Abstract
Neurotropic alphaviruses such as western, eastern, and Venezuelan equine encephalitis viruses cause serious and potentially fatal central nervous system infections in humans and are high-priority potential bioterrorism agents. There are currently no widely available vaccines or licensed therapies for these virulent pathogens. To identify potential novel antiviral drugs, we developed a cell-based assay with a western equine encephalitis virus replicon that expresses a luciferase reporter gene and screened a small molecule diversity library of 51,028 compounds. We identified and validated a thieno[3,2-b]pyrrole compound with a half maximal inhibitory concentration of <10 µmol/L, a selectivity index >20, and potent activity against live virus in cultured neuronal cells. Furthermore, a structure-activity relationship analysis with 20 related compounds identified several with enhanced activity profiles, including 6 with submicromolar half maximal inhibitory concentrations. In conclusion, we have identified a novel class of promising inhibitors with potent activity against virulent neurotropic alphaviruses.
The Alphavirus genus of the Togaviridae family contains ~30 mosquito-borne, enveloped, positive-stranded RNA viruses, one-third of which cause significant diseases in human and animals worldwide [1]. For example, neurotropic alphaviruses such as western, eastern, and Venezuelan equine encephalitis viruses (WEEV, EEEV, and VEEV, respectively) infect the central nervous system (CNS) and can lead to severe encephalitis in humans, with fatality rates of up to 70%, and survivors often bear long-term neurological sequelae [2, 3]. Neurotropic alphaviruses are also important members of the growing list of emerging or resurging public health threats [4] and are listed as Centers for Disease Control and Prevention and National Institute for Allergy and Infectious Diseases category B bioterrorism agents, in part because of numerous characteristics that make them potential biological weapons, namely, (1) high clinical morbidity and mortality, (2) potential for aerosol transmission, (3) lack of effective countermeasures for disease prevention or control, (4) public anxiety elicited by CNS infections, (5) ease with which large volumes of infectious materials can be produced, and (6) potential for malicious introduction of foreign genes designed to increase alphavirus virulence [5].
There are currently no licensed vaccines or antiviral drugs for alphaviruses. Formalin-inactivated vaccines for WEEV or EEEV and a live attenuated VEEV vaccine (TC-83 strain) are available on an investigational drug basis [5], and alternative live attenuated, chimeric, and DNA-based alphavirus vaccines are being actively developed [6–13]. Nevertheless, the broad clinical application of newer-generation vaccines is likely years away. Furthermore, a maximally effective response to an outbreak due to either natural transmission or intentional exposure to a viral pathogen may require both prevention and treatment approaches [14]. Several compounds have been shown to inhibit alphavirus replication in cultured cells, including the nucleoside analogs ribavirin and mycophenolic acid [15], (−)-carbodine [16], triaryl pyrazoline [17], and seco-pregnane steroids from the Chinese herbs Strobilanthes cusia and Cynanchum paniculatum [18]. Furthermore, both (−)-carbodine and seco-pregnane steroids are active in animal models of alphavirus infection and pathogenesis [16, 18]. Nevertheless, there is still a pressing need to identify new antiviral drugs to treat these virulent pathogens. To this end, we developed a cell-based assay amenable to high-throughput screening (HTS) and analyzed a diversity library of >50,000 compounds for activity against WEEV RNA replication. We identified and validated several compounds with potent inhibitory activity against WEEV and related alphaviruses. Furthermore, we conducted limited structure-activity analyses with one of these compounds and identified a series of thieno[3,2-b]pyrrole derivatives as novel inhibitors of neurotropic alphaviruses.
MATERIALS AND METHODS
Cells and viruses
Human neuroblastoma (BE(2)-C), African green monkey kidney (Vero), and baby hamster kidney (BHK) cell lines were purchased from the American Type Culture Collection and cultured in Dulbecco’s Modified Eagle Medium containing 5% bovine grown serum (HyClone), 10 U/mL of penicillin, and 10 µg/mL of streptomycin. BSR-T7/5 cells, which are BHK cells that constitutively express bacteriophage T7 RNA polymerase [19], generously provided by K. Conzelmann (Max von Pettenkofer-Institut [Munich, Germany]), were cultured in Glasgow minimum essential medium containing 10% heat-inactivated fetal bovine serum, 10% tryptose phosphate broth, 1% sodium pyruvate, 0.1 mmol/L of nonessential amino acids, 10 U/mL of penicillin, 10 µg/mL of streptomycin, and 100–500 µg/mL of G418 for selection. BHK cell lines VEErep/SEAP/Pac and EEErep/SEAP/Pac, which stably express double subgenomic VEEV or EEEV replicons with secreted alkaline phosphatase (SEAP) reporter and puromycin resistance genes [20], were generously provided by I. Frolov (University of Texas Medical Branch [Galveston, TX]) and cultured in Dulbecco’s Modified Eagle Medium containing 7% fetal bovine serum, 10 U/mL of penicillin, 10 µg/mL of streptomycin, and 5 µg/mL of puromycin for selection. Infectious WEEV corresponding to strain Cba87 was generated as described elsewhere [21], and all experiments that involved infectious WEEV were conducted under BSL-3 conditions in approved facilities at the University of Michigan (Ann Arbor, MI). Fort Morgan virus (FMV) strain CM4–146 was purchased from the American Type Culture Collection, and Sindbis virus (SINV) strain Toto64 was generously provided by R. Kuhn (Purdue University [West Lafayette, IN]). FMV and SINV stocks were prepared and quantified using Vero cells as described for WEEV [21].
WEEV replicon
We generated the WEEV replicon plasmid pWR-LUC by using the full-length genomic clone pWE2000 [9], generously provided by M. Parker (US Army Medical Research Institute for Infectious Diseases [Frederick, MD]). This cDNA clone contains a T7 polymerase promoter to initiate precise transcription and produce viral RNA with authentic 5' termini. We amplified the gene encoding firefly luciferase (fLUC) from pTRE2hyg-LUC (Clontech) by polymerase chain reaction (PCR) without an ATG initiator codon but with engineered AvrII and BstXI sites. The resultant fragment was inserted into the AvrII-BstXI site of pWE2000. This strategy replaced the majority of the WEEV structural genes with the fLUC reporter gene, but it retained nucleotides encoding the first 27 amino acids of the capsid protein to preserve the predicted stem-loop region in the structural gene translation enhancer that was previously identified in alphaviruses [22]. We further modified pWR-LUC by placing a hepatitis δ virus ribozyme and T7 terminator downstream of the polyadenylation region to ensure efficient transcription termination and produce authentic viral 3' termini (figure 1A). To generate the control plasmid pWR-ΔLUC, we deleted the NheI-NheI fragment to remove the nonstructural protein (nsP) coding region that included the majority of nsP2, 3, and 4.
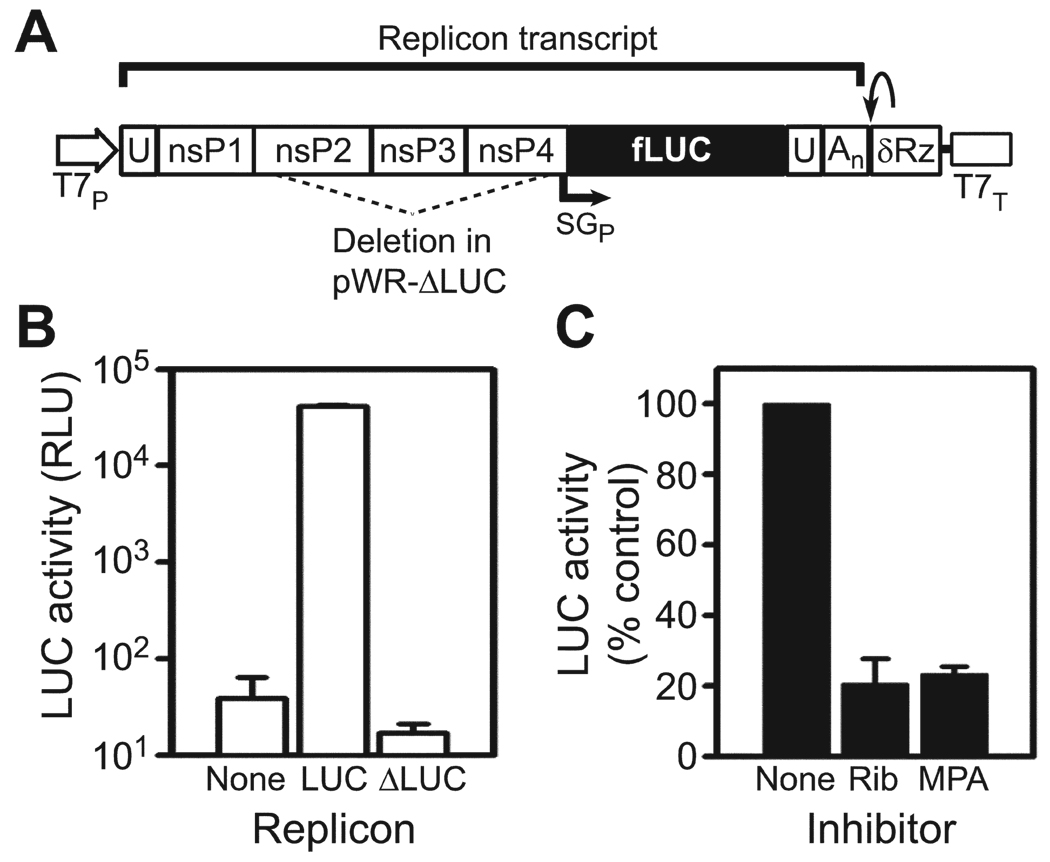
Figure 1.
Cell-based western equine encephalitis virus (WEEV) replicon system for high-throughput screening. A, Schematic of WEEV replicon pWR-LUC. Authentic 5' and 3' viral termini are generated by precise placement of T7 polymerase promoter (T7P) and hepatitis δ virus ribozyme (δRz)/T7 terminator (T7T). The final composition of the replicon transcript is indicated by the bar above the schematic. Transcription of the firefly luciferase (fLUC) reporter gene is controlled by the viral subgenomic promoter (SGP). The region deleted for the control plasmid pWR-ΔLUC is indicated by the dashed lines. An, polyadenylated tail; U, untranslated region. B, fLUC reporter gene activity in BSR-T7/5 cells transfected with empty vector, pWR-LUC, or control pWR-ΔLUC. Results are expressed as relative luciferase units (RLU). C, BSR-T7/5 cells transfected with pWR-LUC were treated with no inhibitor, 50 µmol/L of ribavirin (Rib), or 5 µmol/L of mycophenolic acid (MPA), and fLUC activity was measured after 18 h. Results are expressed as the percentage of fLUC activity relative to that for the untreated control.
Primary HTS, dose-response, and secondary validation of candidate compounds
BSR-T7/5 cells at 60%–70% confluence in 10-cm tissue culture plates (~2 × 106 cells/plate) were transfected with 15 µg of pWR-LUC by using 22 µL of TransIT LT-1 (Mirus), according to the manufacturer’s instructions. Six hours after transfection, cells were detached with 0.05% trypsin and diluted to a concentration of 6.25 × 105 cells/mL, and 20 µL of cell suspension/well was dispensed into 384-well plates preloaded with individual compounds in 30 µL of medium at a concentration of 5–10 µmol/L. All plates contained a series of 32 wells each of negative and positive controls, which consisted of dimethyl sulfoxide and 100 µmol/L of ribavirin, respectively. Plates were cultured at 37°C in 5% CO2 for 18 h, 40 µL of medium was removed and replaced with 10 µL per well of Steady-Glo luciferase reagent (Promega), and luminescence was read on a PHERAstar multimode plate reader (BMG Labtech). Individual compounds identified as primary hits as described below and in table 1 were validated by dose-response analyses, using a similar 384-well format but with 3.3-fold serial dilutions of compounds ranging from 100 µmol/L to 10 n mol/L assayed in duplicate wells. Selected compounds were purchased from the original supplier and were further analyzed by repeat dose-response and toxicity studies, using a 96-well format, in which cell viability was quantitated by a 3-[4,5-dimethylthizol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay as previously described [21] or by Alamar Blue (AbD Serotec) according to the manufacturer’s instructions. For secondary validation, we used VEErep/SEAP/Pac and EEErep/SEAP/Pac cell lines cultured for 24 h with compounds but without puromycin selection and measured SEAP reporter gene expression in supernatants, using Quanti-Blue (InvivoGen) according to the manufacturer’s instructions.
Table 1.
Identification and validation steps in the discovery of novel small molecule compounds that inhibit neurotropic alphavirus replication.
| Step | Experimental system/resource | Criterion | Compounds, no. |
|---|---|---|---|
| Not applicable | CCG chemical diversity library | … | 51,028 |
| HTS | pWR-LUC replicon and BSR-T7/5 cells | (1) fLUC activity >2 SD/plate below the vehicle only (DMSO) negative control or (2) inhibition of fLUC activity >90% per plate of level obtained with ribavirin-positive controla |
196 |
| HTS | pWR-LUC replicon and BSR-T7/5 cells | No activity in previous CCG LUC-based screens | 114 |
| Primary validation | pWR-LUC replicon and BSR-T7/5 cells | Dose-response with IC50 <100 µmol/L | 77b |
| Secondary validation | EEEV/VEEV-SEAP replicon-bearing BHK cells |
Dose-response with IC50 <100 µmol/L | 11 |
| Tertiary validation | Repeat dose-response with pWR-LUC replicon and BSR-T7/5 cells |
Selectivity indexc >5 | 4 |
NOTE. CCG, University of Michigan Center for Chemical Genomics; CC50, 50% cytotoxicity concentration; DMSO, dimethyl sulfoxide; fLUC, firefly luciferase; HTS, high-throughput screening; IC50, half maximal inhibitory concentration.
Compounds that satisfied either criterion were chosen as primary hits.
Primary validated compounds were present in libraries of Chembridge (13 compounds), ChemDiv (25 compounds), Maybridge (38 compounds), and MS Spectrum 2000 (1 compound).
Calculated as CC50/IC50.
Final verification of candidate compound activity with infectious virus
Human BE(2)-C neuroblastoma cells were incubated simultaneously with candidate compounds and infectious WEEV, FMV, or SINV at a multiplicity of infection (MOI) of 1 or 0.1, and MTT viability assays, Northern blots, and infectious virus quantitation by plaque assay were done 6–48 h after infection, as previously described [21]. For reverse transcription–PCR (RT-PCR) analyses, total RNA was isolated 6 h after infection with TRIzol reagent (Invitrogen) in accordance with the manufacturer’s instructions and was digested with RQ1 DNAse (Promega), and RNA concentrations and integrity were determined by spectrophotometry and denaturing agarose gel electrophoresis. First-strand cDNA synthesis was performed with the SuperScript First-Strand Synthesis System (Invitrogen), using equal amounts of total RNA with oligo(dT)12–16 primers. For semiquantitative RT-PCR, we amplified 200–600-bp fragments of the WEEV nsP2 and E1 genes by using cDNA serial dilutions and rRNA as the loading control, and analyzed products by agarose gel electrophoresis and ethidium bromide staining. For quantitative RT-PCR, we amplified ~200-bp fragments of the WEEV or FMV E1 gene and rRNA as an internal control, using iQTM SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions in a 96-well format with triplicate wells. Amplification and detection were done with an iCycler iQ system, and fluorescence threshold values were calculated using SDS 700 system software (Bio-Rad).
RESULTS
Development and validation of WEEV replicon cell-based assay for HTS
The alphavirus life cycle includes the following general steps that are viable targets for antiviral drugs: attachment and entry, genome replication, and encapsidation and release. We focused on the second step, genome replication, in an attempt to identify novel alphavirus inhibitors. The 5' two-thirds of the alphavirus RNA genome encodes nsP1–4, which form an active replication complex that synthesizes a full-length 11–12-kb strand of genomic RNA and a 4-kb strand of subgenomic RNA [1]. The latter RNA segment, which is identical to the 3' one-third of the viral genome, encodes the structural capsid protein and envelope glycoproteins. These viral proteins are not required for genome replication, and therefore their genes can be readily replaced by foreign genes to produce self-replicating alphavirus vectors, termed replicons [23]. We used this approach to generate pWR-LUC, which encodes an fLUC-containing WEEV replicon (figure 1A). To facilitate the host-cell transcription necessary to “launch” the WEEV replicon from a plasmid, we used cells from a highly transfectable BHK cell line derivative, BSR-T7/5, that constitutively express bacteriophage T7 RNA polymerase [19]. One potential complication with using cell-based assays to identify antiviral compounds is the possibility that candidate compounds will induce type I interferon production and hence suppress virus replication indirectly. The use of BSR-T7/5 cells minimizes this potential complication because BHK cells are deficient in both the production and response to type I interferons [24 –26].
BSR-T7/5 cells transiently transfected with the pWR-LUC replicon produced fLUC levels ~3 logs above the background level (figure 1B). Reporter gene expression was dependent on viral RNA replication because essentially no fLUC expression was detected in cells transfected with pWR-ΔLUC, a control plasmid in which the majority of the nsP2–4 region had been deleted (figure 1A and 1B). Furthermore, both ribavirin and mycophenolic acid, which have previously been shown to inhibit SINV replication [15], suppressed fLUC expression in pWR-LUC–transfected BSR-T7/5 cells by ~80% (figure 1C). We concluded from these results that the pWR-LUC:BSR-T7/5 system would function as a convenient and robust platform to identify small molecule inhibitors of WEEV RNA replication.
Primary HTS and validation of candidate antiviral drugs against neurotropic alphaviruses
We initially optimized the pWR-LUC:BSR-T7/5 system to a 384-well HTS format and obtained Z' scores of >0.6. This statistical parameter characterizes the quality and reliability of HTS assays; scores of >0.5 indicate acceptable quality and reliability [27]. We subsequently used the optimized system to screen a diversity library of 51,028 compounds at the University of Michigan Center for Chemical Genomics (CCG). This composite library consisted of compounds from the following smaller collections: Chembridge (13,028 compounds), ChemDiv (20,000 compounds), May-bridge (16,000 compounds), and MS Spectrum 2000 (2000 compounds), the latter of which included drugs approved by the US Food and Drug Administration. Table 1 provides a composite overview of the experimental systems, criteria, and results from the HTS and subsequent validation steps. For the primary HTS, we chose 2 selection parameters to identify compounds that suppressed fLUC activity by ~70% or more and obtained a hit rate of 0.4%. We further excluded 82 compounds that had suppressive activity in previous LUC-based screenings performed at the CCG, thus reducing the potential selection of toxic compounds or those with direct activity against the reporter gene. We subjected the remaining 114 compounds to dose-response analysis for primary validation, where 68% of these compounds had a half maximal inhibitory concentration (IC50) of <100 µmol/L.
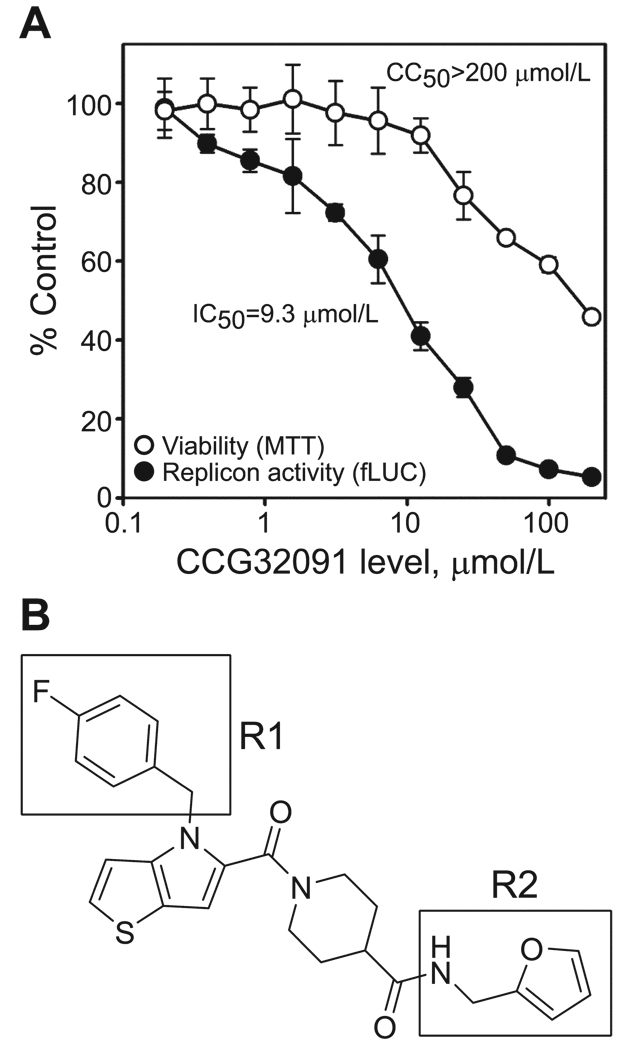
We purchased new material from the original suppliers for 46 available compounds with the lowest IC50 values and conducted secondary validation studies with cell-based replicons derived from VEEV or EEEV that incorporated a SEAP reporter gene rather than the gene encoding fLUC. This step allowed us to further exclude compounds that were active against fLUC but also increased the potential of identifying compounds with broad activity against neurotropic alphaviruses. Eleven compounds showed activity in the secondary validation assays and were evaluated in tertiary validation assays with repeat detailed dose-response and toxicity assessment to calculate the precise 50% cytotoxicity concentration (CC50) and IC50, using the original pWR-LUC:BSR-T7/5 system. Four compounds had a selectivity index (defined as the CC50/IC50 ratio) of >5 and were chosen as candidates for further development as alphavirus inhibitors. One of these compounds, CCG32091, was particularly potent, with a selectivity index of >20 (figure 2A). For comparison, ribavirin had an IC50 of 16.0 µmol/L and a selectivity index of 19 with the pWR-LUC: BSR-T7/5 system. We chose CCG32091, which has a thieno[3,2-b]pyrrole core structure with a 4-fluorobenzyl R1 group attached to the pyrrole nitrogen and a 2-furanylmethylamine R2 group incorporated into the terminal carboxamide (IUPAC name, 1-({4-[(4-fluorophenyl)methyl]-4H-thieno[3,2-b]pyrrol-5-yl}-carbonyl)-N-(furan-2-ylmethyl)piperidine-4-carboxamide), as our initial lead antiviral compound for final verification studies with live virus and structure-activity relationship (SAR) analysis (figure 2B).
Figure 2.
CCG32091 potently inhibits western equine encephalitis virus replicon activity with minimal cytotoxicity. A, Dose-response curves of BSR-T7/5 cells transfected with pWR-LUC and treated with increasing concentrations of CCG32091. Firefly luciferase (fLUC) reporter gene activity (closed circles) was measured by a luciferase assay, and viability (open circles) was measured by MTT assay. Results are expressed as the percentage of untreated controls. Calculated concentrations that produced a 50% inhibition of fLUC activity (IC50) or 50% cytotoxicity concentration (CC50), compared with that of untreated controls, are shown. B, Structure of CCG32091. The R1 and R2 groups central to the structure-activity relationship (see figure 4) are highlighted by boxes.
Verification of CCG32091 antiviral activity with live virus and cultured neuronal cells
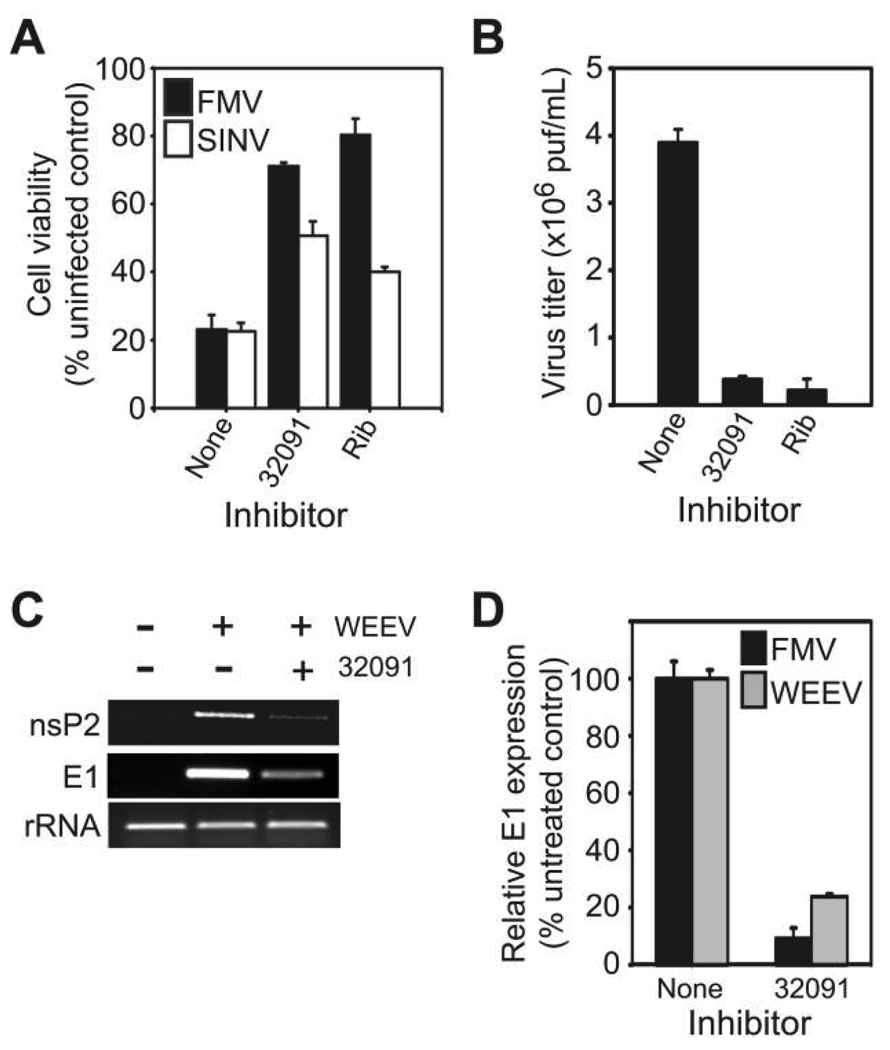
The primary target cell of neurotropic alphaviruses is the CNS neuron [1], and thus we performed a final verification of the antiviral activity of CCG32091 by using an in vitro model with human neuronal cells previously used to study WEEV pathogenesis [21]. For initial experiments with infectious virus, we used FMV, an alphavirus closely related to WEEV, and SINV, the prototypic alphavirus used to study pathogenesis [1]. Both viruses can be handled safely under BSL-2 conditions. One characteristic of alphavirus replication in cultured mammalian cells is the rapid development of cytopathic effect, which is due in part to virus-mediated disruption of host-cell transcription and translation [28–30]. We infected BE(2)-C cells with FMV or SINV in the presence of 12.5 µmol/L of CCG32091 or 50 µmol/L of ribavirin and measured cell viability 48 h after infection, using the MTT assay (figure 3A). Treatment with CCG32091 suppressed virus-induced cytopathic effect and increased cell viability from 20% in infected but mock-treated cells to 50% or 70% in SINV- or FMV-infected cells, respectively. Furthermore, CCG32091 effectively suppressed FMV-induced cytopathic effect at concentrations as low as 3 µmol/L, the lowest concentration that we tested in this assay (data not shown).
Figure 3.
CCG32091 inhibits alphavirus replication in cultured human neuronal cells. A, Human BE(2)-C neuronal cells were infected with Fort Morgan virus (FMV; black bars) or Sindbis virus (SINV; white bars) at a multiplicity of infection (MOI) of 0.1 and simultaneously treated with no inhibitor, 12.5 µmol/L of CCG32091, or 50 µmol/L of ribavirin (Rib), and cell viability was determined 48 h after infection by an MTT assay. B, BE(2)-C cells were infected with FMV at an MOI of 1 and treated with inhibitors as described above, and virus titers in culture supernatants were determined 24 h after infection by plaque assay. C, BE(2)-C cells were infected with western equine encephalitis virus (WEEV) at an MOI of 1 and treated with CCG32091, and viral RNA corresponding to the nsP2 and E1 regions were analyzed by RT-PCR 6 h after infection. rRNA levels are shown as a loading control. D, BE(2)-C cells were infected with FMV (black bars) or WEEV (grey bars) and treated with CCG32091 as described above, and viral RNA levels corresponding to the E1 gene were determined by quantitative RT-PCR.
We also directly assessed the ability of CCG32091 to inhibit virus replication by examining infectious virion production (figure 3B) and viral RNA replication (figure 3C and 3D). CCG32091 suppressed infectious FMV production by >90%, similar to the level of suppression seen with the positive control ribavirin (figure 3B). Furthermore, when we examined viral RNA replication by RT-PCR with either WEEV- or FMV-infected BE(2)-C cells, CCG32091 reduced the accumulation of viral RNAs encoding either nsP2 or E1 by 80%–90% (figure 3C and 3D). Northern blotting confirmed that CCG32091 reduced both genomic and subgenomic RNA accumulation after infection (data not shown). These results demonstrated that CCG32091 suppressed virus replication in infected neuronal cells, inhibited virus-induced cytopathic effect, and had broad activity against several alphaviruses.
SAR analysis with CCG32091
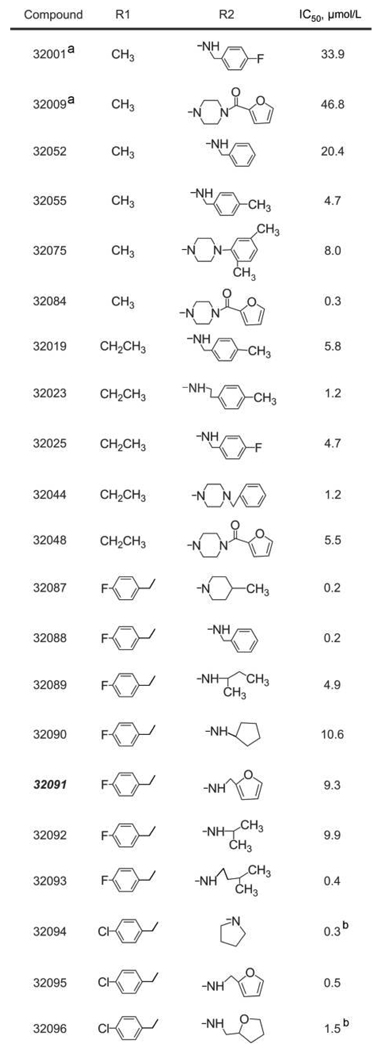
To optimize the therapeutic profile of candidate antiviral drugs in anticipation of future in vivo animal studies, we compared the structure of CCG32091 (figure 2B) with those of compounds in the entire CCG library. We identified 20 compounds that contained a core thieno[3,2-b]pyrrole moiety but had combinations of R1 and R2 groups that differed from those for CCG32091 (figure 4). We previously identified 6 of these compounds as “hits” in the primary HTS and had already completed dose-response analyses for validation (CCG32075, 32089, 32090, 32092, 32095, and 32096). We completed dose-response analysis of the remaining 14 compounds to obtain a limited SAR for CCG32091 (figure 4). This analysis revealed an ~250-fold range of IC50 values, from a high of 46.8 µmol/L to a low of 0.2 µmol/L, and 6 compounds had a submicromolar IC50 (CCG32084, 32087, 32088, 32093, 32094, and 32095). We also completed toxicity studies with these 20 compounds and found that 90% had a CC50 of >100 µmol/L, including 5 of 6 compounds with a submicromolar IC50 (figure 4). Although the data set was limited, several aspects of the SAR could be elucidated. At R1, there appeared to be little difference between methyl and ethyl groups (compare the 4-methylbenzylamides CCG32055 and CCG32019). However, substantially better activity was observed when the small alkyl group at R1 was replaced with 4-fluorobenzyl (compare CCG32088 with CCG32052). A direct comparison between 4-fluorobenzyl and 4-chlorobenzyl at R1 (2-furanylmethyl amides CCG32091 and CCG32095) also suggested that 4-chlorobenzyl may represent a further optimization of R1. Of the amines incorporated at R2, none were clearly superior to the others. In fact, a variety of amines were seen with potent inhibitors, including 4-methylpiperidine (CCG32087), benzyl (CCG32088), isopentyl (CCG32093), and 4-(2-furanylcarboxy)piperazine (CCG-32084). With regard to the internal piperidine carboxamide, activities of the 3-carboxamide analogs CCG32001 and CCG32009 were distinctly inferior to those of the closely related 4-carboxamide analogs CCG32025 and CCG32084, respectively. Overall, these results identified several additional compounds with greater potency than but similar toxicity to the original lead compound CCG32091, and they provide a useful initial data set to begin targeted design for optimized antiviral compound synthesis based on the core thieno[3,2-b]pyrrole structure.
Figure 4.
CCG32091 structure-activity relationship analysis. aCompounds CCG32001 and 32009 have the R2 group attached to a piperidine-3-carboxamide, in contrast to a piperidine-4-carboxamide, which is found in the remaining compounds. bIncreased cytotoxicity, compared with CCG32091, with 50% cytotoxicity concentrations of <100 µmol/L. IC50, half maximal inhibitory concentration.
DISCUSSION
The neurotropic alphaviruses represent emerging pathogens that have the potential for widespread dissemination and the ability to cause substantial morbidity and mortality [4, 5] but for which no licensed therapies currently exist. In this report, we describe the identification of a novel class of thieno[3,2-b]pyrrole compounds with inhibitory activity against WEEV and several related alphaviruses. Heterocyclic compounds that contain a thieno[3,2-b]pyrrole core have been previously identified as possessing physiological activity with potential clinical applications, including uses as anti-inflammatory agents [31], glycogen phosphorylase inhibitors for diabetes treatment [32], and hepatitis C virus (HCV) inhibitors [33]. The latter use is particularly relevant for the work presented in this report, because thieno[3,2-b]pyrroles were identified as allosteric inhibitors of HCV RNA polymerase [33], indicating a plausible potential mechanism of action for their activity against alphaviruses (discussed below). Significantly, the lead compound identified in this report, CCG32091 (figure 2), is a PubChem registered compound (CID, 3240671) and part of the NIH Molecular Libraries–Small Molecule Repository, and it has been identified as an active compound in only 5 of ~250 HTS assays conducted through the NIH Molecular Libraries Screening Center Network. This indicates that the spectrum of its biological activity is fairly narrow, which is a highly desirable attribute in a potential lead compound. In addition, Ilyin et al. [34] recently described a solution-phase strategy for the synthesis of novel combinatorial libraries containing a thieno[3,2-b]pyrrole core, thus providing the opportunity to further explore and potentially exploit these compounds as therapeutics for a range of human diseases, including infections with neurotropic alphaviruses.
The mechanism(s) underlying the antiviral activity of thieno[3,2-b]pyrroles against neurotropic alphaviruses is unknown, but the use of a replicon-based assay for the HTS and validation steps (figure 1 and table 1) implicates viral replicase proteins as potential targets. This hypothesis is supported by the observation that CCG32091 reduced viral RNA accumulation after infection of neuronal cells (figure 3). Furthermore, the broad activity of CCG32091 against infectious virus or replicons derived from WEEV, EEEV, VEEV, FMV, and SINV suggests that a highly conserved viral enzymatic activity may be targeted. Alphavirus nsPs contain several distinct enzymatic activities, including methyltransferase (nsP1) [35], protease and helicase (nsP2) [36, 37], and RNA polymerase (nsP4) [38]. In vitro assays have been developed to detect several of these activities [35, 39, 40], thereby providing a convenient approach to target identification. An alternative approach that takes advantage of the intrinsically high error rate of viral RNA polymerases previously used successfully for antiviral target identification is the isolation and characterization of viral escape mutants [41–43].
The preclinical usefulness of candidate novel antiviral agents rests on their ability to either prevent or treat established disease in animal models while exhibiting minimal toxicity. The treatment of CNS infections presents an additional hurdle to overcome, as the blood-brain barrier represents a formidable obstacle for drug penetration [44]. The blood-brain barrier is a highly effective physiologic barrier whose primary function is to closely regulate access of bloodstream components to the CNS. Although infectious and inflammatory CNS diseases often disrupt blood-brain barrier function and increase permeability, drug penetration remains an important aspect to consider in the development of antiviral agents against neurotropic alphaviruses. Multiple physical-chemical factors, including lipophilicity, ionization properties, molecular flexibility, polar surface area, and size, influence CNS penetration of drugs [45]. The latter 2 properties are particularly important, as studies of marketed CNS and non-CNS drugs indicate that polar surface areas of <60 to 90 Å2 and a molecular weight of <450 Da are required for adequate penetration [46, 47]. The lead thieno[3,2-b]pyrrole compound identified in this report, CCG32091 (figure 2B), has a calculated polar surface area of 67.5 Å2 and a molecular weight of 466 Da (PubChem database; available at: http://pubchem.ncbi.nlm.nih.gov). Several of the compounds identified in the SAR analysis had lower polar surface areas and molecular weights than CCG32091 (figure 4), and we are currently using the SAR results to refine the development of alphavirus inhibitors with reduced toxicity, enhanced potency, and optimal CNS penetration.
Acknowledgments
We thank Michael Parker, Klaus Conzelmann, Ilya Frolov, and Richard Kuhn for providing reagents; Patrick Robichaud and Michael Swanson for HTS and quantitative RT-PCR assistance, respectively; Donna Gschwend for administrative assistance; and all members of the Miller laboratory for helpful comments on the research and manuscript.
Financial support: National Institute of Allergy and Infectious Diseases (grant U54 AI057153, Region V “Great Lakes” Regional Center of Excellence for Biode-fense and Emerging Infectious Diseases Research Career Development Award [to D.J.M.], and grant T32 AI007528 [to D.C.P.]); National Institute of General Medical Sciences (grant T32 GM007863 [to D.C.P.]).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 5th National Regional Centers of Excellence Meeting, Chicago, IL, 7 April 2008.
References
- 1.Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, Griffin DE, et al., editors. Fields virology. 4th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. pp. 917–962. [Google Scholar]
- 2.Deresiewicz RL, Thaler SJ, Hsu L, Zamani AA. Clinical and neuroradiographic manifestations of eastern equine encephalitis. N Engl J Med. 1997;336:1867–1874. doi: 10.1056/NEJM199706263362604. [DOI] [PubMed] [Google Scholar]
- 3.Earnest MP, Goolishian HA, Calverley JR, Hayes RO, Hill HR. Neurologic, intellectual, and psychologic sequelae following western encephalitis: a follow-up study of 35 cases. Neurology. 1971;21:969–974. doi: 10.1212/wnl.21.9.969. [DOI] [PubMed] [Google Scholar]
- 4.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 5.Sidwell RW, Smee DF. Viruses of the Bunya- and Togaviridae families: potential as bioterrorism agents and means of control. Antiviral Res. 2003;57:101–111. doi: 10.1016/s0166-3542(02)00203-6. [DOI] [PubMed] [Google Scholar]
- 6.Barabe ND, Rayner GA, Christopher ME, Nagata LP, Wu JQ. Single-dose, fast-acting vaccine candidate against western equine encephalitis virus completely protects mice from intranasal challenge with different strains of the virus. Vaccine. 2007;25:6271–6276. doi: 10.1016/j.vaccine.2007.05.054. [DOI] [PubMed] [Google Scholar]
- 7.Wu JQ, Barabe ND, Chau D, et al. Complete protection of mice against a lethal dose challenge of western equine encephalitis virus after immunization with an adenovirus-vectored vaccine. Vaccine. 2007;25:4368–4375. doi: 10.1016/j.vaccine.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 8.Nagata LP, Hu WG, Masri SA, et al. Efficacy of DNA vaccination against western equine encephalitis virus infection. Vaccine. 2005;23:2280–2283. doi: 10.1016/j.vaccine.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Schoepp RJ, Smith JF, Parker MD. Recombinant chimeric western and eastern equine encephalitis viruses as potential vaccine candidates. Virology. 2002;302:299–309. doi: 10.1006/viro.2002.1677. [DOI] [PubMed] [Google Scholar]
- 10.Turell MJ, Ludwig GV, Kondig J, Smith JF. Limited potential for mosquito transmission of genetically engineered, live-attenuated Venezuelan equine encephalitis virus vaccine candidates. Am J Trop Med Hyg. 1999;60:1041–1044. doi: 10.4269/ajtmh.1999.60.1041. [DOI] [PubMed] [Google Scholar]
- 11.Wang E, Petrakova O, Adams AP, et al. Chimeric Sindbis/eastern equine encephalitis vaccine candidates are highly attenuated and immunogenic in mice. Vaccine. 2007;25:7573–7581. doi: 10.1016/j.vaccine.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fine DL, Roberts BA, Terpening SJ, Mott J, Vasconcelos D, House RV. Neurovirulence evaluation of Venezuelan equine encephalitis (VEE) vaccine candidate V3526 in nonhuman primates. Vaccine. 2008;26:3497–3506. doi: 10.1016/j.vaccine.2008.04.044. [DOI] [PubMed] [Google Scholar]
- 13.Turell MJ, Parker MD. Protection of hamsters by Venezuelan equine encephalitis virus candidate vaccine V3526 against lethal challenge by mosquito bite and intraperitoneal injection. Am J Trop Med Hyg. 2008;78:328–332. [PubMed] [Google Scholar]
- 14.Bronze MS, Greenfield RA. Therapeutic options for diseases due to potential viral agents of bioterrorism. Curr Opin Investig Drugs. 2003;4:172–178. [PubMed] [Google Scholar]
- 15.Malinoski F, Stollar V. Inhibitors of IMP dehydrogenase prevent Sindbis virus replication and reduce GTP levels in Aedes albopictus cells. Virology. 1981;110:281–289. doi: 10.1016/0042-6822(81)90060-x. [DOI] [PubMed] [Google Scholar]
- 16.Julander JG, Bowen RA, Rao JR, et al. Treatment of Venezuelan equine encephalitis virus infection with (−)-carbodine. Antiviral Res. 2008;80:309–315. doi: 10.1016/j.antiviral.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puig-Basagoiti F, Tilgner M, Forshey BM, et al. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob Agents Chemother. 2006;50:1320–1329. doi: 10.1128/AAC.50.4.1320-1329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Wang L, Li S, et al. Seco-pregnane steroids target the subgenomic RNA of alphavirus-like RNA viruses. Proc Natl Acad Sci U S A. 2007;104:8083–8088. doi: 10.1073/pnas.0702398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM, Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castorena KM, Peltier DC, Peng W, Miller DJ. Maturation-dependent responses of human neuronal cells to western equine encephalitis virus infection and type I interferons. Virology. 2008;372:208–220. doi: 10.1016/j.virol.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol. 1996;70:1182–1190. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frolov I, Hoffman TA, Pragai BM, et al. Alphavirus-based expression vectors: strategies and applications. Proc Natl Acad Sci U S A. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andzhaparidze OG, Bogomolova NN, Boriskin YS, Bektemirova MS, Drynov ID. Comparative study of rabies virus persistence in human and hamster cell lines. J Virol. 1981;37:1–6. doi: 10.1128/jvi.37.1.1-6.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer MJ, Dennin R, Kramer C, et al. Cell and virus sensitivity studies with recombinant human alpha interferons. J Interferon Res. 1983;3:425–435. doi: 10.1089/jir.1983.3.425. [DOI] [PubMed] [Google Scholar]
- 26.Nagai Y, Ito Y, Hamaguchi M, Yoshida T, Matsumoto T. Relation of interferon production to the limited replication of Newcastle disease virus in L cells. J Gen Virol. 1981;55:109–116. doi: 10.1099/0022-1317-55-1-109. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 28.Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J Virol. 2006;80:5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol. 2007;81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar PR, Raju S, Goud PS, et al. Synthesis and biological evaluation of thiophene[3,2-b]pyrrole derivatives as potential anti-inflammatory agents. Bioorg Med Chem. 2004;12:1221–1230. doi: 10.1016/j.bmc.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Whittamore PR, Addie MS, Bennett SN, et al. Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies. Bioorg Med Chem Lett. 2006;16:5567–5571. doi: 10.1016/j.bmcl.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 33.Ontoria JM, Martin Hernando JI, Malancona S, et al. Identification of thieno[3,2-b]pyrroles as allosteric inhibitors of hepatitis C virus NS5B polymerase. Bioorg Med Chem Lett. 2006;16:4026–4030. doi: 10.1016/j.bmcl.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Ilyin AP, Dmitrieva IG, Kustova VA, Manaev AV, Ivachtchenko AV. Synthesis of heterocyclic compounds possessing the 4H-thieno[3,2-b]pyrrole moiety. J Comb Chem. 2007;9:96–106. doi: 10.1021/cc060091h. [DOI] [PubMed] [Google Scholar]
- 35.Ahola T, Laakkonen P, Vihinen H, Kaariainen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez dC, Ehsani N, Mikkola ML, Garcia JA, Kaariainen L. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 1999;448:19–22. doi: 10.1016/s0014-5793(99)00321-x. [DOI] [PubMed] [Google Scholar]
- 37.Hardy WR, Strauss JH. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasiljeva L, Valmu L, Kaariainen L, Merits A. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J Biol Chem. 2001;276:30786–30793. doi: 10.1074/jbc.M104786200. [DOI] [PubMed] [Google Scholar]
- 40.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li ML, Lin YH, Simmonds HA, Stollar V. A mutant of Sindbis virus which is able to replicate in cells with reduced CTP makes a replicase/transcriptase with a decreased Km for CTP. J Virol. 2004;78:9645–9651. doi: 10.1128/JVI.78.18.9645-9651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YH, Yadav P, Ravatn R, Stollar V. A mutant of Sindbis virus that is resistant to pyrazofurin encodes an altered RNA polymerase. Virology. 2000;272:61–71. doi: 10.1006/viro.2000.0329. [DOI] [PubMed] [Google Scholar]
- 43.Scheidel LM, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991;181:490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- 44.Pardridge WM. The blood-brain barrier and neurotherapeutics. NeuroRx. 2005;2:1–2. doi: 10.1602/neurorx.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx. 2005;2:541–553. doi: 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelder J, Grootenhuis PD, Bayada DM, Delbressine LP, Ploemen JP. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res. 1999;16:1514–1519. doi: 10.1023/a:1015040217741. [DOI] [PubMed] [Google Scholar]
- 47.van de Waterbeemd H, Camenisch G, Folkers G, Chretien JR, Raevsky OA. Estimation of blood-brain barrier crossing of drugs using molecular size and shape, and H-bonding descriptors. J Drug Target. 1998;6:151–165. doi: 10.3109/10611869808997889. [DOI] [PubMed] [Google Scholar]