Abstract
Integration of HIV-1 cDNA into the host genome is a crucial step for viral propagation. Two nucleotides, cytosine and adenine (CA), conserved at the 3′ end of the viral cDNA genome, are cleaved by the viral integrase (IN) enzyme. As IN plays a crucial role in the early stages of the HIV-1 life cycle, substrate blockage of IN is an attractive strategy for therapeutic interference. In this study, we used the 2-LTR-circle junctions of HIV-1 DNA as a model to design zinc finger protein (ZFP) targeting at the end terminal portion of HIV-1 LTR. A six-contiguous ZFP, namely 2LTRZFP was designed using zinc finger tools. The designed motif was expressed and purified from E. coli to determine its binding properties. Surface plasmon resonance (SPR) was used to determine the binding affinity of 2LTRZFP to its target DNA. The level of dissociation constant (Kd) was 12.0 nM. The competitive SPR confirmed that 2LTRZFP specifically interacted with its target DNA. The qualitative binding activity was subsequently determined by EMSA and demonstrated the aforementioned correlation. In addition, molecular modeling and binding energy analyses were carried out to provide structural insight into the binding of 2LTRZFP to the specific and nonspecific DNA target. It is suggested that hydrogen-bonding interactions play a key role in the DNA recognition mechanisms of the designed ZFP. Our study suggested an alternative HIV therapeutic strategy using ZFP interference of the HIV integration process.
Keywords: HIV-1, AIDS, zinc finger protein, 2-LTR-circle junctions, molecular modeling
Introduction
Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of acquired immune deficiency syndrome (AIDS). It is one of the greatest health problems facing the world today.1 According to UNAIDS and WHO, global estimates of the number of people living with HIV/AIDS as of the end of 2007 were 33 million adults and children.2 HIV is a member of the family Retroviridae and subfamily lentivirinae. Soon after viral infection, the viral reverse transcriptase synthesizes a double-stranded DNA (ds-DNA). The subsequent insertion of viral genome into the host genome by integrase (IN) is a vital step in the HIV life cycle.3–5 Three forms of unintegrated viral DNA have been identified: a linear form which is flanked by long terminal repeat (LTR) copies at each terminus, and two circular molecules containing either one or two LTRs. The linear form is thought to be the substrate for integration.6–12 Integrase is involved in removing 2 bp from the 3′ ends of viral DNA, the so-called 3′ processing. The second reaction catalyzed by IN, i.e., strand transfer, inserts both 3′ ends of HIV-1 DNA into the host genome.13,14
Current antiretroviral drugs therapy for AIDS involves the use of a multidrug cocktail referred to as highly active antiretroviral therapy (HAART).15 IN inhibitors belong to a novel class of drugs that function by inhibiting integration of proviral DNA into the host cell genome. IN inhibitors can be divided into two groups: 3′ processing and selective strand transfer inhibitors. These drugs are currently undergoing clinical trials.16,17 However, several questions concerning the interaction of IN with its inhibitors remain unclear, and resistant strains have been found.18–20 Therefore, a new approach targeting HIV-1 DNA at the level of the end terminal sequence of LTR by gene therapy may be a useful approach for inhibiting viral replication.
Our strategy herein is based on the specific DNA recognition by Cys2His2 zinc finger protein (ZFP). The Cys2His2 ZFP is the most common DNA-binding domain in the human and metazoan genome and plays a key role as a class of transcription factors. Its structure consists of a simple ββα fold of 30 amino acids in length, which is stabilized by hydrophobic interactions and chelation of a single zinc ion between two histidine residues and two cysteine residues.21,22 Within the residue at the N-terminus of the α-helix, the positions −1, 3, and 6 typically bind to three contiguous base pairs in a major groove of the target DNA.23,24 Therefore, several artificial ZFPs have been designed to target DNA sequences with high specificity and affinity.25–29
The ZF Tools server30 implements an effective algorithm to generate ZFPs for in silico or in vivo applications. The program facilitates web-based design of ZFPs that specifically recognize DNA sequence motifs. In this study, we used the 2-LTR-circle junctions of HIV-1 DNA as a model to design a ZFP targeted at the end terminal part of HIV-1 LTR, which is a region for 3′ processing by IN of HIV-1. We designed a six-contiguous ZFP. Binding affinity was carried out by surface plasmon resonance (SPR) and an electrophoretic mobility shift assay (EMSA). Molecular modeling was also used to gain structural insight into the DNA recognition patterns of the designed ZFP, in the cases of being bound to either a specific or a nonspecific DNA sequence. Relative binding energies of the specific and nonspecific complexes were also calculated.
Results
Procurement of the target sequence and designing the 2LTRZFP
To target the HIV-1 DNA sequence of 2-LTR-circle junctions, the DNA sequence was submitted to the zinc finger tools using “Search DNA Sequence for Contiguous Target Sites” mode. Eight candidate DNA sequences were obtained and the binding prediction scores of these sequences were selected for designing the ZFP. The sequence that had the highest score was 5′ CTAGCAGTACTGGATGGG 3′ (Fig. 1).
Figure 1.
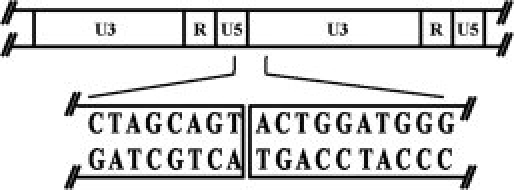
Schematic representation of the circle junction region of HIV-1.45 Predicted organization of U3, R, and U5 in 2-LTR-circle junctions shown above, whereas sequence in box was selected for designing the ZFP.
The design tool generated the full-length amino acid sequence of 2LTRZFP. This protein was composed of 176 amino acids [Fig. 2(A)] for a six zinc finger motif. Each finger was linked with the canonical TGEKP linker.
Figure 2.

(A) Full-length amino acid sequence of 2LTRZFP. The amino acids shown by underline were located at the specific positions of −1, 3, and 6 from the N-terminal of the α-helix, respectively. (B) Full-length optimized nucleotide sequence of 2LTRZFP. (C) Primary sequence alignments of Aart (PDB code 2I13) and 2LTRZFP with 80% sequence identity. The underlined letters represent amino acid residues of the ZFP those are different from the Aart peptide. Positions −1, 3, and 6 above the recognition helix column represent amino acid residues often involved in DNA recognition. Numbers at the end of each line show the running number of the last amino acid residue.
The amino acid sequence of 2LTRZFP was reverse-translated into a nucleotide sequence, and all codons in this sequence were optimized [Fig. 2(B)]. The full-length optimized sequence was sent for full-gene synthesis by Blue Heron Biotechnology. Predicted properties of the resulting proteins were computed using ExPaSy proteomics tools. The resulting theoretical pI/Mw was 9.46/20.1 kDa. Prediction of the protein subcellular localization using the LOCtree program indicated that 2LTRZFP is a nuclear protein and is found in the nucleus.
Construction of pTriEx-4- 2LTRZFP-GFP
The restriction endonucleases XcmI and SmaI in the plasmid vector pTriEx-4-GFP was used as a cloning sites for the 2LTRZFP gene fragment to construct the pTriEx-4-2LTRZFP-GFP containing His6 at N-terminal and fused-GFP at C-terminal under the control of the CMV, T7, or p10 promoters. The plasmid vector pTriEx-4-2LTRZFP-GFP was digested by the restriction endonucleases XcmI and SmaI and the product was subjected to 1% agarose gel electrophoresis and visualized by ethidium bromide staining. Restriction digest analysis showed that the pTriEx-4-2LTRZFP-GFP was constructed successfully at a 547 bp band (data not shown).
Protein expression and purification
2LTRZFP-GFP was expressed in its recombinant forms in E. coli Origami B (DE3) Novagen (Madison, WI). Protein purification was performed using His-Bind column chromatography. The entire expression and purification processes were monitored by SDS-PAGE [Fig. 3(A)]. Comparison between lane 2 [total lysate of E. coli Origami B (DE3)] and lane 3 [Total lysate of E. coli Origami B (DE3) with pTriEx-4-2LTRZFP-GFP was induced by isopropyl β-d-thiogalactopyranoside (IPTG) after OD600 reach 1.0 at 30°C for overnight] indicates that the expression of fusion protein His6-2LTRZFP-GFP (∼50 kDa) was successful. Lane 6 shows that this fusion protein bound to the His-Bind column chromatography, and it can be eluted by elution buffer containing 1M imidazole, 0.5M NaCl, 20 mM Tris-HCl, pH 7.9 (lane 7).
Figure 3.
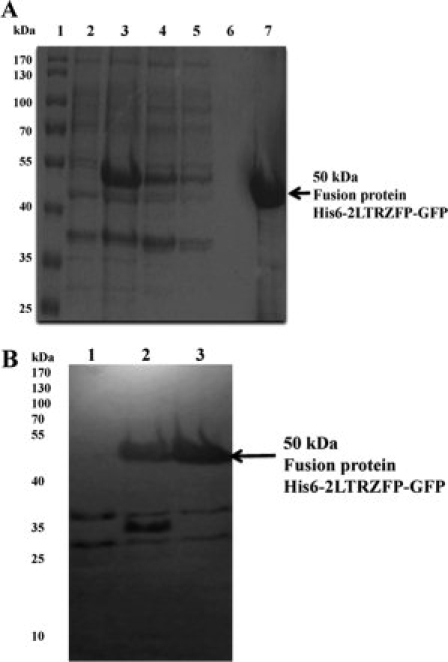
(A) SDS-PAGE analysis of His6-2LTRZFP-GFP at different and purification steps. Lane 1: protein marker, lane 2: total bacterial lysate, lane 3: total bacterial lysate with pTriEx-4-2LTRZFP-GFP after IPTG induction for overnight, lane 4: pass through lysate, lanes 5, 6: solution after washing the column with binding buffer and washing buffer, respectively, lane 7: elution of purified His6-2LTRZFP-GFP. The arrow indicates the size of 50 kDa of His6-2LTRZFP-GFP. Numbers on left show size in kilo Dalton of protein ladder. (B) Western blot analysis of His6-2LTRZFP-GFP at different condition of protein expression. Lane 1: total bacterial lysate, lanes 2, 3: total bacterial lysate with pTriEx-4-2LTRZFP-GFP after IPTG induction for 4 h, and overnight, respectively. The arrow indicates the size of 50 kDa of His6-2LTRZFP-GFP. Numbers on left show size in kilo Dalton of protein ladder.
Western blotting was performed by using anti-His mAb as primary antibody [Fig. 3(B)]; the analysis demonstrated that His6-2LTRZFP-GFP (∼50 KDa) was successfully expressed. To separate GFP and 2LTRZFP, the purified recombinant 2LTRZFP-GFP containing the thrombin recognition sequence was cleaved via thrombin, and 2LTRZFP was purified by using His-Bind column chromatography. (Data not shown).
Expression of 2LTRZFP-GFP in HeLa cells
To test the expression of the 2LTRZFP-GFP recombinant protein in a mammalian system, pTriEx-4-2LTRZFP-GFP was transfected into a HeLa cell line. The emitted green fluorescence was observed under the microscope 24 and 48 h after transfection. The green fluorescence was observed in the nucleus in the pTriEx-4-2LTRZFP-GFP transfection group and in the cytoplasm for pTriEx-4-GFP control group (Fig. 4). This result indicated that 2LTRZFP-GFP recombinant protein is a nuclear protein and confirmed the successful folding of the protein.
Figure 4.
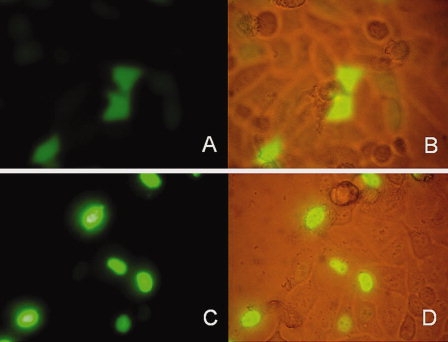
Expression of green fluorescent protein in HeLa cells 24 h after transfection. (A, B) Transfected with recombinant plasmid pTriEx-4-GFP. (C, D) Transfected with recombinant plasmid pTriEx-4-2LTRZFP-GFP.
Evaluating of dissociation constants and competitive DNA binding activity by SPR
To determine real-time binding kinetics between 2LTRZFP-GFP and its target DNA sequence, we used SPR as a tool for qualitative analysis. Different concentrations of 2LTRZFP-GFP were injected into the immobilized chip with specific ds-DNA. The 2LTRZFP-GFP could bind to its target ds-DNA on a nanomolar scale, with Kd = 12.0 nM [Fig. 5(A)]. We also performed a competitive SPR to determine the specificity of binding between 2LTRZFP-GFP and its target DNA. The concentration of 2LTRZFP-GFP (1.2 μM) was used to compete with 26.5 μM of nonbiotinylated target ds-DNA and nonspecific ds-DNA before injection onto the immobilized chip with specific ds-DNA. The 2LTRZFP-GFP could bind to its target DNA sequence [result compared with nonspecific ds-DNA, Fig. 5(B)]. In addition, the specificity of binding activity was also assayed by injection of GFP and bovine serum albumin (BSA) as controls. These results demonstrated that the binding activity was involved with ZFP only [Fig. 5(A)]. To obtain the binding properties, we also tested the binding activity of 2LTRZFP-GFP and its target DNA sequence in zinc buffer containing 1 μM, 1 mM, and 10 mM of EDTA. The binding activity was decreased when the concentration of EDTA was high. These findings supported the conclusion that the binding between 2LTRZFP-GFP and its target DNA sequence depended on zinc ions, and that this metal ion is important for stabilizing ZFP [Fig. 5(C)].
Figure 5.
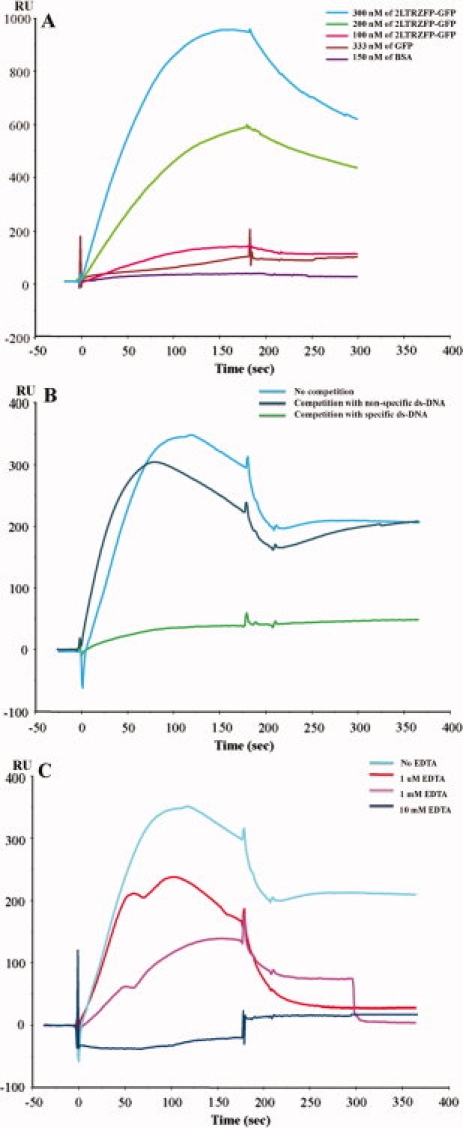
Kinetic analysis between 2LTRZFP-GFP and its target DNA sequence. (A) Binding kinetic of 2LTRZFP-GFP (300, 200, and 100 nM), GFP, and BSA to the immobilized chip with specific ds-DNA. (B) For the competitive SPR, 2LTRZFP-GFP was incubated with nonbiotinylated ds-DNA of its target ds-DNA, and nonspecific ds-DNA in zinc buffer for 15 min before injection. (C) 2LTRZFP-GFP was injected to determine the binding activity with its target DNA sequence in zinc buffer containing 1 μM, 1 mM, and 10 mM of EDTA.
DNA binding activity by EMSA
To confirm the binding activity, EMSA was performed using an EMSA kit as described in the section Materials and Methods. A complete binding complex of 1 μM of 2LTRZFP-GFP and 250 nM of its specific DNA duplex could be observed [see Fig. 6(A), lane 8] until no band of free DNA duplex was seen. At the same time, a faint band of binding complex was observed in the reaction of 2LTRZFP-GFP and nonspecific DNA duplex [Fig. 6(A), lane 9], whereas the intensity of free DNA duplex was not changed [Fig. 6(A), lanes 3 and 6]. This experiment also demonstrated that GFP in 2LTRZFP-GFP recombinant protein was not involved in the binding [lanes 4, 5, and 6 of Fig. 6(A)]. The dilution effect of DNA binding complex can be observed in Figure 6(B,C).
Figure 6.
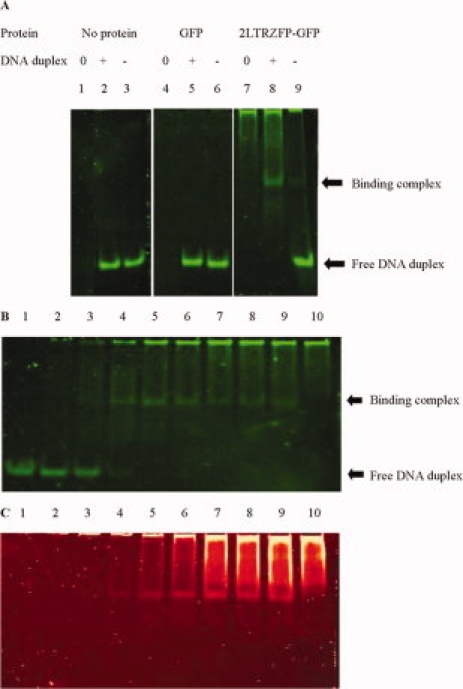
Electrophoretic mobility shift assay (EMSA). (A) The protein used for each reaction is 1 μM, whereas [0], no DNA duplex; [+], specific DNA duplex; [−], nonspecific DNA duplex. The gel was stained with SYBR® Green EMSA stain (B) or with SYPRO® Ruby EMSA stain (C). In panels B and C: lanes 1 to 9, the specific DNA duplex used for each reaction was 250 nM, whereas 2LTRZFP-GFP was varied, the concentration from 0, 0.125, 0.25, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 μM, respectively. Lane 10, 2LTRZFP-GFP was used 3.0 μM, but no DNA duplex.
2LTRZFP-DNA models and their binding energy analyses
Molecular models of the designed 2LTRZFP-DNA complexes, Zif1 and Zif2, were remodeled based on the crystal structure of Aart bound to DNA (PDB code 2I13, see more details in the Materials and Methods), where all backbone atoms were maintained in the same position as found in the crystal structure. Amino acid residues within helical helices were modified solely according to our 2LTRZFP sequence [Fig. 2(C)]. Pair-wise similarity comparison between Aart and 2LTRZFP proteins was also performed using the blastp algorithm of the BLAST tool.31 The result showed a relatively high sequence identity (80%).
After a 2500-cycle minimization for each of Zif1 and Zif2 complex, the root mean square deviation (RMSD) of the minimized structure of each model was measured. The RMSDs of 2LTRZFP and DNA backbones were 0.51 and 0.94 A°, respectively, suggesting that Zif1 and Zif2 are relatively similar in conformation (Fig. 7). As the first crystal structure of Zif268 bound to DNA was obtained, it has been suggested that amino acid positions −1, 3, and 6, with respect to the start of the recognition helix, play specific roles for binding to three contiguous base pairs in each DNA triplet.23,24 The residue-by-residue analysis of 2LTRZFP-DNA interaction for each residue in the recognition helix to each DNA triplet compared to its nonspecific triplet was subsequently explored to understand a DNA recognition pattern for 2LTRZFP.
Figure 7.
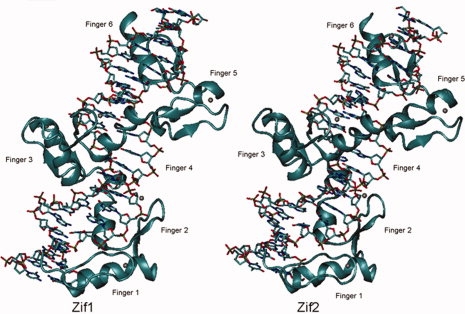
Comparisons of conformation of the recognition binding region in Zif1 (in left panel) and Zif2 (in right panel). The nucleotides are shown in a licorice model and the protein backbone in a cartoon diagram.
Figure 8 demonstrates all possible hydrogen-bonding interaction between each zinc finger motif and DNA residues in both specific (Zif1) and nonspecific (Zif2) complexes, and the distances are listed in Table I. In general, a major contribution to stabilizing the 2LTRZFP-DNA complexes is the electrostatic interactions between amino acid residues in the zinc finger motif and nucleotides in the DNA target, where there is no water-mediated interaction apparent in these complexes.
Figure 8.
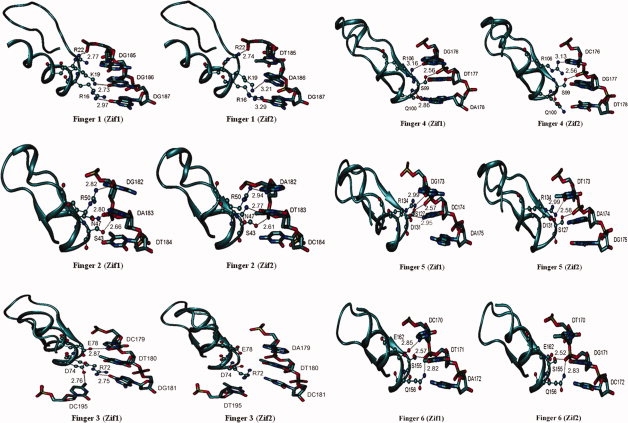
Possible hydrogen bonding between the zinc finger residues and the target DNA triplet within distance < 3.5 A° between H-bond donor and H-bond acceptor associated with the distance.
Table I.
Hydrogen-Bonding Distances between 2LTRZFP Residues and DNA Residues
| 2LTRZFP residue | DNA residue | Distance (Å) | 2LTRZFP residue | DNA residue | Distance (Å) | ||
|---|---|---|---|---|---|---|---|
| Finger 1 | ARG16(NH) | Zif1:DG187(O6) | 2.97 | Finger 4 | SER99(OH) | Zif1:DT177(O2P) | 2.56 |
| Zif2:DG187(O6) | 3.29 | Zif2:DT177(O2P) | 2.56 | ||||
| LYS19(NZ) | Zif1:DG186(O6) | 2.73 | GLN100(O) | Zif1:DA178(N6) | 2.86 | ||
| Zif2:DA186(N6) | 3.21 | Zif2:DT178 | – | ||||
| ARG22(NH) | Zif1:DG185(O2P) | 2.77 | ARG106(NH) | Zif1:DG176(O6) | 3.16 | ||
| Zif2:DT185(O2P) | 2.74 | Zif2:DC176(N4) | 3.13 | ||||
| Finger 2 | SER43(OH) | Zif1:DA183(O1P) | 2.66 | Finger 5 | SER127(OH) | Zif1:DC174(O2P) | 2.57 |
| Zif2:DT183(O1P) | 2.61 | Zif2:DA174(O2P) | 2.58 | ||||
| ASN47(ND) | Zif1:DA183(O2P) | 2.80 | ASP131(OH) | Zif1:DC174(N4) | 2.95 | ||
| Zif2:DT183(O2P) | 2.77 | Zif2:DA174 | – | ||||
| ARG50(NH) | Zif1:DG182(O6) | 2.82 | ARG134(NH) | Zif1:DG173(O6) | 2.99 | ||
| Zif2:DA182(N6) | 2.94 | Zif2:DT173(O4) | 2.99 | ||||
| Finger 3 | ARG72(NH) | Zif1:DG181(O6) | 2.75 | Finger 6 | SER155(OH) | Zif1:DT171(O2P) | 2.57 |
| Zif2:DC181 | – | Zif2:DG171(O2P) | 2.52 | ||||
| ASP74(OH) | Zif1:DC195(N4) | 2.76 | GLN156(NH) | Zif1:DT171(O2P) | 2.82 | ||
| Zif2:DT195 | – | Zif2:DG171(O2P) | 2.83 | ||||
| GLU78(OH) | Zif1:DC179(N4) | 2.87 | GLU162(OH) | Zif1:DC170(N4) | 2.85 | ||
| Zif2:DA179 | – | Zif2:DT170 | – |
Atom names are indicated in parentheses.
In finger 1, although the hydrogen bonding network among ARG16, LYS19, and ARG22 to its DNA target in Zif1 and Zif2 is similar, the hydrogen bonds in Zif2 are weaker than those of Zif1 with respect to its longer bonding distances. The hydrogen-bond distances in finger 2 are very similar in Zif1 and Zif2, whereas they are completely different in finger 3. The amino N atom of ARG72 is bonded to the carbonyl group of DG181 in the Zif1 complex; however, there is no hydrogen bonding between ARG72 and DC181 in Zif2 because of repulsive interaction of the positively charged amino group. The O atom of ASP74 is hydrogen bonded to the N atom of DC195 in Zif1, whereas, in Zif2, the steric effect of the methyl group in DT195 could lead to an improper formation of hydrogen bonds. A hydrogen bond donor at the amino N4 atom of DC179 (Zif1) efficiently establishes a hydrogen bond with GLU78, where the amino group of DA179 (Zif2) is located away from GLU78. Hydrogen-bond patterns in fingers 4, 5, and 6 in Zif1 and Zif2 are similar. The negatively charged amino acids, aspartic acid and glutamic acid, may play an important role in DNA recognition as being involved in selective contacts with cytosine only. Moreover, serine residues in position −2 of the recognition helices are likely to selectively bind to the phosphate backbone of the target DNA as well as arginine residues in position 6 that can bind to all bases of DNA. Finally, from the minimized structure, 2LTRZFP is more probable to establish hydrogen bonds within Zif1 than Zif2, resulting in the more selective binding to the target DNA.
In addition to the structural analysis, the molecular mechanics/generalized Born surface area (MM/GBSA) approach was used to further calculate relative binding energies between the designed peptide and the target DNA compared to its nonspecific sequence (Zif2). The ΔΔGbinding reveals that 2LTRZFP is more favorable for binding to the target DNA in Zif1 than that of Zif2 by −35.0 ± 0.5 kcal/mol (see Table II). This suggests that the Zif1 complex is mainly stabilized by the electrostatic interaction (Ees) as the better hydrogen-bonding network is presented.
Table II.
Estimated Relative Binding Energies of Specific (Zif1) and Nonspecific (Zif2) Complexes from the MM/GBSA Approach
| Sequence | ΔΔEvdW (kcal mol−1) | ΔΔEes (kcal mol−1) | ΔΔGpol (kcal mol−1) | ΔΔGnonpol (kcal mol−1) | ΔΔGbinding (kcal mol−1) |
|---|---|---|---|---|---|
| Zif1-Zif2 | −3.1 ± 1.7 | −255.9 ± 9.0 | 226.3 ± 8.2 | −2.3 ± 0.0 | −35.0 ± 0.5 |
Discussion
The Cys2His2 ZFPs have been demonstrated to be specific binding proteins to the recognition sequences.25–29 In this study, a six-finger protein was constructed specifically to 18 bp of 2-LTR-circle junctions. The first, second, and third fingers can target the terminal sequence of 5′ LTR, whereas the terminal sequence of 3′ LTR can be targeted by the other three fingers. The 2LTRZFP-GFP was expressed and purified from E. coli to investigate its binding properties. The binding activity was on the nanomolar scale, which is similar to the affinity level of HIV-1 IN.32,33 This finding suggested that the HIV-1 IN might be interfered with by the 2LTRZFP as well. The specificity of this binding was obtained by competitive SPR, which indicated that there was specific binding of 2LTRZFP-GFP and its target DNA. However, this binding affinity was decreased when the concentration of EDTA in binding buffer was increased. This data demonstrated the influence of zinc ion on correct folding of 2LTRZFP domains in recognizing a specific DNA sequence. Moreover, 2LTRZFP can specifically bind to its target ds-DNA, whereas binding of GFP in the C-terminal part was not facilitated.
Additionally, theoretical studies were used to investigate at the atomic level the DNA recognition of 2LTRZFP. Although it is well known that amino acid positions −1, 3, and 6 within the recognition helix directly contact the DNA triplet, positions −2, 1, and 5 are also described as playing a role in binding to the phosphate backbone.34 Our study suggests that serine at position −2 and arginine at position 6 are very important in binding to the phosphate DNA backbone and 5′-nucleotide, respectively. However, the roles of amino acid positions −1 and 3 remain unclear. The results herein are particularly interesting because single or multiple mutations of amino acid position −1 and 3 would be able to enhance selectivity of the ZFP motif to the 2-LTR-circle junctions.
Mutation of the IN enzyme encoded by pol gene can occur,18–20 while numerous studies have reported that the important 2–7 base pairs upstream and the well-conserved CA dinucleotide at the 3′ end of viral cDNA play key roles in processing and strand transfer.35–40 Therefore, the advantage of targeting the 3′-end terminal part of HIV-1 LTR to interfere with the IN enzyme is an attractive strategy for a new approach to gene therapy.
Many investigations have targeted viral replication by ZFP recognition of the DNA of viruses. The engineered ZFPs targeting the Sp1 binding site in promoter region of HIV-1 have been reported, and one of them was found to inhibit HIV-1 replication by 75%.41 In 2004, Segal et al. demonstrated that a transcriptional repressor protein, namely KRAB-HLTR3, was able to achieve 100-fold repression of transcription from the HIV-1 promoter. This transcription factor also repressed the replication of several HIV-1 strains 10-to 100-fold in T-cell line and primary blood mononuclear cells with no significant cytotoxicity.42 Recently, ZFPs were designed to bind DNA sequences in the duck hepatitis B virus to inhibit viral transcription in tissue culture. Two candidate ZFPs decreased production of viral products and progeny viral genomes.25 These studies supported the use of ZFPs to inhibit viral replication, opening new avenues in gene therapy. There are a various methods available to deliver a protein for gene therapy. The in vivo adenoviral gene transfer of ZFPs has been performed to induce angiogenesis in a mouse model,43 whereas retroviral and lentiviral gene transfer have been delivered the artificial zinc finger transcription factors to bind sites in the HIV-1 promoter to repress the replication of several HIV-1 strains.42 Recently, various nonbiological and biological carrier systems have been developed for HIV-1 gene therapy, such as nanoparticles, liposomes, or synthetic polymers, which can be taken up by many cell types.44
Our study has demonstrated that 2LTRZFP can specifically bind to the HIV-1 integrase recognition sequence at 2-LTR-circle junctions. The findings may be applied for limiting viral integration to maximize the impact of HIV gene therapy in the future.
Materials and Methods
Zinc finger protein design
Studying order to identify target sites within the DNA sequence, the HIV-1 DNA sequence of 2-LTR-circle junctions45 was submitted to the ZF Tools server of the Barbas Laboratory of the Scripps Research Institute.30 The minimum target size of 18 base pairs was set to obtain the output DNA target sequences and amino acid sequences for the six fingers of the ZFP. This amino acid sequence was then reverse-translated into a nucleotide sequence. Codon usage of the amino acid sequences of the selected ZFP was optimized by using the JAVA Codon Adaptation Tool (JCat)46 and by manual optimization. The designed ZFP was further modified with flanking XcmI and SmaI restriction endonuclease sites. The full-length optimized DNA sequence was sent for full-gene synthesis to Blue Heron Biotechnology (Bothell, WA). Predicted properties of the resulting proteins were computed using ExPaSy proteomics tools.47 Prediction of protein subcellular localization was done using the LOCtree program.48
Plasmid construction
The full-length optimized DNA sequence of 2LTRZFP was synthesized and cloned into pUC19 vectors by Blue Heron Biotechnology. The 2LTRZFP gene fragment was ligated to pTriEx-4-GFP using the flanking XcmI and SmaI sites to construct pTriEx-4–2LTRZFP-GFP, creating an N-terminal His6-fusion protein with GFP in the C-terminal part. This plasmid has a multiple expression system driven by the CMV, T7, or p10 promoters. The ligation product was transformed into the competent E. coli XL-1 Blue cells and plated on Luria-Bertani (LB) agar containing 100 μg/mL of ampicillin. The plasmid miniprep was performed using a QIAGEN Miniprep Kit (Qiagen, Hilden, Germany). The constructed plasmid was preliminary identified by the restriction endonucleases XcmI and SmaI. PCR and DNA sequencing were performed for confirmation.
Expression and purification of His6-2LTRZFP-GFP
We used E. coli Origami B (DE3) as the expression strain. A 10 mL Terrific broth preculture, supplemented with 100 ug/mL ampicillin, 12.5 ug/mL kanamycin, and 12.5 ug/mL tetracycline was incubated at 37°C until reaching an absorbance at 600 nm of about 1.0–1.8. The preculture was diluted 1:100 into 100 mL Terrific broth medium with 100 μg/mL ampicillin, 12.5 ug/mL kanamycin, 12.5 ug/mL tetracycline and supplemented with 100 μM Zn2SO4. The culture was incubated at 37°C. When an absorbance about 1.0 at 600 nm was reached, the bacterial culture was induced by 0.1 mM IPTG at 30°C for overnight. Bacteria were then harvested by centrifugation (7500g at 4°C for 15 min). The bacterial pellets were resuspended in 6.25 mL of B-PER II Bacterial Protein Extraction Reagent (Pierce, Rockford, IL) and lysed by ultrasonication in an ice bath. The lysate was centrifuged at 15,000g for 20 min at 4°C. The supernatant was collected and filtered with microfiltration membranes (0.22-μm pore size). The clear solution containing His6-2LTRZFP-GFP was applied to His-bind column chromatography (Novagen, San Diego, CA) for protein purification. The eluated fraction containing His6-2LTRZFP-GFP was observed using Coomassie brilliant blue R250 stain (Bio-Rad, Hercules, CA). The purified protein was kept in final concentration 25% (W/V) glycerol for long-term storage at −80°C. The protein concentration was quantified by using the Micro-BCA protein assay (Pierce). All the samples were analyzed by SDS-PAGE. Western blot analysis was performed on a Hybond-P polyvinylidene fluoride membrane. (Amersham Bioscience, Piscataway, NJ). After being blocked with 5% skim milk in PBS, the proteins were probed with anti-his-tag monoclonal antibody (Genscript, Piscataway, NJ) as a primary antibody and horseradish peroxidase-labeled goat anti-mouse (Sigma, St Louis, MO) as a secondary antibody, using the ECL system (GE Healthcare, Buckinghamshire, UK).
Cell culture and transfection of HeLa cells
HeLa cervical carcinoma cells were kindly obtained from Dr. A. Lieber, University of Washington, Seattle, WA. HeLa cells (1 × 105) were seeded onto 24-well plate in humidified atmosphere under 5% CO2 at 37°C in Dulbecco's Modified Eagle's medium (DMEM) (Gibco, Grand Island, NY) containing penicillin (100 U/mL), streptomycin (100 μg/mL), and 2 mM l-glutamine, supplemented with 10% fetal bovine serum (FBS) (Gibco) for 24 h before transfection. Transfection mixture was prepared by adding 1 μg of the pTriEx-4-2LTRZFP-GFP or control vector pTriEx-4-GFP and 2 μL of GeneJamer (Stratagene, La Jolla, CA) into DMEM up to 200 μL. Then, the mixture was incubated at room temperature for 10 min. The culture supernatant was discarded and 300 μL of fresh DMEM containing 10% FBS and antibiotics was added. The transfection mixture was added to the cells and incubated at 37°C in 5% CO2 for 5 h. A total of 500 μl of DMEM containing 10% FBS and antibiotics was added and further cultured at 37°C in 5% CO2 for 24–48 h. Green fluorescent protein (GFP) was observed under a fluorescent microscope.
Double-stranded DNA preparation
ds-DNA of specific and nonspecific target DNA was prepared for testing the binding activity of 2LTRZFP-GFP. A pair of specific ds-DNA (sense) was designed as follows: 5′-AAA TCT CTA GCA GTA CTG GAT GGG CTA ATT-3′ and a pair of nonspecific ds-DNA (sense) was also designed as follows 5′-TGA CAG TGC TAG CGT ATC ATC TAG TCG ACG-3′. The specific sequences to 2LTRZFP-GFP are shown in italic. The reaction mixture (100 μL) for annealing was composed of each 600 pmol single-stranded DNA (ss-DNA) and complementary strand in 50 mM NaCl. The mixture was heated at 95°C for 5 min and then slowly cooled to room temperature for 90 min. For immobilization of ds-DNA on a sensor chip SA (Biacore AB, Uppsala, Sweden) for SPR, the 3′ end of the antisense strand of specific target DNA was labeled with biotin.
Surface plasmon resonance
SPR was carried out on a BIACORE 2000™ biosensor (Biacore AB). The target DNA duplexes were immobilized by injecting the biotinilated specific ds-DNA of 0.5 μg/mL in 0.3M NaCl on a Sensor Chip SA (Biacore AB) at a flow rate 5 μL/min in running buffer (HBS-EP), containing 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) surfactant P20 (Biacore AB). Typically 800–1000 RUs of the target ds-DNA were immobilized. After DNA immobilization, the chip was washed with 50 mM NaOH/1M NaCl and then primed with zinc buffer [10 mM Tris-HCl, pH 7.5, 90 mM KCl, 1 mM MgCl2, 90 μM ZnSO4, 5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonylfluoride), which was used as a running buffer for a period of binding analysis. ZFP was diluted in zinc buffer and 60 μL was injected at a flow rate of 20 μL/min, followed by a dissociation phase of 180 s. Before each injection, the baseline stability was achieved by injecting 1M NaCl for 2 min. To calculate the binding constants (Kd), the kinetic parameters were evaluated with BIA evaluation software 3.1 (Biacore AB) using a 1:1 binding model with mass transfer. For competitive SPR, 2LTRZFP-GFP was incubated with different concentrations of nonbiotinylated ds-DNA of its target ds-DNA, and nonspecific ds-DNA in zinc buffer for 15 min before injection.
Electrophoretic mobility shift assay
2LTRZFP-GFP or GFP was incubated with 250 nM of DNA duplex at room temperature for 1 h in zinc buffer. Total volumes were 10 μL per reaction. The reaction mixtures were mixed with 2 μL of 6× EMSA gel-loading solution (component D) of EMSA kit [E33075] (Invitrogen, Paisley, UK) before being loaded on 5% nondenaturing polyacrylamide gels using 100 V, 30 min. Gels were stained by using two fluorescent dyes for detection—SYBR® green EMSA nucleic acid gel stain (component A) and SYPRO® ruby EMSA protein gel stain (component B) by following the protocol from the same kit. The stained gels were imaged at an excited state of 488 nm using a Typhoon Trio phosphorImager (GE Healthcare Biosciences, Piscataway, NJ).
Molecular modeling of 2LTRZFP-DNA complexes
Construction of the 2LTRZFP-DNA complexes was initiated using an X-ray crystallographic structure of a designed six zinc finger motif (Aart) bound to an oligonucleotide from PDB entry 2I1334 as a template. The sequence of the Aart peptide was then modified (using the Discovery Studio version 1.7 program package) in recognition helices according to our designed ZFP resulting from ZF Tools.30 The DNA duplex from the crystal structure of the Aart-DNA complex was further remodeled to 21-mers oligonucleotides of 2-LTR-circle junctions, as follows:
5′-CTCTAGCAGTACTGGATGGGC-3′
3′-GAGATCGTCATGACCTACCCG-5′ termed Zif1 as a specific complex
and
5′-AGTGCTAGCGTATCATCTAGT-3′
3′-TCACGATCGCATAGTAGATCA-5′ termed Zif2 as a nonspecific complex.
All backbone atoms of the protein and DNA sequences for both modified models were maintained as in the crystal structure. The ZFP was thoroughly prepared before performing the energy calculations. Protonation states of ZF residues were decided by reassignments of all histidine residues to δ-histidine (HID) and all cysteine to CYM (cysteine with a negative charge). Finally, a longer range electrostatic model (σ = 1.70 Å, ɛ = 0.67 kcal/mol) of the zinc ion (Zn2+)49 was used instead of the original nonbonded zinc parameters from the AMBER force field, avoiding a disorder of the ZFP during minimizations. The Zn2+ was coordinated within the ZFP by two CYM and two HID residues. A geometry with a tetrahedrally coordinated Zn2+ in the center of the ZFP was subsequently maintained throughout the minimizations. The remaining protein and nucleic acid parameters were determined using the AMBER ff03 force field.50
Both Zif1 and Zif2 models were then solvated with an explicit TIP3P water model51 in a truncated octahedral box with a minimum 10 Å distance between any solute atom and a box edge. Periodic boundary conditions were applied for avoiding the impacts from the atoms on the outer surfaces drifting away from the simulation box. The systems were neutralized with a minimum number of sodium ions and were then energy minimized in two stages (only solvent and ions at first, then all atoms). Each minimization stage was performed using the first 100 steps of a steepest descent algorithm followed up with 2400 steps of conjugate gradient minimization to get closer to an energy minimum. The minimized conformations of Zif1 and Zif2 were visualized and structurally analyzed.
Analysis of the ZFP-DNA interaction energies
To investigate the relative binding energies between the designed ZFP and the target oligonucleotides, a short trajectory of each protein–DNA complex was produced. The system was treated as a ligand/receptor complex, in which the designed ZF motif was the ligand and the target DNA was the receptor. The MM/GBSA approach was then used to calculate the free energies of molecules (Gx) in solution as described in previous work52 [see Eqs. (1) and (2)]. The molecular mechanical energies, the van der Waals interaction (Evdw), electrostatic interaction (Ees), and internal energy (Ein), as well as the free energy of the GB solvated system (Gpol) and the hydrophobic contribution to the solvation free energy (Gnonpol) were calculated with the SANDER program. The GBOBC solvent model53 with igb = 2 was used in this study. The binding energies of ZF motifs to their DNA and the relative free energies between Zif1 and Zif2 are calculated as in Eqs. (3) and (4), averaging over trajectories from the minimizations.
| (1) |
| (2) |
| (3) |
| (4) |
Acknowledgments
The authors thank Professor Carlos F. Barbas III for providing them with EMSA kit and Associate Professor Dr. Prachya Kongtawelert for his assistance to provide the BIACORE 2000™ biosensor and NANOTEC, Thailand for the access to Discovery Studio version 1.7 program package. They thank Dr. Dale Taneyhill for proofreading the manuscript.
Glossary
Abbreviations:
- AIDS
acquired immune deficiency syndrome
- EMSA
electrophoretic mobility shift assay
- GFP
green fluorescent protein
- HIV-1
human immunodeficiency virus type 1
- IN
integrase
- LTR
long terminal repeat
- MM/GBSA
molecular mechanics/generalized Born surface area
- SPR
surface plasmon resonance
- ZFP
zinc finger protein.
References
- 1.Mann JM, Chin J, Piot P, Quinn T. The international epidemiology of AIDS. Sci Am. 1988;259:82–89. doi: 10.1038/scientificamerican1088-82. [DOI] [PubMed] [Google Scholar]
- 2.UNAIDS/World Health Organization (2008) Report on the Global HIV/AIDS Epidemic. August, Mexico City.
- 3.Englund G, Theodore TS, Freed EO, Engelman A, Martin MA. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanstrom R, Delorbe WJ, Bishop JM, Varmus HE. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci USA. 1981;78:124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra SW, Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci USA. 1980;77:3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shank PR, Varmus HE. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978;25:104–114. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshimura FK, Weinberg RA. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell. 1979;16:323–332. doi: 10.1016/0092-8674(79)90009-6. [DOI] [PubMed] [Google Scholar]
- 10.Fritsch E, Temin HM. Formation and structure of infectious DNA of spleen necrosis virus. J Virol. 1977;21:119–130. doi: 10.1128/jvi.21.1.119-130.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guntaka RV, Richards OC, Shank PR, Kung HJ, Davidson N. Covalently closed circular DNA of avian sarcoma virus: purification from nuclei of infected quail tumor cells and measurement by electron microscopy and gel electrophoresis. J Mol Biol. 1976;106:337–357. doi: 10.1016/0022-2836(76)90090-5. [DOI] [PubMed] [Google Scholar]
- 12.Gianni AM, Smotkin D, Weinberg RA. Murine leukemia virus: detection of unintegrated double-stranded DNA forms of the provirus. Proc Natl Acad Sci USA. 1975;72:447–451. doi: 10.1073/pnas.72.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vink C, Plasterk RH. The human immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–438. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 14.Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 15.De Clercq E. New developments in anti-HIV chemotherapy. Curr Med Chem. 2001;8:1543–1572. doi: 10.2174/0929867013371842. [DOI] [PubMed] [Google Scholar]
- 16.Evering TH, Markowitz M. Raltegravir: an integrase inhibitor for HIV-1. Expert Opin Invest Drugs. 2008;17:413–422. doi: 10.1517/13543784.17.3.413. [DOI] [PubMed] [Google Scholar]
- 17.Grant P, Zolopa A. Integrase inhibitors: a clinical review of raltegravir and elvitegravir. J HIV Ther. 2008;13:36–39. [PubMed] [Google Scholar]
- 18.Delelis O, Malet I, Na L, Tchertanov L, Calvez V, Marcelin AG, Subra F, Deprez E, Mouscadet JF. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 2009;37:1193–1201. doi: 10.1093/nar/gkn1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks C, Gulick RM, Roquebert B, Blum L, Collin G, Damond F, Peytavin G, Leleu J, Matheron S, Chene G, Brun-Vezinet F, Descamps D. Raltegravir: the first HIV type 1 integrase inhibitor selection of the Q148R integrase inhibitor resistance mutation in a failing raltegravir containing regimen. Clin Infect Dis. 2009;48:931–939. [Google Scholar]
- 20.Charpentier C, Karmochkine M, Laureillard D, Tisserand P, Belec L, Weiss L, Si-Mohamed A, Piketty C. Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med. 2008;9:765–770. doi: 10.1111/j.1468-1293.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- 21.Miller J, Mclachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 23.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A°: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 24.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A°. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82:8013–8021. doi: 10.1128/JVI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dreier B, Segal DJ, Barbas CF. Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 27.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 28.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF. Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 29.Blancafort P, Segal DJ, Barbas CF. Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361–1371. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 30.Mandell JG, Barbas CF. Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deprez E, Barbe S, Kolaski M, Leh H, Zouhiri F, Auclair C, Brochon JC, Le Bret M, Mouscadet JF. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol Pharmacol. 2004;65:85–98. doi: 10.1124/mol.65.1.85. [DOI] [PubMed] [Google Scholar]
- 33.Bugreev DV, Baranova S, Zakharova OD, Parissi V, Desjobert C, Sottofattori E, Balbi A, Litvak S, Tarrago-Litvak L, Nevinsky GA. Dynamic, thermodynamic, and kinetic basis for recognition and transformation of DNA by human immunodeficiency virus type 1 integrase. Biochemistry. 2003;42:9235–9247. doi: 10.1021/bi0300480. [DOI] [PubMed] [Google Scholar]
- 34.Segal DJ, Crotty JW, Bhakta MS, Barbas CF, Horton NC, Mandell JG, Eberhardy SR, Goncalves J, Coelho S, Berkhout B, Dreier B, Fuller R, Lund CV, Blancafort P, Huber A, Koksch B. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J Mol Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Balakrishnan M, Jonsson CB. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J Virol. 1997;71:1025–1035. doi: 10.1128/jvi.71.2.1025-1035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzman M, Sudol M. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1996;70:9069–9073. doi: 10.1128/jvi.70.12.9069-9073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshinaga T, Fujiwara T. Different roles of bases within the integration signal sequence of human immunodeficiency virus type 1 in vitro. J Virol. 1995;69:3233–3236. doi: 10.1128/jvi.69.5.3233-3236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Ent FM, Vink C, Plasterk RH. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vink C, Van Gent DC, Elgersma Y, Plasterk RH. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafemina RL, Callahan PL, Cordingley MG. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds L, Ullman C, Moore M, Isalan M, West MJ, Clapham P, Klug A, Choo Y. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc Natl Acad Sci USA. 2003;100:1615–1620. doi: 10.1073/pnas.252770699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segal DJ, Goncalves J, Eberhardy S, Swan CH, Torbett BE, Li X, Barbas CF. Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J Biol Chem. 2004;279:14509–14519. doi: 10.1074/jbc.M400349200. [DOI] [PubMed] [Google Scholar]
- 43.Rebar EJ, Huang Y, Hickey R, Nath AK, Meoli D, Nath S, Chen B, Xu L, Liang Y, Jamieson AC, Zhang L, Spratt SK, Case CC, Wolffe A, Giordano FJ. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 44.Lanao JM, Briones E, Colino CI. Recent advances in delivery systems for anti-HIV1 therapy. J Drug Target. 2007;15:21–36. doi: 10.1080/10611860600942178. [DOI] [PubMed] [Google Scholar]
- 45.Whitcomb JM, Kumar R, Hughes SH. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 48.Nair R, Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J Mol Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 49.Stote RH, Karplus M. Zinc binding in proteins and solution: a simple but accurate nonbonded representation. Proteins. 1995;23:12–31. doi: 10.1002/prot.340230104. [DOI] [PubMed] [Google Scholar]
- 50.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 52.Jiranusornkul S, Laughton CA. Destabilization of DNA duplexes by oxidative damage at guanine: implications for lesion recognition and repair. J R Soc Interface 5. 2008;(Suppl 3):S191–S198. doi: 10.1098/rsif.2008.0304.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J Phys Chem B. 2000;104:3712–3720. [Google Scholar]


