Abstract
Human coronavirus OC43 (HCoV-OC43) is one of the causes of the “common cold” in human during seasons of cold weather. The primary function of the HCoV-OC43 nucleocapsid protein (N protein) is to recognize viral genomic RNA, which leads to ribonucleocapsid formation. Here, we characterized the stability and identified the functional regions of the recombinant HCoV-OC43 N protein. Circular dichroism and fluorescence measurements revealed that the HCoV-OC43 N protein is more highly ordered and stabler than the SARS-CoV N protein previously studied. Surface plasmon resonance (SPR) experiments showed that the affinity of HCoV-OC43 N protein for RNA was approximately fivefold higher than that of N protein for DNA. Moreover, we found that the HCoV-OC43 N protein contains three RNA-binding regions in its N-terminal region (residues 1–173) and central-linker region (residues 174–232 and 233–300). The binding affinities of the truncated N proteins and RNA follow the order: residues 1–173–residues 233–300 > residues 174–232. SPR experiments demonstrated that the C-terminal region (residues 301–448) of HCoV-OC43 N protein lacks RNA-binding activity, while crosslinking and gel filtration analyses revealed that the C-terminal region is mainly involved in the oligomerization of the HCoV-OC43 N protein. This study may benefit the understanding of the mechanism of HCoV-OC43 nucleocapsid formation.
Keywords: human coronavirus, OC43 strain, nucleocapsid protein, Ribonucleocapsid, stability, RNA-binding regions, oligomerization domain
Introduction
The strain OC43 of the type II coronavirus family (HCoV-OC43) was first identified in the 1960s and is responsible for ∼20% of all “common colds” in humans.1,2 Although HCoV-OC43 infections are generally mild, more severe upper and lower respiratory tract infections like bronchiolitis and pneumonia have been documented, especially in infants, elderly individuals, and immunocompromised patients.1,3,4 Moreover, there have been reports that clusters of HCoV-OC43 infections cause pneumonia in otherwise healthy adults.2,5 Several studies have reported that both neurotropism and neuroinvasion of HCoV are associated with multiple sclerosis, especially in the case of the OC43 strain.6
CoV particles have an irregular shape, which is determined by an outer envelope with distinctive, “club-shaped.” These peplomers give the virus a crown-like (corona) appearance.7 The viral genome of coronaviruses consists of positive sense, single-stranded RNA of ∼30 kb. The genome contains several genes that encode structural and nonstructural proteins that are required for progeny virion production.1 The virion envelope surrounding the nucleocapsid contains the following structural proteins: S (spike) protein, M (membrane),and E (envelope). A third glycoprotein, HE (hemagglutinin-esterase), is present in most class II coronaviruses.8,9 A helical nucleocapsid exists in the center of the viral particle.10–12 Nucleocapsid protein, the major structural protein of CoVs, binds to the viral RNA genome to form the virion core, which leads to the formation of a ribonucleoprotein (RNP) complex or to a long helical nucleocapsid structure.13,14 The formation of the RNP is important for maintaining the RNA in an ordered conformation suitable for replication and transcription of the viral genome.13,15–17 Previous studies have shown that the CoV N protein is involved in the regulation of cellular processes such as gene transcription, actin reorganization, host cell cycle progression, and apoptosis.18–21 It has also been shown to act as an RNA chaperone.22 Moreover, the N protein is an important diagnostic marker and the most “immuno-dominant” antigen in a host immune response.17,23,24
Previous studies have revealed that the N- and C-terminal domains of the CoV N proteins, including those of SARS-CoV, murine hepatitis virus (MHV), and avian infectious bronchitis virus (IBV), are responsible for RNA binding and oligomerization, respectively.25–30 The central region of the N protein has also been shown to contain an RNA-binding region and the primary sites of phosphorylation.28,31,32 Phosphorylation of the N protein has been shown to play an important role in virus biology.33,34 To clarify the molecular mechanism of ribonucleocapsid formation by CoVs, structures of truncated fragments of N protein including N-terminal and C-terminal domains have been published.26,35–37 Despite the conservation of some motifs, CoV N proteins from different strains often show quite different properties primarily due to their low sequence homology.30 For example, Saikatendu et al. found that the structures of SARS-CoV and IBV N-terminal domains shared several common features, in addition to many subtle structural differences.36 On the basis of the different crystal packing of these two structures, they suggested that the two viruses most likely use different modes of oligomeric self-association during RNP core formation.
The N protein of HCoV-OC43, which has a molecular weight of 50 kDa, is highly basic (pI, 10), and it shows strong hydrophilicity.38 The N protein of HCoV-OC43 shows only 26–30% amino acids similarity to the N protein of other CoV strains.30 Until now, few studies have examined the biochemical properties of the HCoV-OC43 N protein. In this study, we characterized the stability and biochemical properties of the recombinant full-length N protein from HCoV-OC43. RNA-binding and oligomerization regions of the HCoV-OC43 N protein were identified.
Results
Stability studies of recombinant HCoV-OC43 nucleocapsid protein
Conformational changes of the HCoV-OC43 N protein in response to temperatures were monitored using CD spectroscopy. As shown in Figure 1(A), the CD spectra of HCoV-OC43 N protein were scanned from 190 to 250 nm at 20, 45, 60, and 90°C. Analysis by the SELECON program revealed that HCoV-OC43 N protein contains ∼35.1% α-helices, 11.6% β-sheets, 23.8% turns, and 29.5% random coils at room temperature. At high temperatures, the CD spectra of HCoV-OC43 N protein showed decreased intensity between 210 and 220 nm and a large negative peak at 190 nm, which are characteristic of disordered protein. Previous CD spectra of the full-length N proteins from SARS and 229E CoVs indicated that the N protein is relatively disordered and composed of ∼50% nonsecondary structures.17 Our present results suggest that the conformations of HCoV-OC43 N proteins are more ordered than those of HCoV-229E and SARS-CoV N proteins [Fig. 1(A)]. We also measured the melting temperature (Tm) of HCoV-OC43 N proteins using CD [Fig. 1(B)]. Heat denaturation analysis showed that the Tm of HCoV-OC43 N protein is 52°C under our conditions. The thermal denaturation of HCo-OC43 N protein is reversible (data not shown).
Figure 1.
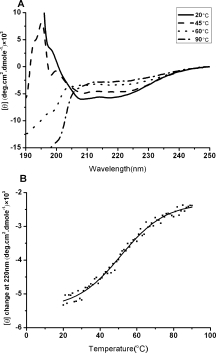
(A) CD spectra of HCoV-OC43 N protein at various temperatures including 20, 45, 60, and 90 °C. The protein concentration was 5 μM and the buffer consisted of 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS. (B) Thermostability measurements of HCoV-OC43 N protein monitored by CD spectra. The protein concentration was 3.8 μM and the buffer consisted of 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS.
Denaturant reagents including urea were used to measure the denaturation of HCoV-OC43 N protein. We used tryptophan (Trp) fluorescence to monitor unfolding because the HCoV-OC43 N protein contains five Trp residues [Fig. 2(A)]. The fluorescence emission spectra showed the maximal emission wavelength of the HCoV-OC43 N protein at ∼340 nm. Urea-induced unfolding caused a significant decrease in FL intensity at 340 nm and a red shift in the maximal emission wavelength. The unfolding of HCoV-OC43 N protein in the presence of urea was observed by monitoring fluorescence spectra at 340 nm, and the denaturation curve was fitted to a two-state model [Fig. 2(B)].39 The denaturation and renaturation curves were completely superimposable (data not shown). The denaturant molarity at the midpoint of the transition (called Cm) was calculated as 4 M and the unfolding free energy change (ΔG0N-U) was determined from the linear extrapolation method to be 14.5 kJ/mol (Supporting Information Figure S1).39
Figure 2.
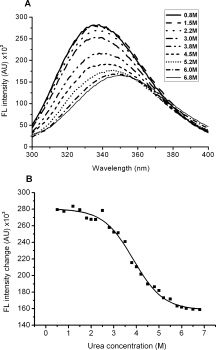
(A) Fluorescence spectra of HCoV-OC43 N protein in Tris-HCl supplemented with different concentrations of urea. The protein concentration was 1 μM and the buffer consisted of 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS. (B) Urea-induced unfolding of HCoV-OC43 N protein monitored by fluorescence emission at 340 nm. The protein concentration was 1 μM and the buffer consisted of 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS.
Studies of nucleic acid binding by HCoV-OC43 nucleocapsid proteins
To measure the binding affinity between the HCoV-OC43 nucleocapsid protein and single-stranded nucleic acids, HCoV-OC43 nucleocapsid protein was allowed to interact with biotin-labeled single-stranded RNA or DNA, and the results were monitored by SPR. Depending on the virus strain, there are two to four UCUAA pentanucleotide repeats, with the last repeat being UCUAAAC and termed the intergenic (IG) sequence at the 3′ end of the leader.40,41 Previous studies showed that HCoV N protein has high affinity for the intergenic sequence.42,43 Therefore, the repeated intergenic sequence of HCoV-OC43, 5′-(UCUAAAC)4-3′, was used as a probe in our SPR and fluorescence experiments. For DNA, uracil was replaced by thymine. The results show that HCoV-OC43 N protein was able to interact with both DNA and RNA. However, HCoV-OC43 N protein showed a higher binding capacity for RNA (∼4989 RU) than for DNA (∼2803 RU) at the same protein concentration [Fig. 3(A)].
Figure 3.
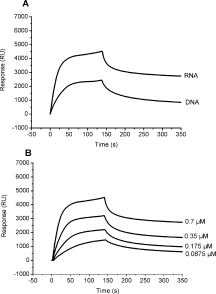
(A) Binding capacity sensorgrams of HCoV-OC43 N protein (0.7 μM) bound to single-stranded RNA or single-stranded DNA. The RNA and DNA sequences immobilized on the SA chip were 5′-(UCUAAAC)4-3′ and 5′-(TCTAAAC)4-3′, respectively. (B) Sensorgrams of HCoV-OC43 N protein bound to single-stranded RNA. HCoV-OC43 N protein was exposed to RNA at concentration of 0.7, 0.35, 0.175, and 0.0875 μM. The buffer consisted of 50 mM Tris-HCl (pH 7.3) containing 150 mM NaCl and 0.1% CHAPS. The association rate increased with an increasing concentration of target N protein and the dissociation rate was independent of concentration. The fitted data was shown in Supporting Information (Figure S3).
Kinetic experiments were carried out by measuring the binding affinity between HCoV-OC43 N protein and single-stranded RNA or DNA. Four concentrations of N protein were used to determine both the ka and kd. Figure 3(B) shows the Biacore SPR traces of N protein bound to RNA. The ka and kd were calculated from the association and dissociation phases of the SPR traces, respectively. The ka and kd of RNA were 4.82 × 104 M–1 s–1 and 5.75 × 10–4 s–1, respectively. The kinetic parameters of DNA showed a lower ka and a higher kd of ∼2.85 × 104 M–1 s–1 and 1.69 × 10–3 s–1, respectively. The dissociation constants (Kd) of HCoV-OC43 N protein with target nucleic acid strands were calculated as kd/ka (in M). The Kd of the N protein bound to RNA was 11.9 nM, which is close to the previously reported Kd of ∼14 nM for the interaction between MHV N protein and RNA.42 As expected, the Kd for N protein and DNA was approximately fivefold higher than that for N protein and RNA.
Identification of the RNA-binding region of HCoV-OC43 nucleocapsid protein
According to the PONDR prediction, HCoV-OC43 N protein contains two structural domains located at the N-terminal and C-terminal regions, and between them lies a central-linker region that is predicted to be disordered [Fig 4(A)].30 To further isolate the RNA-binding region of HCoV-OC43 N protein, we expressed and purified the truncated forms of the N protein: one construct contained only the N-terminal domain (N1–173), another contained only the C-terminal domain (N301–448), and two contained different parts of the central-linker region (N174–232 and N233–300). These domain boundaries are based on the PONDR predictions [Fig. 4(A)]. The binding of each truncated N protein to RNA was analyzed by SPR [Fig. 4(B)]. These studies showed that the N-terminal domain (N1–173) and the central-linker region (N174–232 and N233–300), but not the C-terminal domain, bound to RNA [Fig. 4(B)]. The kinetic constants for association (ka in M−1 s−1) and dissociation (kd in s−1) of the truncated N proteins with respect to singled-stranded RNA have been calculated and listed in Table I. The ka values were essentially the same: 2.18 × 104 and 2.48 × 104 M−1 s−1 for N1–173 and N233–300, respectively, and 7.97 × 103 M−1 s−1 for N174–232. Moreover, the kd values of N1–173 and N233–300 were essentially the same: 1.50 × 10−3 and 1.58 × 10−3 s−1, respectively. These constants were smaller than that of N174–232 (3.53 × 10−3 s−1). The dissociation constants (Kd) of all truncated N proteins with RNA were calculated as kd/ka (in M) and they follow the order: N174–232 > N1–173–N233–300. Our results suggest that the N protein of HCoV-OC43 contains three RNA-binding regions.
Figure 4.
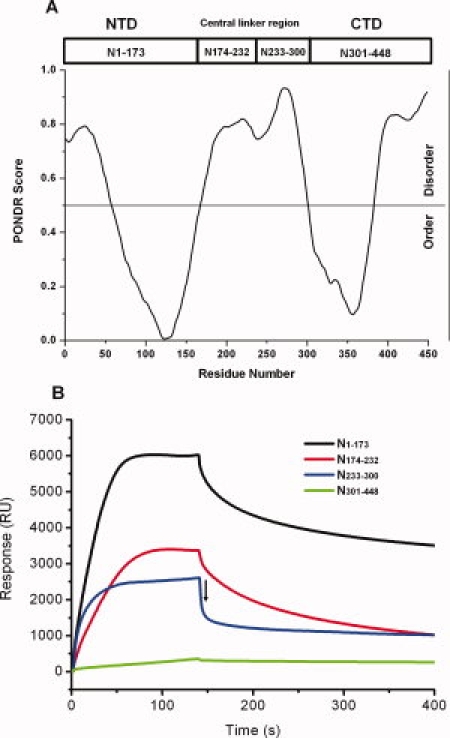
(A) Order/disorder prediction for HCoV-OC43 N protein using the VL3-BA predictor of the PONDR program. (B) Sensorgrams of the truncated forms of HCoV-OC43 N protein including N1–173, N174–232, N233–300, and N301–448 bound to single-stranded RNA. Protein was tested at concentrations of 2 μM in buffer containing 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS. The RNA immobilized on the SA chip was 5′-(UCUAAAC)4-3′. In N233–300, bulk effects caused by the differences in the refractive index of the running buffer and sample solution caused a sharp descent in the curves from 140 s to 142 s; hence, the dissociation measurements were started at 142 s (marked by the arrow).
Table I.
The Kinetic Parameters of the Binding of Full-Length and Truncated N Proteins to Single-Stranded RNA, 5′-(UCUAAAC)4-3′
| Construct | ka (M−1 s−1) | kd (s−1) | aKd (M) |
|---|---|---|---|
| Full-length | 4.82 ± 0.21 × 104 | 5.75 ± 0.23 × 10−4 | 11.9 ± 1.0 × 10−9 |
| N1–173 | 2.18 ± 0.34 × 104 | 1.5 ± 0.21 × 10−3 | 68.9 ± 8.4 × 10−9 |
| N174–232 | 7.97 ± 0.62 × 103 | 3.53 ± 0.32 × 10−3 | 44.4 ± 3.7 × 10−8 |
| N233–300 | 2.48 ± 0.28 × 104 | 1.58 ± 0.24 × 10−3 | 64.1 ± 7.2 × 10−9 |
| N301–448 | b— | — | — |
Kd values were obtained by dividing kd by ka.
No significant binding was observed.
Oligomerization of the HCoV-OC43 nucleocapsid protein
The self-association characters of SARS-CoV and HCoV-229E N proteins has been reported previously.17,44 We further characterized the oligomerization of full-length HCoV-OC43 N proteins using crosslinking assays. As shown in Figure 5(A), HCoV-OC43 N protein was able to form dimers, trimers, tetramers, and higher-order multimers as the glutaraldehyde concentrations was increased. To evaluate whether electrostatic interactions stabilize the compact conformations of higher-order multimers, the oligomeric behaviors of N protein at different ionic strengths were studied. Briefly, the purified full-length N proteins, which were in a buffer containing sodium chloride, were cross-linked by 0.04% glutaraldehyde. In addition, we analyzed the oligomerization of N protein using analytical gel filtration chromatography [Fig. 5(B)]. N protein eluted with a retention volume of ∼49.5 mL, corresponding to an apparent molecular weight of 621 kDa, based on the standard curve. This implies an oligomer containing ∼12 molecules.
Figure 5.
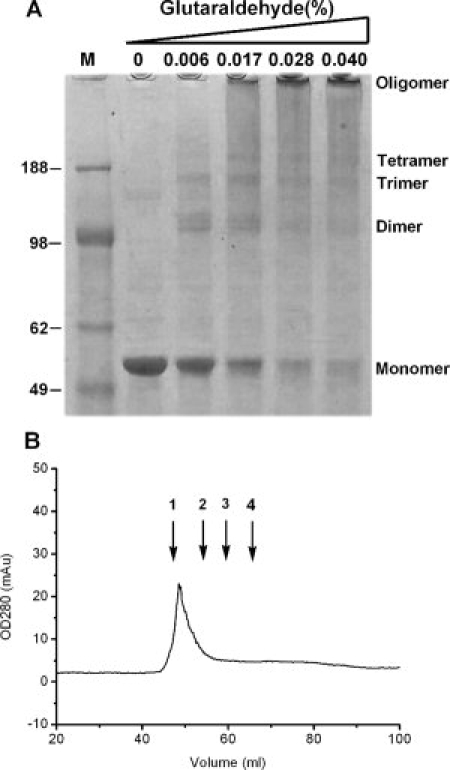
(A) Crosslinking assay of full-length N protein in the presence of various concentrations of glutaraldehyde. The concentration of the protein was 2 μM. (B) Gel filtration chromatography-based analysis (Superdex 300 XK16/70) of N protein dissolved in 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS. The concentration of the target protein was 3 μM. Standard proteins of 669 kDa, 158 kDa, 67 kDa, and 43 kDa are denoted as 1, 2, 3, and 4, respectively.
Identification of the oligomerization region of HCoV-OC43 nucleocapsid protein
To determine the regions that are of greatest importance for HCoV-OC43 N protein oligomerization, several regions of N protein were purified and tested using crosslinking assays and analytical gel filtration. As shown in Figure 6(A), crosslinking studies of N233–448 detected dimers, tetramers, and oligomers. A shorter form of the protein, N301–448, also formed dimers, trimers, and oligomers [Fig. 6(B)]. Gel filtration analysis showed a peak of N301–448 at high molecular weight (225 kDa), consistent with a ∼12-mer [Fig. 6(C)]. The crosslinking assay was also conducted to investigate oligomerization of the N-terminal (N1–173) and middle segment (N174–232) of N protein. Both truncated N proteins remained as monomers under these experimental conditions [Fig. 7], suggesting that these regions are not involved in the oligomerization. Our data showed that the 148 amino acids at the C-terminus were the oligomerization domain of HCoV-OC43 N protein.
Figure 6.
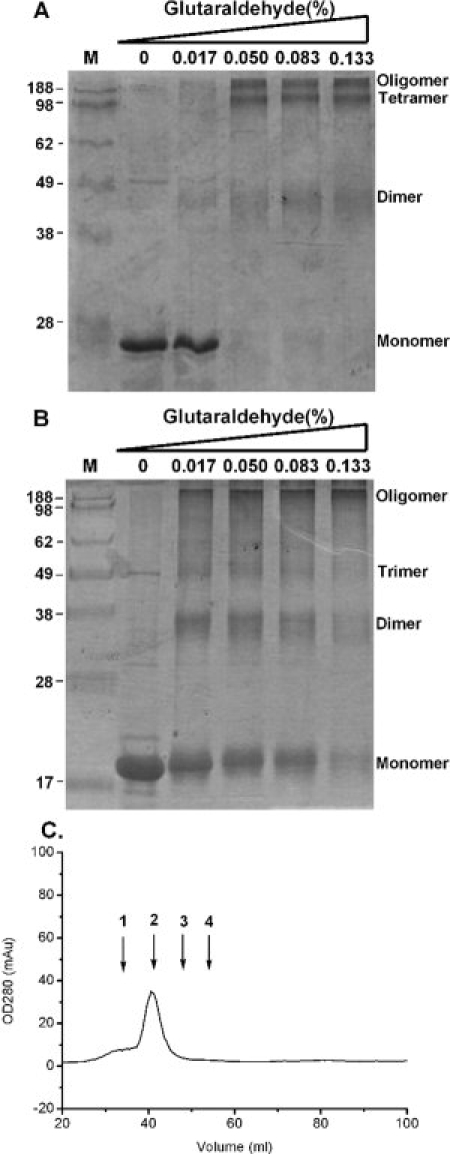
(A) Crosslinking assay of N233–448 in the presence of glutaraldehyde at various concentrations. The concentration of target protein was 2 μM. (B) Crosslinking assay of N301–448 in the presence of the indicated concentrations of glutaraldehyde. The concentration of target protein was 3 μM. (C) Gel filtration chromatography based analysis (Superdex 200 XK16/70) of N301–448 buffered by 50 mM Tris-HCl with 150 mM NaCl and 0.1% CHAPS at pH 7.3. The concentration of target protein was 5 μM. The protein markers of 669 kDa, 223 kDa, 67 kDa, and 43 kDa are denoted as 1, 2, 3, and 4, respectively.
Figure 7.
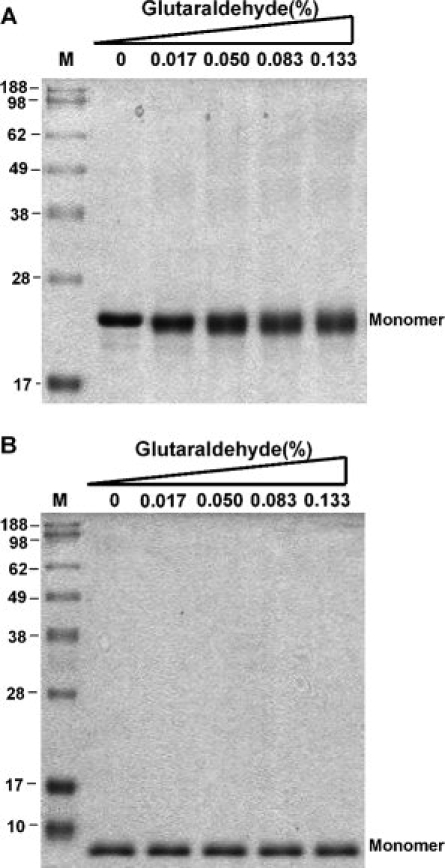
(A) Crosslinking assay of N1–173 in the presence glutaraldehyde at various concentrations. The concentration of target protein was 2 μM. (B) Crosslinking assay of N174–232 in the presence of the indicated concentrations of glutaraldehyde. The concentration of target protein was 1.5 μM.
Discussion
We have previously analyzed the full-length SARS-CoV and HCoV-229E N proteins by CD and found them to be relatively disordered, showing nearly 50% disordered structures.17 Here, we showed that the HCoV-OC43 N protein possesses a more ordered structure than other human coronavirus N proteins, including those of SARS and 229E, which feature 35% α–helix content (Supporting Information Figure S2). Unlike SARS-CoV and HCoV-229E N proteins, the recombinant HCoV-OC43 N protein is highly resistant to proteolysis.17 According to previous heat and urea denaturation analyses, the Tm and Cm values of SARS-CoV N protein are 38°C and 2.77 M, respectively, implying a low stability of the SARS-CoV N protein.45 In this work, the Tm and Cm of the HCoV-OC43 N protein were found to be 52°C and 4 M, respectively. Our results suggest that the HCoV-OC43 N protein is more stable than the SARS-CoV N protein.45
In this study, we have shown that the recombinant HCoV-OC43 N protein expressed in E. coli binds to the intergenic sequence of the RNA genomic leader. This binding demonstrates that the obtained recombinant N protein was properly folded. We examined the binding of recombinant HCoV-OC43 protein for nucleic acids. Our kinetic data showed that HCoV-OC43 N protein binds preferentially to single-stranded RNA over the corresponding single-stranded DNA. The decreased affinity for DNA may be due to two factors. First, the 2′-hydroxyl group of ribose in RNA may form hydrogen bonds with residues of the N protein. Second, in DNA, the methyl group on the C5 of thymine may create a steric clash with residues within the RNA-binding groove. The Hiscox group suggested that phosphorylation of the N protein determined the recognition of virus RNA because phosphorylation may alter the conformation of N protein, and thus affect its RNA-binding sites.12,33,34 In addition, Mohandas et al. have reported that dephosphorylation of MHV N protein by cellular phosphoprotein phosphatase may facilitate the infectious process.46 However, the role of phosphorylation in the RNA-binding sites of HCoV-OC43 N protein was not investigated in our study because the N protein was expressed in E coli. It would be interesting to study the effects of phosphorylation on the conformation and RNA-binding properties of HCoV-OC43 N protein in the future.
Previous studies have reported that the N-terminus of SARS-CoV N protein provides a scaffold for RNA binding.29,36,47 X-ray analysis revealed that the fold of the N-terminal domain of N protein is essentially conserved across the various CoV groups. It has a U-shaped structure with the two arms rich in basic residues and the flexible loops well-ordered around the β-sheet core of the N-terminal domain.26,36 Spencer and Hiscox found that the N-terminal region of IBV protein facilitates long-range, nonspecific interactions between N protein and viral RNA, thus leading to the formation of the ribonucleocapsid via a lure and lock mechanism.29 Similar to the N-terminal domain of SARS-CoV and IBV N proteins, the N-terminal domain of HCoV-OC43 N protein contains many positively charged residues and has been shown by SPR to be responsible for RNA binding. In addition to the N-terminal domain (N1–173), two highly positively charged portions of the central linker region, N174–232 and N233–300, also show RNA-binding activity. Given their high pI values, these RNA-binding regions may rely on electrostatic interactions to interact with RNA. Compared with N1–173 and N233–300, N174–232 has lower RNA-binding affinity and, therefore, appears to play an auxiliary role in RNA binding, despite the presence of a serine/arginine motif (SR-rich motif) that is usually involved in RNA recognition.48 In a previous study, the SR-rich motif of MHV N protein was shown to be the RNA-binding region.28,42 In addition, the C-terminal domain of SARS-CoV N protein has been proposed to bind nucleic acids and to participate in genomic RNA-binding.25,49 However, our studies suggest that the C-terminal domain (N301–448) of the HCoV-OC43 N protein does not bind RNA.
A previous study showed that SARS-CoV N protein forms high-order oligomers through the action of its C-terminal 138 amino acids. These oligomers exist predominantly as dimers.44,50 The N-terminal domain allows for the association of dimers to form higher-order oligomers. Crystal structures of the C-terminal domain of SARS-CoV and IBV N proteins show a similar general polypeptide fold, which strongly suggests that the dimerized N protein is the functional unit in vivo for the four groups of coronaviruses.27,35 The crystal structure of the C-terminal domain shows a tightly intertwined, twofold symmetric C-terminal domain dimer with a β-hairpin (β1 and β2) from one subunit extending into the cavity of the opposite subunit; this forms an antiparallel β-sheet with hydrogen bonds across the dimer interface. Luo et al.49 have reported that this dimer association does not involve electrostatic forces. These authors have further suggested that the SR-rich motif of the SARS-CoV N protein binds to the central region and thereby triggers the multimerization of dimers.51 In addition, Fan et al. showed that the N-terminal region of the IBV N protein is involved in the oligomerization of N protein.26 Here, we analyzed the self-association properties of the HCoV-OC43 N protein and found it to form oligomers. We identified the C-terminal 148 amino acids of HCoV-OC43 N protein to be the oligomerization domain. When expressed in isolation, this region of the C-terminus formed oligomers at only micromolar concentrations, and this oligomerization occurred in the absence of the N-terminal domain. The other truncated constructs of HCoV-OC43 N protein, N1–173 and N174–232, were found to remain as monomers under all of the tested experimental conditions, suggesting that these two regions are not involved in oligomerization during ribonucleocapsid formation.
In summary, we found the HCoV-OC43 N protein exhibited specific properties compared with other CoVs. For example, HCoV-OC43 N protein is more highly ordered and stabler than the SARS-CoV protein previously studied.17,45 We also showed that HCoV-OC43 N protein is composed of three RNA-binding regions lying within the N-terminal region (residues 1–173) and central-linker region (residues 174–232 and 233–300) that bind to RNA in a cooperative manner. Although the C-terminal region (N301–448) lacks RNA-binding activity, this region plays an important role in the oligomerization of the HCoV-OC43 N protein. This study may benefit the development of drugs to disrupt the binding of viral N protein with RNA and viral assembly.
Materials and Methods
Drugs and reagents were purchased from Sigma Chemical Co. All oligoribonucleotides and oligodeoxyribonucleotides were synthesized using an automated DNA synthesizer and purified by gel electrophoresis. The biotin-linked oligomers were synthesized by incorporating the biotin synthon at the 5′-end of the oligomers; oligomers were then immobilized to the streptavidin-coated biosensor chip used for the surface plasmon resonance (SPR) experiments.
Expression and purification of the full-length and truncated N proteins
The templates for the HCoV-OC43 N protein were provided by the Institute of Biological Chemistry, Academia Sinica (Taipei, Taiwan). To generate both full-length and truncated forms of the recombinant N proteins, the N protein gene was amplified by polymerase chain reaction (PCR) from plasmid pGENT using various primers. The PCR products were digested with NdeI and XhoI, and the DNA fragments were cloned into pET28a (Novagen) using T4 ligase (NEB). Bacteria transformed with the resultant plasmid were grown in culture. Protein expression was induced by supplementing the culture medium with IPTG to 1 mM followed by incubation at 10°C for 24 h. After harvesting the bacteria by centrifugation (3500g, 30 min, 4°C), the bacterial pellets were lysed with lysis buffer (50 mM Tris-buffered solution, pH 7.3, 150 mM NaCl, 0.1% CHAPS, and 15 mM imidazole). Soluble proteins were obtained from the supernatant following centrifugation (15,000 rpm, 30 min, 4°C) to remove the precipitate. Full-length and truncated N proteins carrying a His6-tag at their N-termini were purified using a Ni-NTA column (Novagen) with an elution gradient ranging from 15 to 300 mM imidazole. The pure fractions were collected and dialyzed against low-salt buffer. Since the N protein is positively-charged, it was further purified by SP cation exchange chromatography using a gradient from 0.05M to 1.5M NaCl in 50 mM Tris at pH 7.3, 0.1% CHAPS. The protein concentrations were determined using the Bradford method with Bio-Rad protein assay reagents.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were obtained using a JASCO-815 CD spectropolarimeter. Temperature was controlled by circulating water at the desired temperature in the cell jacket. Each protein were dissolved in 50 mM Tris-buffered solution, pH 7.3, 150 mM NaCl, and 0.1% CHAPS. The CD spectra were collected between 250 and 190 nm with 1 nm bandwidth at 1 nm intervals. All spectra were obtained from an average of five scans. The photomultiplier absorbance did not exceed 600 V during the analysis. CD spectra were normalized by subtraction of the background scan with buffer alone. The mean residue ellipticity, [θ], was calculated based on the equation, [θ] = MRW×θλ/10×l×c, where MRW is the mean residue weight, θλ is the measured ellipticity in milidegrees at wavelength λ, l is the cuvette pathlength (0.1 cm), and c is the protein concentration in g/mL. The results were analyzed using the SELCON3 program to calculate the percentage of each type of secondary structure.52 In addition, Tm was determined from the polynomial fitting of the observed curve and taken as the temperature corresponding to half denaturation of the N protein. The first derivative of absorption with respect to temperature, dA/dT, of the melting curve was computer-generated and used for determining the Tm.
Fluorescence spectroscopy
In the urea-induced unfolding experiment, N protein was added to a final concentration of 1 μM to buffer containing various concentrations of urea, and the samples were incubated at 25°C for 30 min. The buffer consisted of 50 mM Tris, pH 7.3, and 0.1% CHAPS. Tryptophan fluorescence measurements were made using an F-4500 fluorescence spectrophotometer (Hitachi) equipped with a cuvette with a 1 cm light path. The excitation wavelength was 288 nm, and the emission data were collected between 300 and 400 nm.
Surface plasmon resonance binding experiments
The affinity, association, and dissociation of N proteins and RNA were measured in a BIAcore 3000A SPR instrument (Pharmacia, Uppsala, Sweden) equipped with a SensorChip SA5 from Pharmacia; the apparatus measured binding by monitoring the refractive index change of the sensor chip surface. These changes, recorded in resonance units (RU), are generally assumed to be proportional to the mass of the molecules bound to the chip. The surface was first washed three times by injecting 10 μL of 100 mM NaCl solution with 50 mM NaOH. To control the amount of RNA (or DNA) bound to the SA chip surface, the biotinylated oligomer was immobilized manually onto the surface of a streptavidin chip until a signal of 1200 RU was achieved in the first cell. The chip surface was then washed with 10 μL of 10 mM HCl to eliminate non-specific binding. The second flow cell was unmodified and served as a control. The appropriate N proteins were dissolved in 50 mM Tris (pH 7.3) with 150 mM NaCl and 0.1% CHAPS, and passed over the chip surface for 140 s at a flow rate of 30 μL/min to reach equilibrium. Blank buffer solution was then passed over the chip to initiate the dissociation reaction; this step was continued for an additional 600 s to complete the reaction. After 600 s, the surface was recovered by washing with 10 μL of 0.1% SDS for each single-stranded RNA (or DNA). Before fitting to the 1:1 Langmuir model, binding data were corrected by subtraction of the control to account for simple refractive index differences. Sensorgrams for interactions between RNA (or DNA) and protein were analyzed using BIA evaluation software (version 3). To verify if mass transport effects may arise during the interaction between N protein and immobilized RNA, binding experiments were performed at different flow rates and different surface binding capacity. It was found that the binding interaction between N protein and RNA is not a mass transport-limited process because the derived kinetic parameters (especially the association rate constant, ka) are independent of the flow rate and binding capacity.
Chemical crosslinking assay
To investigate the oligomerization features of N proteins, a chemical crosslinking experiment was performed. A series of protein solutions containing N proteins was supplemented with various concentrations of glutaraldehyde, and each reaction mixture was incubated at room temperature for 5 min. The reaction was stopped by the addition of 1M Tris. pH 7.3 (0.5%, v/v, final concentration) and incubation on ice. The sample solution was then analyzed by SDS-PAGE.
Analytical gel filtration chromatography
Gel filtration experiments were performed using fast protein liquid chromatography (Amersham Biosciences) equilibrated with buffer containing 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, and 0.1% CHAPS at a flow rate of 0.5 mL/min. Blue dextran was used to determine of the void volume (V0). Several proteins of known molecular weight were used as standards (thyroglobulin, 669 kDa; catalase, 232 kDa; aldolase, 158 kDa; albumin, 67 kDa; ovalbumin, 43 kDa; chymotrypsinogen A, 25 kDa and ribonuclease A, 13.7 kDa, purchased from GE Healthcare), and their elution volumes (Ve) were determined. The standard curve was plotted as the logarithm of the molecular weight against Ve/V0 for the standard proteins.
Acknowledgments
We thank Dr. Tai-Huang Huang (IBS, Academia Sinica) for his helpful discussions and guidance. This manuscript was edited by American Journal Expert (AJE).
Glossary
Abbreviations:
- CoV
coronavirus
- E
envelope
- HCoV
human coronavirus
- IBV
avian infectious bronchitis virus
- M
membrane
- MHV
murine hepatitis virus
- N protein
nucleocapsid protein
- RNP
ribonucleoprotein
- S
spike
- SARS
severe acute respiratory syndrome
- SR-rich
serine and arginine-rich
- Tm
melting temperature.
References
- 1.St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy. Fr Clin Infect Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagneur A, Sizun J, Vallet S, Legr MC, Picard B, Talbot PJ. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J Hosp Infect. 2002;51:59–64. doi: 10.1053/jhin.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel RP, Hendley JO, Davies JA, Gwaltney JM., Jr Coronavirus infections in military recruits. Three-year study with coronavirus strains OC43 and 229E. Am Rev Respir Dis. 1974;109:621–624. doi: 10.1164/arrd.1974.109.6.621. [DOI] [PubMed] [Google Scholar]
- 6.Arbour N, Day R, Newcombe J, Talbot PJ. Neuroinvasion by human respiratory coronaviruses. J Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng Q, Bentwich Z. A novel coronavirus and SARS. N Engl J Med. 2003;349:709. doi: 10.1056/NEJMc031427. [DOI] [PubMed] [Google Scholar]
- 8.Hogue BG, Brian DA. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiscox JA. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You JH, Reed ML, Dove BK, Hiscox JA. Three-dimensional reconstruction of the nucleolus using meta-confocal microscopy in cells expressing the coronavirus nucleoprotein. Adv Exp Med Biol. 2006;581:313–318. doi: 10.1007/978-0-387-33012-9_55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stohlman SA, Fleming JO, Patton CD, Lai MM. Synthesis and subcellular localization of the murine coronavirus nucleocapsid protein. Virology. 1983;130:527–532. doi: 10.1016/0042-6822(83)90106-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masters PS, Parker MM, Ricard CS, Duchala C, Frana MF, Holmes KV, Sturman LS. Structure and function studies of the nucleocapsid protein of mouse hepatitis virus. Adv Exp Med Biol. 1990;276:239–246. doi: 10.1007/978-1-4684-5823-7_33. [DOI] [PubMed] [Google Scholar]
- 14.Masters PS, Sturman LS. Background paper. Functions of the coronavirus nucleocapsid protein. Adv Exp Med Biol. 1990;276:235–238. doi: 10.1007/978-1-4684-5823-7_32. [DOI] [PubMed] [Google Scholar]
- 15.Pyrc K, Jebbink MF, Berkhout B, Van Der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, Stohlman SA. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang TK, Wu MP, Chen ST, Hou MH, Hong MH, Pan FM, Yu HM, Chen JH, Yao CW, Wang AH. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du L, Zhao G, Lin Y, Chan C, He Y, Jiang S, Wu C, Jin DY, Yuen KY, Zhou Y, Zheng BJ. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surjit M, Liu B, Chow VT, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuniga S, Sola I, Moreno JL, Sabella P, Plana-Duran J, Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan KH, Cheng VC, Woo PC, Lau SK, Poon LL, Guan Y, Seto WH, Yuen KY, Peiris JS. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin Diagn Lab Immunol. 2005;12:1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mourez T, Vabret A, Han Y, Dina J, Legrand L, Corbet S, Freymuth F. Baculovirus expression of HCoV-OC43 nucleocapsid protein and development of a Western blot assay for detection of human antibodies against HCoV-OC43. J Virol Methods. 2007;139:175–180. doi: 10.1016/j.jviromet.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, Hsiao CD, Huang TH. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan H, Ooi A, Tan YW, Wang S, Fang S, Liu DX, Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayaram H, Fan H, Bowman BR, Ooi A, Jayaram J, Collisson EW, Lescar J, Prasad BV. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid proteimplications for nucleocapsid formation. J Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson GW, Stohlman SA. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J Gen Virol. 1993;74:1975–1979. doi: 10.1099/0022-1317-74-9-1975. (Part 9) [DOI] [PubMed] [Google Scholar]
- 29.Spencer KA, Hiscox JA. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 2006;580:5993–5998. doi: 10.1016/j.febslet.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, Lin CH, Huang TH. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters PS. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch Virol. 1992;125:141–160. doi: 10.1007/BF01309634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng D, Koetzner CA, Mcmahon T, Zhu Y, Masters PS. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J Virol. 1995;69:5475–5484. doi: 10.1128/jvi.69.9.5475-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer KA, Dee M, Britton P, Hiscox JA. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology. 2008;370:373–381. doi: 10.1016/j.virol.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Gill A, Dove BK, Emmett SR, Kemp CF, Ritchie MA, Dee M, Hiscox JA. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu IM, Oldham ML, Zhang J, Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J Biol Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saikatendu KS, Joseph JS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda M, Chang CK, Ikeya T, Guntert P, Chang YH, Hsu YL, Huang TH, Kainosho M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J Mol Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamahora T, Soe LH, Lai MM. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989;12:1–9. doi: 10.1016/0168-1702(89)90048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. II. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988;27:8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang X. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J Virol. 2000;74:10571–10580. doi: 10.1128/jvi.74.22.10571-10580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson GW, Stohlman SA, Tahara SM. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 43.Welter LM, Stohlman SA, Deans RJ. MHV leader RNA secondary structure affects binding to the nucleocapsid protein. Adv Exp Med Biol. 1990;276:247–254. doi: 10.1007/978-1-4684-5823-7_34. [DOI] [PubMed] [Google Scholar]
- 44.Yu IM, Gustafson CL, Diao J, Burgner JW, Li Z, Zhang J, Chen J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J Biol Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Wu X, Wang Y, Li B, Zhou H, Yuan G, Fu Y, Luo Y. Low stability of nucleocapsid protein in SARS virus. Biochemistry. 2004;43:11103–11108. doi: 10.1021/bi049194b. [DOI] [PubMed] [Google Scholar]
- 46.Mohandas DV, Dales S. Endosomal association of a protein phosphatase with high dephosphorylating activity against a coronavirus nucleocapsid protein. FEBS Lett. 1991;282:419–424. doi: 10.1016/0014-5793(91)80528-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 48.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 49.Luo H, Chen J, Chen K, Shen X, Jiang H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid proteself-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 50.Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang CF, Huang TH. The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett. 2005;579:5663–5668. doi: 10.1016/j.febslet.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo H, Ye F, Chen K, Shen X, Jiang H. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 52.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]


