.1242/dev.126.18.4165. [DOI] [PubMed] [Google Scholar]
- Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753–64. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Matsuda Y, Ishimoto A. Biochemical characterization of the Drosophila axin protein. FEBS Lett. 2000;474:189–94. doi: 10.1016/s0014-5793(00)01601-x. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–44. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–7. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1
Western blot analysis reveals similar levels of construct expression. Expression was induced in females and eggs collected to assess maternally and zygotically expressed protein. Axin constructs were detected by their FLAG tag and α-catenin was used as input control.
Supplemental Fig. 2.
Mutant Axin proteins are not temperature-sensitive. Axin mutant embryos expressing the Axin point mutants AxinL501P, AxinH504L, AxinK446E, AxinL447S or AxinL447Q developed at the indicated temperature. The phenotypic score of cuticle preparations was then determined. Error bars represent standard deviation. Note that the 1–10 scale is not linear but rather magnifies subtle changes (scores 4–6, loss of single denticles rather than denticle belts), which is reflected in larger error bars.
Supplemental Fig. 3.
AxinH504L rescues zygotic axin mutants to adulthood without visible defects (though it fails to rescue maternal/zygotic axin mutants). tub>AxinH504L expression was induced in flies receiving maternal wild type Axin with one quarter of the flies homozygous mutant for axin. Two tub>AxinH504L; axin/axin rescued flies are shown. (A) Male fly displaying fully formed legs with tarsal claws at their distal tip (arrowheads) and wild type wings including an intact wing margin (arrows). (B) The head of the fly shown in (A) has wild type antenna (arrowhead) and eyes (arrow) with the regular array of lenses. (C) The ventral abdomen of a rescued female shows wild type sternites (arrowheads) with regular bristles (arrows). Also visible is the wild type wing margin (large arrowhead). Blank areas, e.g. adjacent to the legs, are artifacts of the projection.
Supplemental Fig. 4
Subtle defects are observed in a rare zygotically expressing tub>AxinL447Q escaper. (A) Ventral view of the fly shows wild type legs (black arrows) and wings with intact margins (white arrows) (the wings are curved up, due to the dominant marker Curly). (B) On the head of the fly, with wild type antenna (arrowhead) and a compound eye (arrow) with regular arrays of lenses is visible, indicating normal differentiation. (C) The only defects observed are in the ventral abdomen where sternites (black arrow) and their bristles (white arrows) require Wg signaling and are absent (large arrowheads) or partially formed (small arrowheads). Z stacks were collected and projected, which results in some unstructured areas with sharp boundaries, for example below the head in (B).