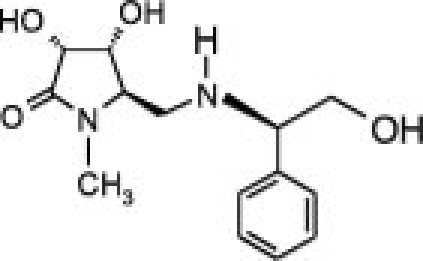
Abstract
Two structurally-related members of the lysosomal mannosidase family, the broad substrate specificity enzyme human lysosomal α-mannosidase (hLM, MAN2B1) and the human core α-1, 6-specific mannosidase (hEpman, MAN2B2) act in a complementary fashion on different glycosidic linkages, to effect glycan degradation in the lysosome. We have successfully expressed these enzymes in Drosophila S2 cells and functionally characterized them. hLM and hEpman were significantly inhibited by the class II α-mannosidase inhibitors, swainsonine and mannostatin A. We show that three pyrrolidine-based compounds designed for selective inhibition of Golgi α-mannosidase II (GMII) exhibited varying degrees of inhibition for hLM and hEpman. While these compounds inhibited hLM and GMII similarly, they inhibited hEpman to a lesser extent. Further, the two lysosomal α-mannosidases also show differential metal dependency properties. This has led us to propose a secondary metal binding site in hEpman. These results set the stage for the development of selective inhibitors to members of the GH38 family, and, henceforth, the further investigation of their physiological roles.
Keywords: mannosidase, family GH38, inhibitors, glycosidase, Drosophila S2, enzyme kinetics
Introduction
Lysosomal α-mannosidases, LMIIs (EC 3.2.1.24) are members of the glycoside hydrolase family 38 (GH38, Class II).1 They are involved in the catabolism of Asn-linked glycans of glycoproteins and play a vital role in maintaining cellular homeostasis. LMIIs catalyze the hydrolysis of α-1,2-, α-1,3- and α-1,6-glycosidic bonds with retention of configuration of the anomeric carbon of the released mannose residue. Genetic deficiency of LMII leads to the accumulation of nondegraded oligosaccharides in the lysosome and, consequently, symptoms consistent with the lysosomal storage disease, α-mannosidosis.2–4 The presence of a novel lysosomal α-mannosidase that was not associated with genetic α-mannosidosis was first described in human fibroblasts5 and partially purified from human spleen6 and rat liver.7 This enzyme catalyzes the hydrolysis of only the core α-1,6-mannose linkage. Subsequently, the enzyme was also purified from porcine epididymal fluid,8 (hence the designation Epman) and the porcine cDNA cloned.9 Later, the cDNA encoding the human orthologue (hEpman) was cloned.10 The α-1,6-specific human lysosomal mannosidase (hEpman, MAN2B2) and the broad specificity human lysosomal α-mannosidase (hLM, MAN2B1) act in a complementary fashion to effect glycan degradation. Besides showing distinct substrate specificity, hEpman has only 28% sequence identity with hLM.
The oligosaccharide structure on an individual glycoprotein contributes to cell adhesion during development, viral infection, immune response and metastasis of oncogenically transformed cells.11 Complex-type branched oligosaccharides have been associated with an increase in malignant transformation and cancer metastasis. By virtue of its inhibition of the GH38 N-glycosylation protein, Golgi α-mannosidase II (GMII), swainsonine, a plant indolizidine alkaloid has been under investigation as an antimetastatic agent in a murine melanoma cell model,12 as well as in human clinical trials.13,14 However, as swainsonine is also an effective inhibitor of LMII, there is the potential for side-effect symptoms resembling those of lysosomal storage disease. To facilitate the search for highly selective inhibitors of GMII, we sought to initiate a structure/function analysis of LMs.
Based on our success with GMII,14 we turned to the Drosophila S2 expression system to achieve higher yields of LMs for structural and functional studies. The yields and purity of hLM and hEpman isolated from tissue extracts have been inconsistent.6,15 Previously, hLM has been cloned and expressed in Pichia pastoris,16 CHO cells15 and in mammalian HEK293 cells.10 hEpman has been cloned and expressed in a mammalian expression system.10 However, these did not result in quantities sufficient for full structural analysis. A crystal structure of bovine LMII (bLMII) was determined to 2.7 Å using the previously solved structure of dGMII as a molecular replacement starting point.17 This enzyme however was purified directly from tissue and is not available in amounts required for further analysis or amenable to site-directed mutagenesis.
High expression levels of the two lysosomal α-mannosidases have allowed us to test promising compounds as inhibitors and in the future, will facilitate the search for more selective antimetastatic inhibitors for GMII. For example, 5α-substituted analogs of swainsonine have been shown to be as potent as swainsonine when evaluated against jack bean mannosidase (a plant enzyme with lysosomal-like properties).18 Greater diversity in the synthesis of C(5)-substituted swainsonine analogs could afford selectivity for GMII. Here, we have tested hLM and hEpman with three pyrrolidine compounds with substitution at the C(5) position of the pyrrolidine ring and show that GMII selectivity is partially achieved with respect to one of the LMII enzymes.
Results
Stable cell line expression in Drosophila S2 cells
Human lysosomal α-mannosidase (hLM, MAN2B1) and the core α-1, 6 specific human lysosomal mannosidase (hEpman, MAN2B2) were cloned into the Drosophila pMT/BiP vector containing an upstream metallothioneine promoter, a BiP secretion signal and a hexahistidine tag at the carboxy-terminus for efficient purification. These plasmid constructs were transfected into Drosophila S2 cells.19 Transient expression in S2 cells showed enzyme activity and protein expression in the culture media at the expected molecular size. From the stable cell line expression, the single cell clones that had optimal expression level and α-mannosidase activity were selected and scaled up (see Supporting Information Fig. S1). These were gradually adapted to grow in serum-free media for large-scale expression of protein. This step was necessary as the serum in the medium sometimes hindered column chromatography purification. The above procedure is summarized briefly in Figure 1.
Figure 1.
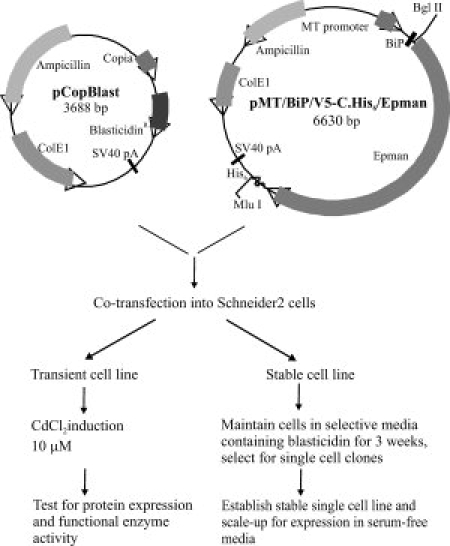
Summary of steps involved in protein expression in Drosophila S2 system.
Purification of hEpman
hEpman was purified from the secreted medium successively by dye-affinity chromatography using Cibacron blue F3GA resin, followed by a cobalt chelating Sepharose step, cation-exchange and size exclusion chromatography (Fig. 2). Most proprietary serum-free media contain components that interfere with binding to affinity chromatography columns by blocking the column matrix. The Cibacron blue binding step eliminated interfering media components, greatly reduced the sample volume, and afforded 30-fold purification (see Table I). Use of cobalt chelating Sepharose as a second step in the purification proved effective in enhancing the purification to 70-fold. This step utilized the hexahistidine tag present at the C-terminus of the expressed protein. Earlier studies of the mammalian expressed hEpman10 had reported that the His6-tag was undetectable by immunoblotting. However, for our construct, we were able to use immunoblotting to detect the carboxy-terminal His6-tag throughout transient transfection, single cell selection and during the purification steps.
Figure 2.
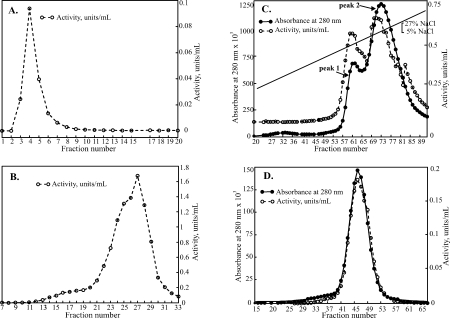
Steps involved in the purification of hEpman: Graphs show α-mannosidase activity, units/mL (open circles, dashed line) and absorbance at 280 nm (filled circles, solid line). (A) Cibacron Blue F3GA step eluted with 500 mM NaCl. Fractions 1-10 were pooled. (B) Pooled fractions bound to cobalt chelating Sepharose and eluted with pH 5.0 acetate buffer. Fractions 21–30 were pooled for the next step (C) Cation exchange chromatography on SP-HiTrap with an applied 0.05–0.27 M salt gradient. Fractions from peaks 1 and 2 were pooled separately (D) Final purification step of peak 2 on a Superdex 200 16/60.
Table I.
Purification of hEpman from 2.3 L of Culture Medium of Stably Transfected Cellsa
| Fraction | [Protein], (mg/mL) | Total protein (mg) | Total enzyme (units) | Specific activity (U/mg) | Yield % | Purification fold |
|---|---|---|---|---|---|---|
| Culture supernatant | 2.2 | 4945 | 24.1 | 0.0049 | 100 | 1 |
| Cibacron blue | 0.1 | 112 | 17.8 | 0.16 | 74 | 33 |
| Chelating Sepharose | 0.6 | 45.0 | 15.6 | 0.35 | 65 | 71 |
| Cation-exchange | ||||||
| Peak 1 | 2.4 | 11 | 5.3 | 0.50 | 22 | 102 |
| Peak 2 | 4.3 | 32 | 15.6 | 0.49 | 65 | 100 |
| Superdex 200 | ||||||
| Peak 1 | 19 | 10 | 6.3 | 0.66 | 26 | 135 |
| Peak 2 | 16 | 25 | 22 | 0.9 | 91 | 182 |
Each step in the purification was assayed using 3 mM pNP-Man as substrate. Activities fluctuate slightly as the enzyme was inhibited by salt present in the elution buffers.
Cation exchange chromatography resolved hEpman into two peaks [Fig. 2(C)]. On an SDS-PAGE gel, both these peaks ran at ∼120 kDa (Supporting Information Fig. S2, lanes 5 and 6). Also, MALDI-TOF mass spectrometry showed the molecular weight to be the same (123.6 kDa). The presence of two distinct peaks may be as a result of subtle differences in glycosylation or other post-translational modification not detectable at the resolution range of the mass spectrometry. Size-exclusion chromatography was carried out as the final step of purification [Fig. 2(D)].
In the end, hEpman was purified 100 to 200-fold with a specific activity of 0.7–1.0 units mg−1 (Table I). The final purified yields of the enzyme are shown in Supporting Information Figure S2 (lanes 7 and 8). We were able to obtain yields of 15 mg L−1 of active hEpman. As compared to the mammalian system which gave 11% yield, we have been able to achieve 90% recovery of the initial activity in the Drosophila S2 system.
Purification of hLM
hLM was purified in a similar manner as hEpman without the initial Cibacron blue binding. Stepwise elution with buffers of decreasing pH after cobalt chelating Sepharose binding resulted in about 50-fold enrichment of the enzyme (see Supporting Information Fig. S4, lanes 6 and 7. Cation-exchange chromatography proved to be an effective second step in the purification of hLM. Three distinct protein peaks were observed over a gradient of 0.3–0.6 M NaCl (Supporting Information Fig. S3). Only the third peak showed enzyme activity which was subjected to size-exclusion chromatography and concentrated to 0.5 mg mL−1.
During the final concentration step, the 110 kDa hLM underwent proteolytic processing into a 62 kDa and 48 kDa polypeptide (see lanes 6 and 7, Supporting Information Fig. S4). N-terminal sequencing of these two polypeptides showed the 62 kDa species to be GGYET (start of the N-terminus of hLM after the signal sequence peptide is cleaved) and the 48 kDa to be APQPI. The APQPI site of cleavage could be as a result of a flexible disordered loop in hLM that is readily subject to proteolysis. Structural comparison with bovine lysosomal α-mannosidase, bLMII,17 (sharing 80% sequence identity with hLM) shows that there is very little sequence conservation in this region and that it is also probably quite flexible. Similar proteolytic clipping has been reported earlier for hLM15 and also in bovine LM20 and feline LM.21
The final purified hLM was enriched almost 300-fold from the crude culture medium with a specific activity of 3.0 units mg−1 (Supporting Information Table I). The final purified yields of active hLM were 2.2 mg L−1 of culture medium. We have been able to achieve 30% recovery of the initial activity in the S2 expression system as compared to 7% yield (0.287 units mg−1) in Pichia.16 However, the yields obtained in CHO cells were much higher (43 units mg−1, 11 mg L−1).15
Characterization of enzymes
The amino terminal sequence of lysosomal enzymes is known to contain a cleavable signal sequence that is lost as the protein traverses through the secretory pathway in the ER.22 N-terminal sequencing results for hLM and hEpman showed that the start sequences at the amino terminus were GGYET and AGPIRAF for hLM and hEpman respectively. This was in agreement with the amino terminal signal sequence predicted by SignalP 3.0.23 The same signal sequence cleavage position was observed for hLM purified from CHO cells.15
Both purified hLM and hEpman were characterized by MALDI-TOF mass spectrometry for their molecular weights which were 123.6 kDa for hEpman (expected 112.7 kDa) and 118.68 kDa for hLM (expected 109.98 kDa). We infer that the difference in the mass from the expected size of the translated protein sequence is a result of glycosylation (eleven predicted glycosylation sites for hEpman and ten for hLM, http://www.cbs.dtu.dk/services/NetNGlyc).24
pH optimum of enzyme activity
To characterize the enzymatic properties of hLM and hEpman, we first determined their optimal pH using the artificial substrate, p-nitrophenyl-α-d-mannopyranoside (pNP-Man). hLM and hEpman showed activity between pH 3.6 and 5.5 with their optimal activity at pH 4.2 for hLM and pH 4.0 for hEpman. This is consistent with the lysosomal localization of these two enzymes. hLM and hEpman expressed in the Drosophila expression system thus had a similar pH profile to those expressed in Pichia or mammalian systems.10,16
Enzyme kinetics: Km estimation
The Km of hLM and hEpman for the substrate, pNP-Man was determined to be 1.8 ± 0.4 mM and 23 ± 2 mM respectively (errors indicating the standard deviation of the data). The Km of hLM conforms to earlier studies.16 However, to the best of our knowledge, the Km of hEpman has not been reported previously.
hLM has a higher affinity in comparison to hEpman towards its unnatural substrate, pNP-Man. This is expected considering that hEpman is highly specific in its cleavage of the α-1,6 mannosidic linkage as compared to the broader substrate specificity for hLM.
Inhibition studies with pyrrolidine compounds
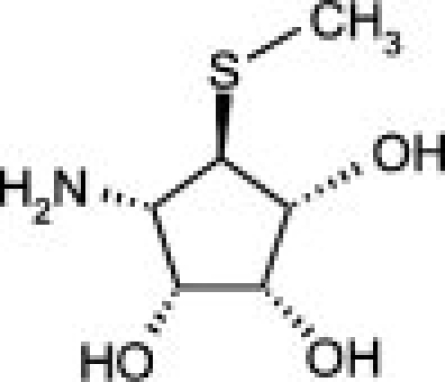
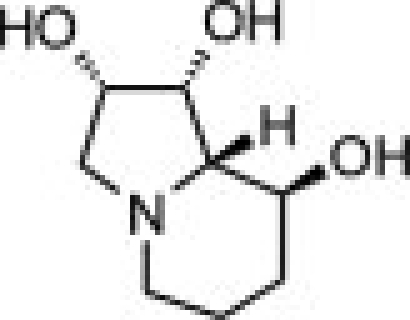
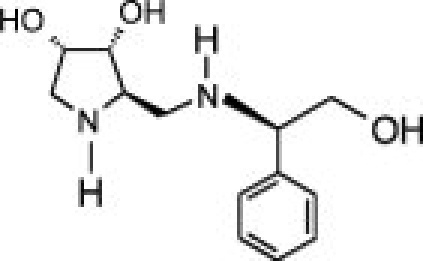
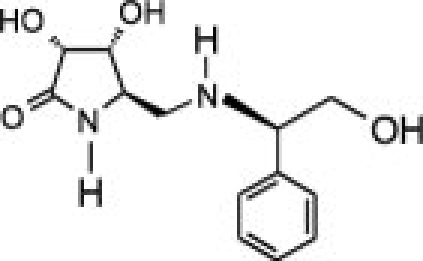
The availability of high yields of hEpman and hLM allowed us to perform comparative inhibition studies in the search for selective compounds towards GMII. The Class II α-mannosidase inhibitors, swainsonine and mannostatin A significantly inhibited hLM and hEpman (Supporting Information Fig. S5). The Ki values for swainsonine and mannostatin A were both determined to be 0.4 μM for hLM. For hEpman, Ki was 4 μM for swainsonine and 0.6 μM for mannostatin A (Table II).
Table II.
Summary of Inhibition Data that Distinguished hLM from hEpman. Errors Indicate the Standard Deviation of the Data, Ki Values for dGMII are Indicated for Comparison with the LMs.
Three pyrrolidine-derivatives based on the swainsonine structure (compounds 1, 2 and 3) that had been previously tested with the Drosophila Golgi α-mannosidase II26,27 were investigated for their inhibitory potential on the two lysosomal α-mannosidases. We hoped that the synthesized pyrrolidine compounds would be more selective for dGMII than swainsonine itself.
As shown in Table II and Supporting Information Figure S6, the compounds inhibited hLM and hEpman to a varying extent. In the case of hLM, compound 1 was the least potent while 3 was the best inhibitor. Substitution of the pyrrolidine system in 1 by a pyrrolidinone in 2 resulted in a moderate 1.7-fold increase in inhibition potency. Replacement of the lactam moiety by a methyl group in 3 significantly improved the inhibition potency over 2 by 25-fold. 3 was nearly as potent as swainsonine and mannostatin A against hLM. The three pyrrolidine-derivatives were less active against hEpman, although the pattern of inhibition was similar. 1 showed a very significant decrease in inhibitory activity towards hEpman, being 20 times less potent than for hLM. 2 was sixfold more inhibitory than 1, while the addition of a methyl group in 3 resulted in a further eightfold improvement in activity against hEpman.
Metal cation dependency
The crystal structures of both dGMII and bLMII have shown the presence of a Zn2+ cation in the active site.14,17 Both hLM and hEpman were subjected to a 1 mM metal cation screen for their metal dependency properties (Fig. 3).
Figure 3.
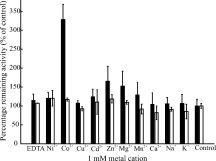
Metal cation dependency for hLM and hEpman. The bar graph represents a 1 mM metal cation screen for hEpman (black bars) and hLM (white bars). Error bars indicate the variance of the data. The activity of the enzyme after adding the metal ion is expressed as a percentage of the respective control enzyme in the absence of metal.
Neither hLM nor hEpman were affected by 1 mM EDTA. Further, the metal cation screen showed that while hLM had no sensitivity to any metal ion, hEpman was activated threefold with Co2+ and stimulated 1.25 to 1.5-fold with Zn2+, Mg2+, and Mn2+. The results are consistent with previously published data.10
Since hEpman was activated by Co2+, we used inductively-coupled plasma atomic emission spectroscopy (ICP-AES) to test for the presence of the intrinsic metal cation in its active site. For hEpman, purified by Ni-NTA chromatography in the absence of any exogenously added metals, ICP-AES analysis showed the presence of stoichiometric Zn2+ and the absence of Co2+ (data not shown). However, if hEpman was exposed to Co2+ during the course of cobalt chelating Sepharose chromatography, followed by extensive dialysis, cation-exchange chromatography and gel filtration, the presence of both stoichiometric Zn2+ and Co2+ was observed by ICP-AES. We interpret this result by invoking a second metal binding site in hEpman for Co2+ (discussed later).
It is possible that the presence of Co2+ stimulated the activity of hEpman through a slight local conformational change in its active site, which might affect the binding interaction with inhibitors. However, we observed no evidence of a change in inhibitory effects of the compounds in the presence of 1 mM CoCl2 (data not shown).
Discussion
We have shown in this article that consistent good yields of active hEpman (15 mg L−1) and hLM (2 mg L−1) can be expressed in Drosophila S2 cells. The S2 expression system is well suited for large eukaryotic proteins that require post-translational modifications such as glycosylation28 and amidation.29 The Drosophila S2 expression system has been employed successfully in our group for expressing large eukaryotic enzymes such as Drosophila GMII,14 human maltase glucoamylase,30 both requiring glycosylation, and small eukaryotic proteins containing a large number of disulfide bonds including fish antifreeze proteins31 and insect odorant receptors (E. Rossi, D.A. Kuntz, D.R. Rose, unpublished). Such modifications are also achieved in mammalian cells, but S2 cells provide easy maintenance and high density growth in serum-free media. With the incorporation of an appropriate signal sequence in the construct, the protein is secreted out of the host cells and does not accumulate in the cytoplasm. This eliminates the need to lyse the cells before purification, reducing the number of contaminating proteins and proteases.
A high yielding expression system with functionally active hLM and hEpman would thus be useful in enzyme replacement therapeutic studies for patients suffering from lysosomal storage disorder. The development of a good expression system also opens the door to detailed structural and kinetic analyzes on these enzymes.
Inhibitor analysis have indicated marked differences in the inhibition profiles of two members of the same LM family. Our results show that the two lysosomal α-mannosidases (that differ in their substrate specificity) display varying inhibitory potential towards the pyrrolidine-derivatives of swainsonine. Compound 3 with a pyrrolidinone system and a methyl group in the lactam moiety has shown significant improvement in its inhibition potency for both hLM and hEpman in comparison to 1 and 2, although hEpman showed 20-fold less inhibition than hLM. Knowledge of the crystal structures of these compounds with dGMII in conjunction with our inhibition data allowed us to infer the differences/similarities in the inhibition characteristics between the three members of family 38 enzymes.
The crystal structure of dGMII with compound 1 had shown that the presence of polar groups in the C5 position of pyrrolidine ring would introduce additional polar interactions with the active site residues in the enzyme.27 Based on this, compounds 2 and 3 were designed by replacing the pyrrolidine system in 1 with a pyrrolidinone system.26 Compounds 2 and 3 allowed for a new hydrogen bond formation around the active site and in addition the methyl group in 3 formed a hydrophobic interaction with a hydrophobic pocket in the active site of dGMII.26 The key residues in the active site pocket and the zinc co-ordination involved in inhibitor binding are strikingly conserved in dGMII, hLM and hEpman. Trp95 in dGMII (residue numbering corresponds to the crystal structure, the corresponding gene TrEMBL accession number Q24451 numbering is Trp158) which forms pi-interactions with both the phenyl ring and pyrrolidine group of 127 is conserved in both LMs. dGMII Asp472 and Tyr727, which form hydrogen bonds with compounds 2 and 3, and Phe206, Trp415, which along with Tyr727 contribute to the hydrophobic interaction formed by the methyl group of 3 are completely conserved in the three enzymes.
Because the three enzymes have conserved catalytic site residues, we expected that the binding of the inhibitors targeting this region would be similar. Sequence similarity in the active site cleft of dGMII, hLM and hEpman would explain the formation of correspondingly similar polar and hydrophobic interactions. While the inhibition constant Ki determined for hLM agrees with that of dGMII, hEpman was not strongly inhibited by the pyrrolidine compounds. This behavior could be attributed to the differences in the metal dependency of hEpman (see below) and provides a strong basis for determining the crystal structure of hEpman.
Our studies on the effect of metal cations on hEpman showed that it was activated threefold with 1 mM CoCl2 and to a lesser extent by Zn2+, Mg2+, and Mn2+. Another member of the cobalt-activated family 38 cytosolic class II α-mannosidase, Man2C1 has been shown to contain Co2+ in its active site.32 Interestingly, like hEpman, Man2C1 is inhibited comparatively poorly by swainsonine.33
Analysis of the residues in close proximity (<3.3 Å) to those residues involved in co-ordination with zinc ion (His90, Asp92, Asp204, and His471 in dGMII) showed that except for Phe342 all these “secondary” residues are well conserved in the two LMs. In hEpman, this phenylalanine is substituted by a lysine (Lys290). The phenyl ring of Phe342 in dGMII is 3.25 Å away from Asp92 and could make a stacking interaction. An analysis of representative crystal structures indicates that Phe342 does not get displaced in dGMII upon binding to the inhibitors.26 However, a charged lysine at this position might affect the metal ion co-ordination in hEpman and thus induce different metal dependency properties.
We propose the presence of a secondary binding site for cobalt in hEpman on the basis of the following observations. Firstly, hEpman was not sensitive to EDTA and further showed no loss in enzyme activity up to 5 mM EDTA. This would indicate that the intrinsic zinc ion in the active site is tightly coordinated, and is unlikely to be displaced by Co2+. Secondly, ICP-AES data for hEpman, purified in the absence of added cobalt did not show the presence of Co2+ and only detected Zn2+. However, once exposed to cobalt and further purified, equimolar stoichiometric Zn2+ and Co2+ were detected. Taken together these results are indicative of the presence of two metal binding sites.
We then examined the residues in the substrate binding sites of hEpman to seek support for the notion of a potential second metal ion binding site. Residues 339-343 in dGMII take part in interactions in the saccharide “holding” site.34 This sequence of GD340DFR in dGMII corresponds to GC288DKQ in hEpman. dGMII Asp341 (hEpman Asp289) is the presumptive acid-base catalyst in the mannosidase reaction and any alteration of its environment is expected to have profound effects on enzymatic activity. Cys288 in hEpman could contribute a potential coordinating side chain for Co2+ in conjunction with hEpman Cys217 (Asp270 in dGMII), and neighboring conserved hEpman Asp289 and Tyr216 residues. In a simple substitution model of hEpman, the Cys288 is close enough to Cys217 to favor formation of a solvent-exposed disulfide bond (Fig. 4). Interestingly, such bonds have been shown to be susceptible to rupture by Co2+.35 In hEpman, Co2+-induced disulfide rupture would result in spatial rearrangement of neighboring Asp289 and Tyr216, which could then participate in coordinating with Co2+. Asp and Tyr residues are known to coordinate with cobalt in glutamate mutase and methionine synthase,36,37 Asp and Cys residues with zinc in β-lactamase38 and Cys with cobalt in nitrile hydrolase.39
Figure 4.
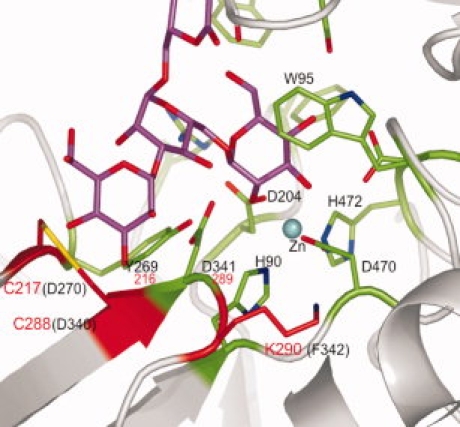
Changed residues in the active site of hEpman which may be responsible for cobalt activation. The active site region of dGMII containing bound Man5 (from PDB 3CV5) was altered to show the mutations in the region of the presumptive acid-base catalyst D341 (residue numbering in the dGMII crystal). Residues conserved in dGMII, hLM and hEpman are colored green, residues altered in hEpman are colored red, Man5 is colored magenta, and the active site zinc is represented as a cyan sphere. Black lettering corresponds to the dGMII crystal structure sequence, while red denotes the corresponding hEpman residues.
The presence of a Co2+ binding site in this region for hEpman, together with the dGMII Phe342 to hEpman Lys290 change, may well preclude any sugar binding at all in the region corresponding to the dGMII “holding” site. Understanding any other functional significance of a secondary binding site, such as some sort of regulatory role, will require further study.
Crystallization trials and optimization are currently underway for hEpman which shows 26% sequence identity to the previously solved crystal structure of bLMII.17 Comparison of the active site and its secondary interactions in hEpman with dGMII, which is not cobalt-activated, will reveal differences that can be exploited in specific inhibitor design. Design of novel inhibitors as selective, antimetastatic agents for GMII will benefit from this work. Towards this goal, we have partially achieved our objective of selective inhibition of GMII with respect to hEpman, though the compounds do not show greater selectivity than its natural inhibitor swainsonine. Since such C5-substituted analogs of swainsonine are nearly as potent as swainsonine against dGMII, this could be used as a lead towards extending the substituent groups in swainsonine and exploiting the differences in the binding sites of Golgi and lysosomal enzymes.
Materials and Methods
Cloning
hLM and hEpman (Genbank accession sequence number U68567 for hLM and NM015274 for hEpman respectively) were cloned into the Drosophila expression vector pMT-BiP-V5-HisA (see Supporting Information).
Expression in Drosophila S2 cells
Proteins were expressed using the Drosophila expression system (DES, Invitrogen). Transient transfection was achieved by cotransfection of S2 cells (3 × 106 cells mL−1) with 19 μg of the hLM/hEpman construct and 1 μg of blasticidin resistance plasmid pCopBlast (Invitrogen) using the calcium phosphate procedure. A stably transfected cell population was obtained after 3 weeks of growth of transfected cells in S2 medium containing 16 μg mL−1 blasticidin (Invitrogen). Approximately five stably transfected cells were added to each well of a 96-well plate containing 100 μL of blasticidin in S2 medium with nontransfected S2 cells (2.5 × 105 cells per well) as a feeder layer. The selected single cell clones were then scaled up to Fernbach flasks by adapting them to grow in serum-free insect cell medium (Hyclone). Protein expression was induced with 4 μM and 5 μM CdCl2 for hLM and hEpman respectively. After 72 h, the medium containing the secreted protein was harvested.
Purification of hLM and hEpman
The cells and medium were separated by centrifugation at 2000g for 10 min. The medium was further clarified by centrifugation at 12,000g for 20 min. PMSF (1 mM) and 0.02% w/v sodium azide were added to the supernatant. The purification steps were carried out at 4°C except where stated otherwise.
hEpman purification
The supernatant was first batch-bound to Cibacron Blue F3GA (Sigma) at a ratio of 10 mL of supernatant per mL of beads at 20°C for 2 h. The resin was washed with 10 mM Tris pH 8.0, 20 mM NaCl and eluted with high salt buffer (10 mM Tris pH 8.0, 500 mM NaCl). Fractions containing active hEpman were incubated with 2.5 mM CoCl2 for 30 min at 20°C and then batch-bound to chelating Sepharose (GE Healthcare) for 2 h at a ratio of ∼20 μL resin to 1 mL of pooled fractions. Protein was eluted using Tris buffer (10 mM Tris, 500 mM NaCl) of decreasing pH in a stepwise manner. The column was first washed with Tris buffer at pH 8.0 followed by Tris buffer at pH 7.0. hEpman was eluted with acetate buffer (20 mM sodium acetate pH 5.0, 500 mM NaCl). Eluted fractions were pooled and dialyzed against 50 mM MES pH 6.0, 15 mM NaCl and loaded onto a cation exchange SP HiTrap (5 mL, GE HealthCare). A gradient of 5–27% buffer B (50 mM MES pH 6.0, 1 M NaCl) over 90 mL was applied to elute hEpman. The final step involved size exclusion chromatography on a Superdex 200 (16/60, GE Healthcare) and the enzyme was concentrated to 20 mg/mL.
hLM purification
The supernatant containing hLM was first batch-bound to chelating Sepharose and purified in a similar manner as carried out for hEpman, then dialyzed into 50 mM MES pH 5.8, 15 mM NaCl and further purified by cation-exchange on a 5 mL SP HiTrap column. Protein was eluted with a gradient of 20–60% buffer B (50 mM MES pH 5.8, 1 M NaCl) over 60 mL. Pooled fractions were concentrated and resolved on a 16/60 Superdex 200. The enzyme was concentrated to 0.5 mg/mL.
Enzyme kinetics
The activity of hLM and hEpman were assayed using p-nitrophenyl-α-d-mannopyranoside (pNP-Man, Sigma) as the substrate. The colorimetric assay of crude culture media, column fractions and the purified protein were carried out at 37°C for 25 min in a 50 μL total reaction volume containing 3 mM substrate and 100 mM sodium acetate (pH 4.0). The reaction was stopped with 50 μL of 0.5 M sodium carbonate and the absorbance was quantified on a Spectramax Plus 384 plate reader (Molecular Devices) at 405 nm. One unit of enzyme activity is defined as the amount of enzyme that liberates 1 μmol of para-nitrophenol in 1 min at 37°C and at pH 4.0. A molar extinction coefficient of 18 mM−1 cm−1 for para-nitrophenol was used for calculating the specific activity.40
Progress curves were carried out at varying substrate concentrations and with a fixed enzyme concentration,41 and it was noted that the reaction rate was linear for at least 40 min. The incubation time for reactions was thus set as 25 min, well within the linear portion of the progress curve. Enzyme concentrations used in the assay were 95 nM and 0.63 μM for hLM and hEpman respectively. The final concentration of DMSO in the reactions was 3% for hLM and 20% for hEpman. Data points were collected in triplicate. Data were fitted by nonlinear regression analysis using Grafit v 4.05 (Erithacus software)42 to obtain Km and Vmax. Km was calculated from a series of eight separate data sets and averaged. The IC50 is the concentration of the inhibitor that inhibits 50% of the enzyme activity was determined graphically. The Ki values for the inhibitors were determined at three different concentrations of inhibitor. Lineweaver-Burke plots were used to check for competitive inhibition. The Ki for competitive inhibition was calculated using
| (1) |
where, Kmobs is the observed Km determined for the enzyme in the presence of inhibitor and I is the inhibitor concentration used. Swainsonine and mannostatin A were made as stock solutions of 100 mM in DMSO and further dilutions were made in water. The 100 mM stock solutions of compounds 1, 2, and 3 were made in methanol.
Effect of metal cations
The enzyme assay reaction was carried out in the presence of 1 mM of ZnCl2, CuCl2, CdCl2, MgCl2, CaCl2, NiCl2, CoCl2, NaCl, KCl, and EDTA.
ICP-AES
ICP-AES was carried out using 6 μM of hEpman on a Perkin–Elmer Optima 3000 dual view ICP optical emission spectrometer. The metal content in hEpman was determined relative to the amount in the protein buffer.
Western blot analysis
Immunoblotting was carried out using penta-His antibody (See Supporting Information).
Acknowledgments
The authors thank Anton Dobrin for technical assistance. They thank Mr. Rey Interior (amino acid sequencing facility, Hospital for Sickkids) for Edman sequencing, Ms. Ying Li (Department of Chemistry) for MALDI-TOF, Mr. Dan Mathers (ANALEST, University of Toronto) for ICP-AES analysis, Dr. K.W. Moremen (University of Georgia) for the mammalian clones of hLM and hEpman and Dr. Hélène Fiaux and Dr. Sandrine Gerber-Lemaire (EPF, Lausanne, Switzerland) for the pyrrolidine inhibitors.
Glossary
Abbreviations
- CHO
Chinese hamster ovary
- DES
Drosophila expression system
- dGMII
Drosophila Golgi α-mannosidase II
- hEpman
human epididymal-specific α-1,6-lysosomal manno- sidase
- hLM
human lysosomal α-mannosidase
- ICP-AES
inductively-coupled plasma atomic emission spectroscopy
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- PMSF
phenylmethanesulfonyl fluoride
- pNP-Man
p-nitrophenyl-α-d-mannopyranoside
- S2
Schneider 2.
References
- 1.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucl Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawley AC, Walkley SU. Developmental analysis of CNS pathology in the lysosomal storage disease α-mannosidosis. J Neuropathol Exp Neurol. 2007;66:687–697. doi: 10.1097/nen.0b013e31812503b6. [DOI] [PubMed] [Google Scholar]
- 3.Novikoff PM, Touster O, Novikoff AB, Tulsiani DP. Effects of swainsonine on rat liver and kidney: biochemical and morphological studies. J Cell Biol. 1985;101:339–349. doi: 10.1083/jcb.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tulsiani DR, Touster O. Swainsonine, a potent mannosidase inhibitor, elevates rat liver and brain lysosomal α-d-mannosidase, decreases Golgi α-D-mannosidase II, and increases the plasma levels of several acid hydrolases. Arch Biochem Biophys. 1983;224:594–600. doi: 10.1016/0003-9861(83)90247-3. [DOI] [PubMed] [Google Scholar]
- 5.Daniel PF, Evans JE, De Gasperi R, Winchester B, Warren CD. A human lysosomal α(1–6)-mannosidase active on the branched trimannosyl core of complex glycans. Glycobiology. 1992;2:327–336. doi: 10.1093/glycob/2.4.327. [DOI] [PubMed] [Google Scholar]
- 6.De Gasperi R, Daniel PF, Warren CD. A human lysosomal α-mannosidase specific for the core of complex glycans. J Biol Chem. 1992;267:9706–9712. [PubMed] [Google Scholar]
- 7.Haeuw JF, Grard T, Alonso C, Strecker G, Michalski JC. The core-specific lysosomal α(1–6)-mannosidase activity depends on aspartamidohydrolase activity. Biochem J. 1994;297:463–466. doi: 10.1042/bj2970463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura N, Dacheux F, Venien A, Onoe S, Huet JC, Dacheux JL. Localization of a maturation-dependent epididymal sperm surface antigen recognized by a monoclonal antibody raised against a 135-kilodalton protein in porcine epididymal fluid. Biol Reprod. 1992;47:1040–1052. doi: 10.1095/biolreprod47.6.1040. [DOI] [PubMed] [Google Scholar]
- 9.Okamura N, Tamba M, Liao HJ, Onoe S, Sugita Y, Dacheux F, Dacheux JL. Cloning of complementary DNA encoding a 135-kilodalton protein secreted from porcine corpus epididymis and its identification as an epididymis-specific α-mannosidase. Mol Reprod Dev. 1995;42:141–148. doi: 10.1002/mrd.1080420203. [DOI] [PubMed] [Google Scholar]
- 10.Park C, Meng L, Stanton LH, Collins RE, Mast SW, Yi X, Strachan H, Moremen KW. Characterization of a human core-specific lysosomal α1,6-mannosidase involved in N-glycan catabolism. J Biol Chem. 2005;280:37204–37216. doi: 10.1074/jbc.M508930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Humphries MJ, Matsumoto K, White SL, Molyneux RJ, Olden K. Augmentation of murine natural killer cell activity by swainsonine, a new antimetastatic immunomodulator. Cancer Res. 1988;48:1410–1415. [PubMed] [Google Scholar]
- 13.Tulsiani DR, Broquist HP, Touster O. Marked differences in the swainsonine inhibition of rat liver lysosomal α-D-mannosidase, rat liver Golgi mannosidase II, and jack bean α-D-mannosidase. Arch Biochem Biophys. 1985;236:427–434. doi: 10.1016/0003-9861(85)90643-5. [DOI] [PubMed] [Google Scholar]
- 14.Van Den Elsen JM, Kuntz DA, Rose DR. Structure of Golgi α-mannosidase II: a target for inhibition of growth and metastasis of cancer cells. EMBO J. 2001;20:3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg T, King B, Meikle PJ, Nilssen O, Tollersrud OK, Hopwood JJ. Purification and characterization of recombinant human lysosomal α-mannosidase. Mol Genet Metab. 2001;73:18–29. doi: 10.1006/mgme.2001.3173. [DOI] [PubMed] [Google Scholar]
- 16.Liao YF, Lal A, Moremen KW. Cloning, expression, purification, and characterization of the human broad specificity lysosomal acid α-mannosidase. J Biol Chem. 1996;271:28348–28358. doi: 10.1074/jbc.271.45.28348. [DOI] [PubMed] [Google Scholar]
- 17.Heikinheimo P, Helland R, Leiros HK, Leiros I, Karlsen S, Evjen G, Ravelli R, Schoehn G, Ruigrok R, Tollersrud OK, McSweeney S, Hough E. The structure of bovine lysosomal α-mannosidase suggests a novel mechanism for low-pH activation. J Mol Biol. 2003;327:631–644. doi: 10.1016/s0022-2836(03)00172-4. [DOI] [PubMed] [Google Scholar]
- 18.Fujita T, Nagasawa H, Uto Y, Hashimoto T, Asakawa Y, Hori H. Synthesis of the new mannosidase inhibitors, diversity-oriented 5-substituted swainsonine analogues, via stereoselective Mannich reaction. Org Lett. 2004;6:827–830. doi: 10.1021/ol049947m. [DOI] [PubMed] [Google Scholar]
- 19.Hauser H, Wagner R. Mammalian cell biotechnology in protein production. Berlin: Walter de Gruyter & Co; 1997. [Google Scholar]
- 20.Tollersrud OK, Berg T, Healy P, Evjen G, Ramachandran U, Nilssen O. Purification of bovine lysosomal α-mannosidase, characterization of its gene and determination of two mutations that cause α-mannosidosis. Eur J Biochem. 1997;246:410–419. doi: 10.1111/j.1432-1033.1997.00410.x. [DOI] [PubMed] [Google Scholar]
- 21.Berg T, Tollersrud OK, Walkley SU, Siegel D, Nilssen O. Purification of feline lysosomal α-mannosidase, determination of its cDNA sequence and identification of a mutation causing α-mannosidosis in Persian cats. Biochem J. 1997;328:863–870. doi: 10.1042/bj3280863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 23.Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002;7:310–322. [PubMed] [Google Scholar]
- 25.Kawatkar SP, Kuntz DA, Woods RJ, Rose DR, Boons GJ. Structural basis of the inhibition of Golgi α-mannosidase II by mannostatin A and the role of the thiomethyl moiety in ligand-protein interactions. J Am Chem Soc. 2006;128:8310–8319. doi: 10.1021/ja061216p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiaux H, Kuntz DA, Hoffman D, Janzer RC, Gerber-Lemaire S, Rose DR, Juillerat-Jeanneret L. Functionalized pyrrolidine inhibitors of human type II α-mannosidases as anti-cancer agents: optimizing the fit to the active site. Bioorg Med Chem. 2008;16:7337–7346. doi: 10.1016/j.bmc.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Englebienne P, Fiaux H, Kuntz DA, Corbeil CR, Gerber-Lemaire S, Rose DR, Moitessier N. Evaluation of docking programs for predicting binding of Golgi α-mannosidase II inhibitors: a comparison with crystallography. Proteins. 2007;69:160–176. doi: 10.1002/prot.21479. [DOI] [PubMed] [Google Scholar]
- 28.Johansen H, Van Der Straten A, Sweet R, Otto E, Maroni G, Rosenberg M. Regulated expression at high copy number allows production of a growth-inhibitory oncogene product in Drosophila Schneider cells. Genes Dev. 1989;3:882–889. doi: 10.1101/gad.3.6.882. [DOI] [PubMed] [Google Scholar]
- 29.Aldecoa A, Gujer R, Fischer JA, Born W. Mammalian calcitonin receptor-like receptor/receptor activity modifying protein complexes define calcitonin gene-related peptide and adrenomedullin receptors in Drosophila Schneider 2 cells. FEBS Lett. 2000;471:156–160. doi: 10.1016/s0014-5793(00)01387-9. [DOI] [PubMed] [Google Scholar]
- 30.Rossi EJ, Sim L, Kuntz DA, Hahn D, Johnston BD, Ghavami A, Szczepina MG, Kumar NS, Sterchi EE, Nichols BL, Pinto BM, Rose DR. Inhibition of recombinant human maltase glucoamylase by salacinol and derivatives. FEBS J. 2006;273:2673–2683. doi: 10.1111/j.1742-4658.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- 31.Scotter AJ, Kuntz DA, Saul M, Graham LA, Davies PL, Rose DR. Expression and purification of sea raven type II antifreeze protein from Drosophila melanogaster S2 cells. Protein Expr Purif. 2006;47:374–383. doi: 10.1016/j.pep.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Yamashiro K, Itoh H, Yamagishi M, Natsuka S, Mega T, Hase S. Purification and characterization of neutral α-mannosidase from hen oviduct: studies on the activation mechanism of Co2+ J Biochem. 1997;122:1174–1181. doi: 10.1093/oxfordjournals.jbchem.a021878. [DOI] [PubMed] [Google Scholar]
- 33.Haeuw JF, Strecker G, Wieruszeski JM, Montreuil J, Michalski JC. Substrate specificity of rat liver cytosolic α-d-mannosidase. Novel degradative pathway for oligomannosidic type glycans. Eur J Biochem. 1991;202:1257–1268. doi: 10.1111/j.1432-1033.1991.tb16498.x. [DOI] [PubMed] [Google Scholar]
- 34.Shah N, Kuntz DA, Rose DR. Golgi α-mannosidase II cleaves two sugars sequentially in the same catalytic site. Proc Natl Acad Sci USA. 2008;105:9570–9575. doi: 10.1073/pnas.0802206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Torres E, Mendiola MA, Pastor CJ. Crystal structures of triazine-3-thione derivatives by reaction with copper and cobalt salts. Inorg Chem. 2006;45:3103–3112. doi: 10.1021/ic052009d. [DOI] [PubMed] [Google Scholar]
- 36.Chen HP, Marsh EN. How enzymes control the reactivity of adenosylcobalameffect on coenzyme binding and catalysis of mutations in the conserved histidine-aspartate pair of glutamate mutase. Biochemistry. 1997;36:7884–7889. doi: 10.1021/bi970169y. [DOI] [PubMed] [Google Scholar]
- 37.Liptak MD, Datta S, Matthews RG, Brunold TC. Spectroscopic study of the cobalamin-dependent methionine synthase in the activation conformation: effects of the Y1139 residue and S-adenosylmethionine on the B12 cofactor. J Am Chem Soc. 2008;130:16374–16381. doi: 10.1021/ja8038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J Mol Biol. 2005;345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 39.Miyanaga A, Fushinobu S, Ito K, Wakagi T. Crystal structure of cobalt-containing nitrile hydratase. Biochem Biophys Res Commun. 2001;288:1169–1174. doi: 10.1006/bbrc.2001.5897. [DOI] [PubMed] [Google Scholar]
- 40.Bessey OA, Lowry OH, Brock MJ. A method for the rapid determination of alkaline phosphatase with five cubic millimeters of serum. J Biol Chem. 1946;164:321–329. [PubMed] [Google Scholar]
- 41.Segel IH. Enzyme kinetics: behaviour and analysis of rapid equilibrium and steady state enzyme systems. New York: John Wiley & Sons; 1975. [Google Scholar]
- 42.Leatherbarrow RJ. GraFit Version 4.0, Erithacus Software Ltd. Staines, UK: 1998. [Google Scholar]