Abstract
In eukaryotes the proteasome is responsible for the degradation of many proteins that are targeted for turnover by post-translational modification with ubiquitin. A similar system was identified in Mycobacterium tuberculosis and has shown to be essential for the pathogenesis of this bacterium. Here, we overview the current information of the Mtb proteasome and discuss the role of this protease in pathogenesis.
Keywords: Proteasome, prokaryotic ubiquitin-like protein, Pup, tuberculosis
Introduction
Mycobacterium tuberculosis (Mtb) is among the leading causes of death in the world and as a consequence, it is a priority for investigators to find new targets for chemotherapy of increasingly drug-resistant strains. In order to identify cellular pathways that could be targeted for drug development, a screen of Mtb transposon mutants for genes encoding proteins that provide defense against nitric oxide (NO) and other reactive nitrogen intermediates (RNI) was performed [1]. RNI are produced by macrophages to slow the growth of invading pathogens and are thought to inhibit microbial growth by damaging nucleic acids, proteins, and lipids (reviewed in [2]). Importantly, the production of NO by the inducible nitric oxide synthase (iNOS) in macrophages is essential to control Mtb growth in mice [3]. Although wild type mice survive much longer than iNOS-deficient mice after Mtb infection, bacteria are rarely sterilized from these immunocompetent animals. This observation suggested that Mtb contains defenses to resist eradication by NO. The screen for NO sensitive Mtb mutants identified two proteins putatively associated with proteasome function: Mpa (Mycobacterium proteasome ATPase) and PafA (proteasome accessory factor A). Significantly, several groups also determined that mutants of the proteasome pathway are severely attenuated in mice [1, 4–7]. These results provided evidence that the bacterial proteasome is essential for resisting the toxic effects of NO and causing disease in mice, making the mycobacterial proteasome a plausible candidate for the rational development of drugs targeting tuberculosis.
In this review we discuss the most recent literature concerning the Mtb proteasome system, and speculate how regulated proteolysis by this highly conserved protease may be required for Mtb pathogenesis.
Proteasomes: chambered proteases found in all domains of life
Proteasomes are found in all eukaryotes and Archaea but only in some bacteria of the order Actinomycetales. In eukaryotes the 26S proteasome is composed of two functionally distinct sub-complexes: the 20S core particle (CP), where proteins are degraded, and the 19S regulatory particle (RP) that binds ubiquitylated substrates to be degraded by the 20S CP [8, 9] The 20S CP is a self-compartmentalized broad-spectrum protease that has a cylindrical structure [9]. It is composed of four stacked rings with catalytic activity located within the center of the cylinder. The two inner rings are composed of seven distinct catalytic β-subunits while the outer two rings are composed of seven distinct α-subunits[9] The β-subunits have post-acidic, tryptic, and chymotryptic activities, imparting the capacity of the proteasome to cleave most types of peptide bonds. The main function of the α rings is to form a gated channel that controls the passage of unfolded substrates and cleaved peptides in and out of the proteolytic cylinder. The α rings also serve as a docking surface for protein complexes such as the 19S RP, which interacts with either entrance into the 20S CP [9, 10]
The 19S RP is responsible for recruiting substrates and regulating the opening of the 20S CP. It is composed of multiple ATPase (Rpt) and non-ATPase (Rpn) subunits. The ATPase subunit is formed by six distinct ATPases associated with different cellular activities (AAA+) that form a ring and interacts with the α-subunits of the 20S CP [8]. Most importantly, the 19S RP contains numerous proteins that bind and remove Ub from substrates prior to degradation by the 20S CP.
20S CPs in Archaea and Actinomycetes are similar in sequence and structure to those in eukaryotes, but are composed of homo-heptameric β-subunit rings stacked between homo-heptameric rings of α-subunits [reviewed in [11, 12]]. In bacteria, 20S CPs have been characterized for the genera Frankia [13], Rhodococcus [14], Streptomyces [15], and Mycobacterium [16]. With one exception to be discussed, the presence of only one type of β-subunit limits the 20S CP to chymotryptic activity.
The best-characterized prokaryotic AAA+ ATPases is PAN (proteasome activating nucleotidase) from the Archaeon Methanococcus janaschii [17]. PAN can associate with Thermoplasma 20S CP in vitro and induce gate opening, unfolding and translocation of substrates to the catalytic domains of the proteasome [17–19]. In contrast, no bacterial proteasome has ever been shown to interact with its cognate ATPase or promote the degradation of a polypeptide. The mechanism by which substrates are recognized for degradation is still unknown because regulatory complexes that associate with PAN or other prokaryotic proteasome-associated ATPases have not been identified.
The structure of the Mtb 20S CP has been solved and, like other prokaryotic proteasomes, is composed of only one type of α-subunit encoded by prcA and one type of β-subunit encoded by prcB [20]. These subunits interact with each other to form a canonical 20S CP [16, 20]. Using different peptide substrates Lin and colleagues demonstrated that the Mtb proteasome has post-acidic, tryptic and chymotryptic protease activity [16]. This broad specificity resembles that of eukaryotic proteasomes rather than the limited chymotryptic specificity present in other prokaryotic proteasomes. However, it is not yet known how a single type of β-subunit is able to confer such a broad spectrum of protease activity. Importantly, proteolytic activity was best observed upon deletion of the N-terminal eight amino acids of the α-subunit [16]. This suggested that the α-ring effectively blocks access of even small peptide substrates to the 20S CP. This observation strongly supports the hypothesis that an activating protein or proteins are needed to open the α-ring for substrate degradation.
Proteasomes are essential for eukaryotic life, but its requirement in bacterial functions and/or survival appears to be varied. For example, it has been reported that deletion of the prcBA genes from Mycobacterium smegmatis (M. smegmatis), a non-pathogenic relative of Mtb, did not severely impact bacterial growth [21, 22]. In contrast, chemical or genetic silencing of Mtb proteasome activity dramatically slowed bacterial growth in vitro and in vivo [1, 6]. Interestingly, unlike M. smegmatis, Mtb lacks Lon, another ATP-dependent chambered protease, which may explain the non-essentiality of the M. smegmatis proteasome.
Proteins associated with Mtb proteasome function
The mpa and pafA genes were inferred as proteasome associated given their close vicinity to the 20S CP prcBA genes, and that they are present only in bacteria containing proteasomes [23]. Furthermore, mutations in either gene resulted in identical NO-sensitive and in vivo attenuated phenotypes, suggesting that both of them participate in the same pathway.
Mycobacterium proteasome ATPase (Mpa)
Mpa is an ATPase similar to ATPases present in the eukaryotic 19S RPs [4]. Studies using cryo-electron microscopy revealed that Mpa forms hexameric rings similar to other AAA ATPases [4, 24]. Additionally, Mpa contains Walker A and B motifs involved in the binding and hydrolysis of ATP [24, 25]. Mutations within these motifs or S-nitrosylation of Mpa inhibited the ability of Mpa to hydrolyze ATP in vitro [4, 26]. Most importantly, it has been shown that the ATPase activity is essential for RNI resistance [4].
Although structural analysis predicts that Mpa physically interacts with the proteasome 20S CP and plays a role in binding, unfolding and translocating substrates into the 20S CP, this has not yet been experimentally shown. Interestingly, mpa607, an mpa mutant identified in the NO screen with a transposon insertion in its penultimate codon, had the same phenotype as the mpa null strain; it was sensitive to RNI in vitro and highly attenuated in the mouse model of infection [1, 4]. The mpa607 mutant lacks two terminal amino acids, tyrosine (Tyr) and leucine [1]. Further analysis revealed that the penultimate Tyr is also found in several ATPase subunits of the eukaryotic 19S RP, most Archaeal proteasome-associated ATPases and all bacterial proteasome-associated ATPases [5]. In Archaea, the penultimate Tyr of PAN was shown to be essential for activating the opening of the 20S CP [27]. When the penultimate Tyr of Mpa was mutagenized to either phenylalanine or glutamate (Glu), Mtb was sensitized Mtb to RNI in vitro to a similar degree as the mpa null mutant [5]. Taken together, these studies suggest that the C-terminus of Mpa may interact with the 20 CP in order to open and deliver proteins into the catalytic core for degradation.
Prokaryotic ubiquitin-like protein (Pup)
Until very recently it was a mystery how proteins were targeted to prokaryotic proteasomes since ubiquitin (Ub) or ubiquitin-like modifiers (Ubl) had not been conclusively found in prokaryotes. Most proteins degraded by the eukaryotic proteasome are first modified at one or more lysines through the covalent attachment of Ub, a 76 amino acid protein that tags proteins for selective turnover. Ub is ligated to substrates in a tightly regulated, multi-step ATP-dependent cascade involving activation (E1) and conjugation (E2, E3) enzymes (reviewed in [28]). But attachment of a single Ub to a substrate Lys is not generally sufficient to initiate degradation; a number of Ubs need to be added, therefore, after attachment of an initial Ub moiety to a substrate, additional activated Ubs are ligated to lysines (usually Lys48) of the previously conjugated Ub molecule. These polyUb chains serve as an unambiguous trigger for proteolysis by the 26S proteasome [8].
Using an E. coli bacterial two-hybrid screen of an Mtb genomic DNA library searching for potential binding partners of Mpa, Pearce and colleagues discovered the first prokaryotic ubiquitin-like protein, Pup, a 64 amino acid protein that modifies and targets mycobacterial proteins to the proteasome for degradation [29]. BLASTP analysis [30] identified Pup homologues only in Actinobacteria. Furthermore, pup is part of a putative operon together with the proteasomal genes prcBA, supporting its association with proteasome function. Although Pup and Ub are similar in size, they lack sequence and structural homology [31, 32]. Furthermore, the last three C-terminal residues of Pup are Gly-Gly-glutamine (Gln), while the last two C-terminal residues of proteolytically activated ubiquitin and other ubiquitin-like modifiers is Gly-Gly. Pearce and colleagues showed that Pup interacts both non-covalently and covalently with Mpa (which is also a degradation substrate), and covalently with the degradation substrate FabD. Mass spectrometry analysis revealed that Pup formed an isopeptide bond between its C-terminus and the ε-amino group of a specific lysine residue (Lys173) of FabD. Surprisingly, the C-terminal residue of Pup participating in the linkage was glutamate (Glu), indicating that the C-terminal Gln was deamidated before or during conjugation to FabD. Purification of recombinant Pup from mycobacteria and E. coli revealed that the C-terminal Gln of Pup was specifically deamidated in mycobacteria, suggesting the presence of a specific Pup deamidase [29].
Further studies showed that multiple “pupylated” proteins accumulated in the Mtb mpa mutant strain, indicating that Pup-dependent proteasomal degradation was not limited to FabD. In a parallel study, Burns and colleagues showed that M. smegmatis Pup was conjugated to superoxide dismutase (SodA) and myoinositol-1-phosphate synthase (Ino1) using the same Gly-Gly-Glu~Lys linkage, and that pupylated proteins also accumulated in a prcBA mutant compared to wild type M. smegmatis [21]. Deletion of the Pup C-terminal Gln inhibited “pupylation”, confirming the importance of this amino acid in Pup conjugation [21].
These exciting findings showed that pupylation is functionally similar to ubiquitylation in that substrates are covalently conjugated with a small protein modifier at lysines in order to target them for degradation. However, this process differs from ubiquitylation in that Pup is not proteolytically activated to reveal a C-terminal Gly-Gly motif, but is rather activated and conjugated to target proteins via a C-terminal glutamate.
Proteasome accessory factor A (PafA) and de-amidase of Pup (Dop)
Enzymes similar to those involved in the activation and conjugation of Ub to proteasome substrates have not been described, suggesting that different enzymatic activities participate in pupylation. Pup-conjugated substrates could not be detected in an Mtb pafA mutant, demonstrating that PafA is essential for pupylation [29]. A bioinformatic analysis found that PafA is likely to be structurally related to γ-glutamyl-cysteine synthetase-2, suggesting that it is member of the carboxylate-amine ligase superfamily [33, 34]. Based on this observation it was proposed that PafA phosphorylates the γ-carboxyl group of the C-terminal Glu of de-amidated Pup in an ATP-dependent manner; this “activated” form of Pup would then react with the amino group of the target Lys to form an isopeptide bond [33]. However, it is not known whether the last Glu of Pup attaches by its α- or γ-carboxylate. In addition, the mechanism of Glu~Lys bond formation is not yet understood.
The observation that most Pup molecules were deamidated at the C-terminus when purified from mycobacteria but not E. coli suggested the presence of a specific enzyme required for this process [29]. Using Pup as bait for in vitro pull down experiments with Mycobacterium bovis lysates, Striebel and colleagues identified Dop (deamidase of Pup) that converted the C-terminal Gln of Pup to Glu [35]. Dop was predicted to be involved in proteasomal protein degradation because it is highly similar to PafA and encoded immediately upstream of pup and the 20S CP genes (reviewed in [34]). Interestingly Dop required the presence of ATP for the deamidation of Pup, but ATP hydrolysis was not detected [35]. This was supported by the observation that ATPγS, a non-hydrolyzable ATP analogue, also allowed the deamidation to proceed, but at a slower rate. Similar to PafA, the catalytic mechanism of Pup deamidation by Dop remains to be characterized.
A model for proteasomal degradation in Mtb
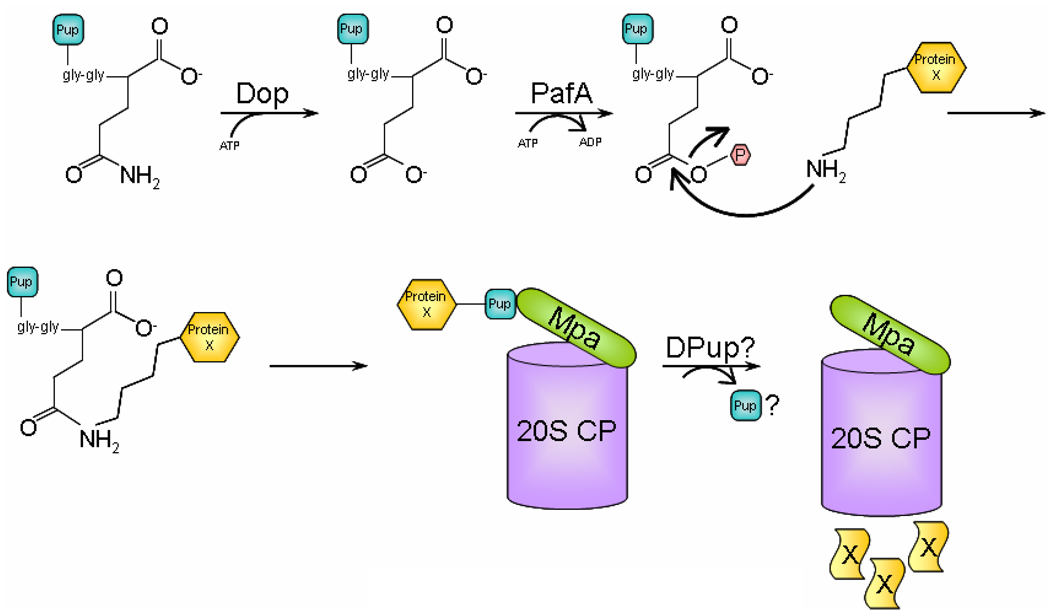
Based on the current data, we propose the following model for pupylation: Pup is synthesized with a Gln residue at the C-terminus, which is deamidated by Dop, resulting in a Glu residue at the C-terminus of Pup (Fig. 1). Subsequently, PafA phosphorylates the γ-carboxylate of the Glu residue, forming an intermediate that would favor nucleophilic attack by the amino group of a Lys residue present in the substrate destined to be degraded. The active site amino acids in Dop and PafA involved in catalysis remain unknown. Pup then interacts with Mpa, facilitating the delivery of substrate to the 20S CP. It is not yet known if Pup, like Ub, is removed prior to degradation or is also degraded.
Fig. 1. Model for protein degradation by the Pup-proteasome system (PPS).
Tagging of a proteasome substrate, such as Protein X, is initiated with deamidation of Pup by Dop and ATP binding. Deamidated Pup then binds to PafA that with ATP hydrolysis catalyzes conjugation of a Pup monomer to Protein X. PafA facilitates the nucleophilic attack by an amino group present in Protein X to the γ-carboxylate of the C-terminal Glu residue in Pup. As a result, Protein X gets mono-pupylated; it is unknown if poly-Pup chains can form. Subsequently, Pup binds to Mpa, which might promote unfolding and translocation of Protein X into the 20S CP. During translocation, Pup might be removed by de-pupylating (DPUP) enzymes. Finally, Protein X is degraded to small peptides by the proteolytic activity in the central chamber of the 20S CP.
Striebel and colleagues noted that Dop is present in bacteria encoding Pup that naturally terminates in Glu [35]. This suggests that Dop has another function in addition to Pup deamidation. Perhaps Dop is required, along with PafA, to ligate Pup to substrates. In vitro data suggest that this is not the case because PafA was sufficient to conjugate recombinant Pup terminating with Glu to FabD [35]. Analysis of a dop mutant will be required to ultimately determine if it indeed has functions in addition Pup deamidation. Finally, several bacterial species encode PafA and Dop but do not have conspicuous Pup or proteasome homologues [34]. It is not known if pafA and dop are expressed in these bacteria, or if they have pupylation or proteasome independent functions.
The link to pathogenesis?
It is likely that the proteasome is required for the virulence of Mtb for numerous reasons. One possibility is that the bacterial proteasome is required to degrade one or many proteins that are toxic after nitrosative or oxidative damage. Another possibility is that certain proteasomal substrates must be degraded in order to allow other proteins to participate in NO detoxification or to repair macromolecules damaged by NO. These may include transcriptional repressors of genes that synthesize DNA repair enzymes or other proteins that must replace essential proteins damaged by RNI.
It is important to note that RNI are not the only stresses that Mtb encounters in vivo. Macrophage acidification [36], the production of reactive oxygen species [37] and even eukaryotic Ub peptides [38] can participate in the control of Mtb growth. The Mtb proteasome may regulate the expression of genes that counteract these host defenses. Mtb has also long been believed to suppress the host immune response to favor its persistence [39, 40]. It would not be surprising if the Mtb proteasome transcriptionally or post-translationally regulated the production of secreted or surface exposed proteins that modulate host immunity. Because all ATP-dependent proteases, including ClpP, Lon, FtsH, and HslV, degrade transcription factors in numerous non-pathogenic and pathogenic bacteria (reviewed in [12, 41]), it is possible that the Mtb proteasome is involved in the regulation of several pathways. Transcriptome and proteome analysis of proteasome-deficient strains will begin to test this hypothesis.
It is notable that only a handful of pupylated substrates have been identified so far, and there is no obvious link between them and Mtb virulence. The identification of the “pupylome” using epitope tagged Pup and high-resolution mass spectrometry should reveal how many proteins rely on Pup for regulation or turnover. The identity of these proteins may ultimately begin to explain why defects in protein degradation attenuate Mtb virulence in vivo.
Conclusions
The discovery of proteasomal degradation as a requirement for Mtb virulence has spawned several exciting areas of research, in particular, the understanding of how this activity participates in pathogenesis and the characterization of a novel post-translational modification system. It is likely that the disruption of proteasome function has pleiotropic effects and it will take time to determine which are responsible for the attenuation of Mtb in mice. In contrast, there has been remarkable progress in defining the biochemistry of the Pup-proteasome system (PPS). Yet, there is much more to learn about this system. Although it is functionally similar to the eukaryotic Ub-proteasome system (UPS), the biochemistry that targets proteins for degradation appears quite different. As a result, many important questions associated with crucial steps in the path to the proteasome have arisen. For one, how does PafA select proteins for pupylation? Higher eukaryotes have hundreds if not over 1,000 E3 ligases that provide specificity to the UPS, how is this achieved in Mtb? PafA could possibly form dimers or hetero-dimers with Dop [33, 42]. If this were true, we could speculate that these proteins form a complex that binds both the substrate and Pup for conjugation, and additional proteins may bind to impart an E3-like specificity to the PPS.
It is also unknown if Pup chains are formed on certain substrates in a manner akin to Ub chains. To date, there is no evidence that Pup conjugates to other Pup molecules or onto multiple sites on a single substrate. Another question of interest is whether or not “de-pupylating” enzymes exist. Ub is removed for recycling before the degradation of a substrate, is Pup recycled as well? If so, is a DUB-like protease involved, or are we facing a new enzymatic reaction as seen for the attachment of Pup to substrates?
Now that several components involved in protein degradation are known, it is expected that rapid progress will be made in characterizing these proteins, and in the identification of new players as well. Elucidation of the PPS will not only give us numerous fascinating challenges to ponder, but it will also provide critical data for ultimately developing therapeutics to combat one of the world’s deadliest diseases.
Acknowledgements
We thank K. Burns and S. Quezada for critical review of this manuscript. K.H.D is supported by NIH grant HL092774 and is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darwin KH, Ehrt S, Weich N, Gutierrez-Ramos J-C, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh MU, Nathan CF. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr Opin Microbiol. 2000;3:35–42. doi: 10.1016/s1369-5274(99)00048-x. [DOI] [PubMed] [Google Scholar]
- 3.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darwin KH, Lin G, Chen Z, Li H, Nathan C. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- 5.Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamichhane G, Raghunand TR, Morrison NE, Woolwine SC, Tyagi S, Kandavelou K, Bishai WR. Deletion of a Mycobacterium tuberculosis proteasomal ATPase homologue gene produces a slow-growing strain that persists in host tissues. J Infect Dis. 2006;194:1233–1240. doi: 10.1086/508288. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 9.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 11.Groll M, Bochtler M, Brandstetter H, Clausen T, Huber R. Molecular machines for protein degradation. Chembiochem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 12.Butler SM, Festa RF, Pearce MJ, Darwin KH. Self-compartmentalized Bacteria Proteases and Pathogenesis. Mol. Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 13.Pouch M-N, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol. Microbiol. 2000;35:368–377. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 14.Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, De Mot R, Baumeister W. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 15.Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J. Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C. Mycobacterium tuberculosis prcBA genes encode a gated proteasome. Mol. Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 17.Benaroudj N, Goldberg AL. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol. 2000;2:833–839. doi: 10.1038/35041081. [DOI] [PubMed] [Google Scholar]
- 18.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 19.Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP Binding to PAN or the 26S ATPases Causes Association with the 20S Proteasome, Gate Opening, and Translocation of Unfolded Proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Hu G, Lin G, Wang M, Dick L, Xu R-M, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol. Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 21.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE. 3rd, Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 23.Nagy I, Geert S, Vanderleyden J, De Mot R. Further sequence analysis of the DNA regions with the Rhodococcus 20S proteasome structural genes reveals extensive homolgy with Mycobacterium leprae. DNA Seq. 1997;7:225–228. doi: 10.3109/10425179709034040. [DOI] [PubMed] [Google Scholar]
- 24.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;1999:27–43. [PubMed] [Google Scholar]
- 25.Walker J, Saraste M, Runswick M, Gay N. Distantly related sequences in the α- and β- subunits of ATP synthase myosin, kinases and other ATP-requireing enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 29.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009 doi: 10.1042/BJ20090738. in press. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein Pup is intrinsically disordered. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.07.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darwin KH. Prokaryotic Ubiquitin-Like Protein, Proteasomes, and Pathogenesis. Nat. Rev. Microbiol. 2009 doi: 10.1038/nrmicro2148. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 36.Vandal OH, Nathan CF, Ehrt S. Acid resistance in Mycobacterium tuberculosis. J Bacteriol. 2009 doi: 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 38.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ting LM, Kim AC, Cattamanchi A, Ernst JD. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 40.Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, Bloom BR, Rubin EJ, Ernst JD. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172:6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 41.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann C, Doseeva V, Pullalarevu S, Krajewski W, Howard A, Herzberg O. YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–383. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]