Abstract
RNA polymerase (RNAP) is an essential and highly conserved enzyme in all organisms. The process of transcription initiation is fundamentally different between prokaryotes and eukaryotes. In prokaryotes, initiation is regulated by σ factors, making the essential interaction between σ factors and RNAP an attractive target for antimicrobial agents. Our objective was to achieve the first step in the process of developing novel antimicrobial agents, namely to prove experimentally that the interaction between a bacterial RNAP and an essential σ factor can be disrupted by introducing carefully designed mutations into σA of Bacillus subtilis. This disruption was demonstrated qualitatively by Far-Western blotting. Design of mutant σs was achieved by computer-aided visualization of the RNAP-σ interface of the B. subtilis holoenzyme (RNAP + σ) constructed using a homology modeling approach with published crystal structures of bacterial RNAPs. Models of the holoenzyme and the core RNAP were rigorously built, evaluated, and validated. To allow a high-quality RNAP-σ interface model to be constructed for the design of mutations, a crucial error in the B. subtilis σA sequence in published databases at amino acid 165 had to be corrected first. The new model was validated through determination of RNAP-σ interactions using targeted mutations.
Keywords: RNA polymerase, sigma factor, homology model, antimicrobials, protein-protein interactions
Introduction
RNA polymerase (RNAP) is the enzyme responsible for transcription in all organisms. Although RNAP is structurally conserved among all domains of life,1 there is considerable difference in how the enzyme functions in eukaryotes and prokaryotes. The active site is highly conserved but the factors that regulate transcription are very different.
Bacterial RNAP core enzyme consists of five subunits: two α, and single β, β′, and ω subunits. A sixth subunit, σ, binds to the core to form what is known as the holoenzyme (HE) enabling the enzyme to bind to specific promoter sequences on the DNA and to initiate transcription.2 This is the first step of the transcription cycle which consists of three stages: initiation, elongation, and termination. Upon initiation of transcription, RNAP frequently produces short RNA transcripts, two to nine nucleotides in length, without clearing the promoter region (abortive initiation). Once the RNA chain reaches ∼12 nucleotides, the transcription complex becomes more stable allowing σ to stochastically dissociate and RNA chain elongation to continue.3–5 σ Factors are a large family of proteins which are essential for initiation and specificity of transcription. In addition to a “housekeeping” σ factor, several additional σ factors may be present in a cell at any one time. The additional σ factors generally become activated due to specific environmental signals, leading to the transcription of specific genes, such as those involved in the stress response/pathogenicity. In Bacillus subtilis, the primary “housekeeping” σ factor is σA.6
High resolution structures of different forms of RNAP have been solved, including core,7,8 HE,3,9 and an elongation complex.10 Although these high resolution structures are all from extremophiles, due to high levels of sequence conservation, the information obtained is transferable to other bacterial RNAPs. The structure and function of bacterial RNAP HEs have been reviewed recently.11 The highly conserved bacterial σ factors consist of four major conserved regions, designated 1–4. There are many contacts between σ and the core enzyme, but the most extensive interaction occurs between the polar surface of σ region 2.2 and a solvent exposed coiled-coil region of the RNAP β′ subunit.3,9
Because RNAP is an essential and highly conserved enzyme, it has become an attractive target for the development of new antimicrobial drugs. The mechanism of initiation, as regulated by σ factors, is unique to prokaryotes, so their interaction with RNAP represents an excellent target. This target is exploited by natural anti-sigma factors in Gram positive and negative organisms, and also in drug discovery programs.12,13B. subtilis has been extensively studied as a paradigm for σ-factor regulated gene expression (particularly during sporulation) (e.g., as shown in Ref.14). It is a harmless soil bacterium but is closely related to many important pathogens in the low G+C group of Gram positive bacteria (firmicutes). Creation of high-quality models of the RNAP HE enables their use as a tool to design transcription initiation inhibitors. Key residues involved in the interaction between σ factors and the RNAP core can then be specifically targeted which may lead to improved models and aid rational design of inhibitors of this essential interaction as potential antibiotics with a novel mechanism of action.
B. subtilis RNAP core and HE have been modeled previously.15 The interface between β′ and σA was investigated and peptide mimics proposed to compete with σA for binding to the core enzyme. Because of software limitations and sequencing errors in σA used in the previous study, we have re-investigated this essential interaction. We show that the previous (incorrect) sequence of σA resulted in a flawed analysis of its interaction with RNAP. Our new model, and the importance of the corrected amino acid sequence, is supported by a protein–protein interaction assay. Our investigation of the interface between RNAP and σA will now permit accurate rational design of inhibitors of this interaction.
Results
sigA sequencing
When sequencing multiple clones containing B. subtilis sigA or regions of sigA (data not shown), a “mutation” was always observed in region 2.2. To investigate this further, sigA from B. subtilis strains 168, 168CA, and SB19 (refer Table SI in Supporting Information) was sequenced. All three strains showed the same sequence, all with a single amino acid different to that found in the B. subtilis 168CA genome sequence SubtiList web server (http://genolist.pasteur.fr/SubtiList/). An example of a sequencing run is shown in the Supporting Information (Fig. S1). A substitution of G for T gives rise to a glutamine (CAG) in position 165, which was previously thought to be a histidine (CAT).16
To further support our data, a sequence alignment of sigma factors from a diverse range of Gram positive and negative bacteria was performed (σ region 2.2 shown in Fig. 1). The histidine (H) present in the B. subtilis sigA database sequence is highlighted with an asterisk. In all other sequences, including the less closely related alternative factors σF (sigF, B. subtilis) and σ32 (rpoH, Escherichia coli), the amino acid at the equivalent position is always glutamine (Q). Given our sequencing results and the alignments, we propose that B. subtilis σA also contains a glutamine at amino acid position 165. Subsequent projects to re-sequence the B. subtilis 168 genome have confirmed our results.17 Comparison of the other RNAP subunits showed there were no other errors in sequencing.
Figure 1.

Alignment of region 2.2 of 15 σ factors. Three-letter organism codes are as follows: Bsu, B. subtilis; Bha, Bacillus halodurans; Sau, Staphylococcus aureus; Mtu, Mycobacterium tuberculosis; Sco, Streptomyces coelicolor; Taq, Thermus aquaticus; Tth, Thermus thermophilus; Cmu, Chlamydia muridarum; Eco, Escherichia coli; Pae, Pseudomonas aeruginosa; Ret, Rhizobium etli; Tma, Thermatoga maritima; Mge, Mycoplasma genitalium. The asterisk indicates the incorrect amino acid for B. subtilis in sequence databases.
B. subtilis RNAP model construction and structure validation
Homology models of RNAP core and HE subunits were created individually and then combined to form the complete enzyme. The crystal structures of Thermus aquaticus core8 and Thermus thermophilus HE3 were used as templates. The major subunits (α, β, β′) of B. subtilis RNAP share about 45% identity and 65% similarity with the corresponding T. aquaticus and T. thermophilus subunits (refer Table SII in the Supporting Information for a full identity and similarity list).
The enzyme models were subjected to structure clean-up and optimization. After each minimization set, the quality of the model was examined by Ramachandran analysis18–20 (refer Fig. S2 in the Supporting Information). A check of the templates showed that only 54% of T. aquaticus core residues were in the favored regions of the Ramachandran plot (81% in the allowed regions), while for T. thermophilus HE 86% of residues were in the favorable region (97% in the allowed regions). This very low score for the T. aquaticus core is not ideal for a template, but this is the only core RNAP structure available that is not in complex with an antibiotic. The T. thermophilus HE score of 86% is very high considering the large size of the enzyme. The scores for the homology models are comparable to the crystal structure scores, suggesting the models are of good quality. These scores were obtained for models refined by minimization of the sidechains with the backbone constrained. Further minimization without constraints led to deterioration of the backbone secondary structures for both our models and the crystal structures. Only models minimized with the backbone constrained were used in further analysis. An alternative approach to enzyme construction was also undertaken to investigate if there was a difference between minimizing the subunits first and then putting together the enzyme, or putting the enzyme together from unminimized structures and then minimizing. This second procedure was performed to account for possible changes in the structure of each subunit under the influence of the surrounding subunits. However, the same result was obtained (data not shown).
The final B. subtilis RNAP models, with sidechains minimized, are shown in Figure 2. Panel A shows the core enzyme while Panel B shows the HE. Parts of the β and β′ subunits where there are gaps in the template structure (>10 amino acids) are not displayed, because the models are not reliable under these circumstances. Sequence alignments available as Supporting Information (A1–A4) show these regions. Both models show the classic crab claw shape made with the two α subunits (cyan and pink) at the back, and the β (green) and β′ (red) subunits forming the two halves of the DNA-binding channel. ω (cream) is positioned on top of β′ and σA (blue) is on the top front of the HE only and protrudes across the channel. The region where the major interaction between σA and the core occurs is shown in the insets of Panels A and B. The interacting helix-turn-helix (HtH) motif of β′(red) can be seen clearly in the Panel B inset.
Figure 2.
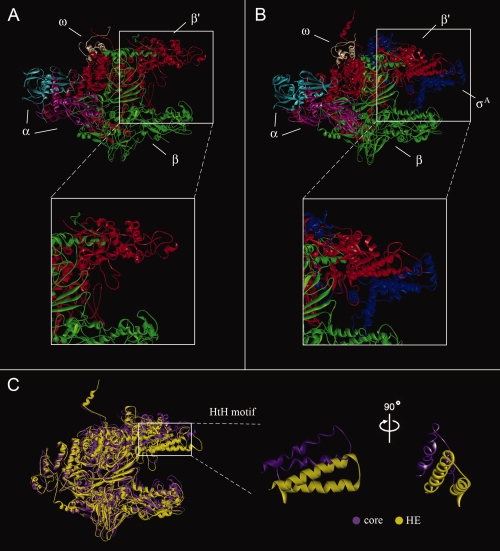
Homology models of B. subtilis RNAP core (A) and RNAP HE (B). The inset panels show regions of RNAP that change upon binding of σ, from core to HE. The enzyme subunits are color coded as indicated. Panel C: Superimposition of the two RNAP models, using the α subunits. Only the β and β′ subunits are shown.
RNAP is a dynamic structure: Changes from core to HE
Apart from the obvious addition of σA, the core enzyme undergoes a number of changes when forming the HE. The major change is highlighted in Panel C of Figure 2. The top half of the “claw” of the HE (yellow) has closed down to hold σA compared to the more open channel in the core enzyme (purple). Showing the HtH motif only from two different angles emphasizes the fact that this region of the HE has moved down in comparison to the core, rather than rotated. The presence of more disordered regions with less secondary structure in the core enzyme may be explained by the lower resolution and quality of the core crystal structure template. To analyze the flexibility of RNAP in more detail, all subunits were subjected to distance difference matrix (DDM) analysis.21 This allows evaluation of relative changes in the position of each amino acid Cα atom with respect to all other Cα atoms in each subunit. As has been suggested before (reviewed in Ref.11), our models confirm that the two α subunits, the ω subunit and the C-terminal halves of the β and β′ subunits move as rigid bodies upon binding of σ. This leads to DDMs with no significant changes within any of the subunits. The DDMs of the N-terminal halves of β and β′ (refer Fig. S3 in the Supporting Information) did show changes between the core and HE. However, the largest of these changes are due to the insertions in the B. subtilis sequence when compared to the template sequences, and are thus not meaningful.
The interface between β′ and σA
We minimized the B. subtilis homology model and the T. thermophilus crystal structure in exactly the same way, so any differences observed are meaningful. The major interaction between β′ and σA is between a coiled-coil region of β′(comprising a HtH motif) and region 2.2 of σA (a single helix), highlighted red and blue, respectively, in the center of Figure 3. Panels A–D of Figure 3 show the amino acids forming hydrogen bonds (A and C) and hydrophobic interactions (B and D) in both the B. subtilis and T. thermophilus structures. Our B. subtilis model has an extensive network of proposed hydrogen bonds as shown in Panel A. Interestingly, one of these is formed by σA Q165, which was previously thought to be a histidine (refer previous section), and when the enzyme is modeled with H in position 165, this hydrogen bond is not formed (data not shown). The hydrogen bond network seen in the T. thermophilus crystal structure [Fig. 3(C)] is even more extensive, with more amino acids involved. It has been suggested, that no single molecular cause can explain the greater thermal stability of thermophilic proteins, and that they rely instead on combinations of stabilizing effects.22 On the other hand, older studies proposed that a greater number of specific interactions, particularly electrostatic interactions and hydrogen bonds, are largely responsible for the higher stability of thermophilic proteins.23,24 The alignment in Panel E summarizes the amino acids involved in hydrogen bonds (green dots) and shows that corresponding amino acids in B. subtilis and T. thermophilus are implicated, with T. thermophilus having an extra four amino acids from β′ and an extra two from σA involved. A detailed list of predicted hydrogen bonds is given in Table I, the predicted hydrogen bond lengths are listed as an indication of the likely strength of the bonds. While more hydrogen bonds are predicted for T. thermophilus, most of these are expected to be weak (>2 Å).
Figure 3.
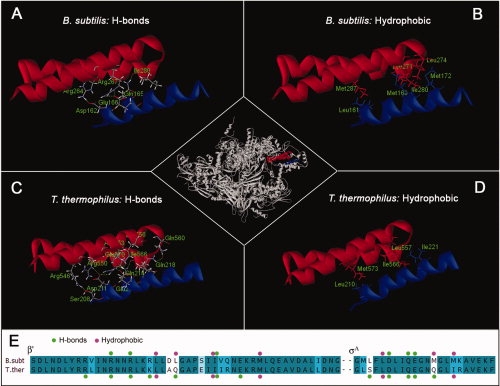
The major interaction interface between β′ and σA. The center shows the RNAP HE with the β′ coiled-coil in red and σA region 2.2 in blue. The amino acids forming the hydrogen bond network in the B. subtilis HE model and the T. thermophilus crystal structure are shown in Panels A and C, hydrophobic interactions in B. subtilis and in T. thermophilus are in Panels B and D. A summary of the interactions is shown in the alignment in E, with hydrogen bonds and hydrophobic interactions marked with green and pink dots, respectively.
Table I.
Summary of Amino Acids and Atoms Involved in the Predicted Hydrogen Bond Network Between β′ and σA in B. subtilis and T. thermophilus
|
B. subtilis |
T. thermophilus |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| σA amino acid | Atom | β′ amino acid | Atom | Distance (Å) | σA amino acid | Atom | β′ amino acid | Atom | Distance (Å) |
| D162 | OD2 | R264 | HH21 | 1.8 | S208 | OG | R546 | HH12 | 1.7 |
| E166 | OE2 | R267 | HE | 1.9 | D211 | OD1 | R550 | HE | 2.2 |
| E166 | OE2 | R267 | HH21 | 2.4 | E215 | OE2 | R553 | HE | 2.3 |
| Q165 | HE22 | I280 | O | 1.9 | D211 | OD1 | R553 | HH12 | 2.4 |
| E215 | OE2 | K556 | HZ2 | 2.3 | |||||
| Q214 | HE21 | E570 | OE2 | 2.5 | |||||
| Q214 | HE22 | I566 | O | 1.9 | |||||
| Q218 | HE21 | Q560 | OE1 | 2.4 | |||||
Figure 3 (Panels B and D) shows the residues involved in hydrophobic interactions between β′ and σA. They are illustrated as sidechains from each subunit extending towards each other. This occurs in two areas along the interface in both the B. subtilis model (B) and the T. thermophilus crystal structure (D), with more B. subtilis amino acids predicted to be involved in hydrophobic interactions. In T. thermophilus, three residues from β′ and two from σA interact and in B. subtilis, two additional amino acids from each chain are involved (Panel E, pink dots). The extra amino acids involved in B. subtilis are σA M169 and β′ L274. Interestingly, the corresponding amino acid in T. thermophilus at both these positions is a polar glutamine, and these two amino acids are instead involved in hydrogen bonds.
Comparison of new HE model to previous modeling
When comparing the new B. subtilis holoenzyme model from this study to a previous model,15 several important interactions not observed previously were found between the residues at the β′-σA interface, as summarized in Table SIII in the Supporting Information. One difference in particular is the presence of σA Q165 in the predicted hydrogen bond network of the model created in this study. Q165 is the residue previously thought to be a histidine. With the correct amino acid now in this position, a more extensive hydrogen bond network is predicted. The interface in our new HE model is also more similar to the interface present in the T. thermophilus crystal structure (as shown in Fig. 3). This probable similarity between distantly related organisms is significant and we suggest it further validates the use of a non-pathogenic model organism closely related to pathogens for experimental design of inhibitors.
Design of peptides to inhibit HE formation
A necessary first step to test the theory that disrupting binding of σA to β′ will inhibit transcription initiation, is to create mutants of σA in the 2.2 region by mutating the residues that are proposed to be important in the interaction (as shown in Fig. 3) and observing their effect on binding. Inhibitors were designed based on σA and not β′, as σA is limiting in the cell. An initial set of mutations were chosen to have either minimal (D162G, Q165A, and M172A) or dramatic (D162K, Q165K) effects on binding, as illustrated in Figure 4(A).
Figure 4.
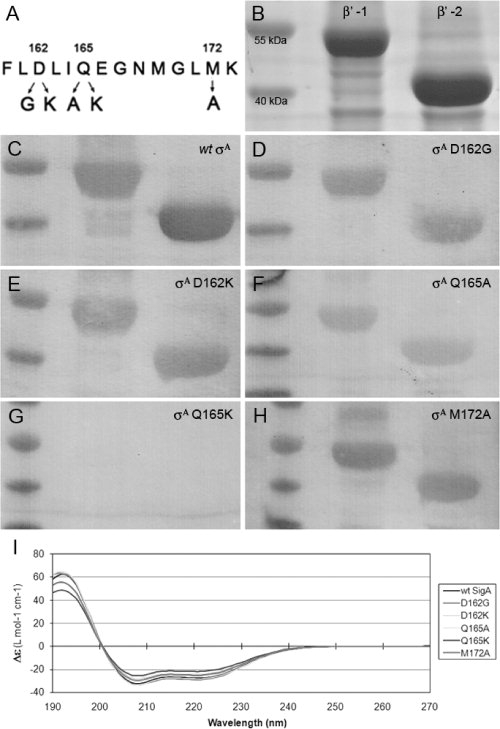
Evaluation of the RNA polymerase binding capacity of σA mutants. (A) Schematic showing the different σA mutations generated. (B) Coomassie blue stain of the β′ fragments used in the Far-Western blots. Far-Western blots show binding of wild-type σA (C), σA D162G (D), σA D162K (E), σA Q165A (F), σA Q165K (G), and σA M172A (H) to the β′ fragments. Each blot shows a molecular weight ladder in the first lane followed by two lanes of β′-bound σA. (I) Circular dichroism analysis of σA and mutants.
σA Mutants have altered binding capacity for β′
Mutations were introduced into B. subtilis sigA using site-directed mutagenic PCR. The mutated and wild-type genes were inserted into a T7 expression vector and overproduction of proteins carried out in E. coli. In contrast to well established protocols used to renature σ expressed as an insoluble form in inclusion bodies,25 we optimized conditions so that wild-type and mutant σA proteins were overproduced in soluble form. Wild-type σA produced by our optimized methods has been shown to be functional in transcription assays.26
Binding of σA proteins to β′ was tested by Far-Western blotting. Two fragments of β′27 were used to test the binding of σA and its mutant forms. These fragments overlap, so both contain the coiled-coil region of interest, as shown in Figure S4 of the Supporting Information. Far-Western blotting is a well established method for identifying protein–protein interactions and has been used in previous studies to identify where σ70 binds to RNAP in E. coli28 and where NusA binds to RNAP in B. subtilis.27
The two β′ fragments are shown in Figure 4(B), with molecular weights of ∼40 and 55 kDa when compared with the molecular weight marker (first lane of each panel). Upon addition of wild-type σA to the membrane, binding to both β′ fragments was observed as shown in Figure 4(C) (second and third lanes). By comparing the wild-type data to that of the mutant proteins, qualitative comparisons can be made as to their ability to bind to β′. σA mutations D162G, D162K, and M172A (Panels D, E, and H) do not appear to have a large effect on the ability of σA to bind to β′. On the other hand, mutating amino acid 165 drastically reduces the capacity for σA binding. Q165K (Panel G) abolishes binding altogether, while Q165A (Panel F) clearly shows a reduced level of binding.
To confirm that the altered binding was due to the change in the σA-β′ interface and not due to protein misfolding, circular dichroism (CD) was performed to compare the structures of the σA proteins. All mutants and the wild-type σA displayed the same CD spectra (Panel I, Fig. 4) indicating that all proteins were folded correctly.
Modeling of σA mutants
Mutations were introduced into the B. subtilis HE model to observe their effects on the σA -β′ interface. In wild-type σA Q165 is proposed to dip into a hydrophobic pocket [Fig. 5(A)] and form a hydrogen bond with the mainchain oxygen of β′ I280 (refer also Fig. 3 and Table I). With lysine (K) in its place, the sidechain does not appear to be able to fit into the pocket [Fig. 5(B)], which would lead to a loss of the hydrogen bond, as well as the hydrophobic interactions between the sidechain of Q165 and the pocket. Our model also suggests the possibility of a clash with β′ I280 [Fig. 5(C)]. When alanine (A) is at this position, decreased binding was seen rather than complete abolishment of the interaction [Fig. 4(F)]. Alanine has a short, hydrophobic sidechain, so some hydrophobic interactions with the pocket would be possible, but the hydrogen bond would be lost.
Figure 5.
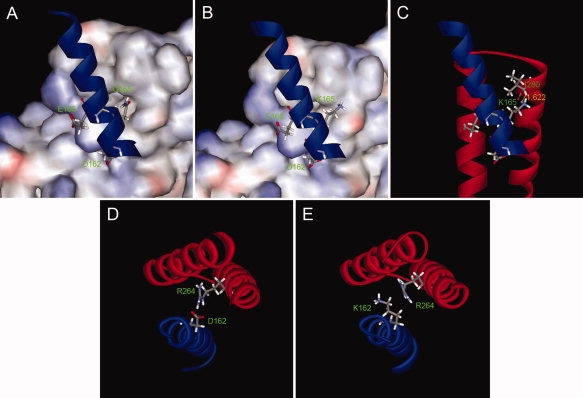
Modeling of the interaction of mutant σA with RNAP. σA region 2.2 is shown in blue and the surface of β′ is colored according to the electrostatic potential in Panels A and B. The interacting region of the β′ subunit is represented by a red ribbon in Panels C–E. Wild-type Q165 fits into a hydrophobic pocket on the surface of β′ (A). With K165 modeled in its place, the sidechain cannot fit into the pocket (B) and there is a clash with I280 of β′ (C, clash shown in yellow). Wild-type D162 forms a hydrogen bond with β′ R264 (D). With K modeled into position 162 (E) binding can still occur because the sidechain bends away from the interface.
Far-Western blots also showed that mutating the very highly conserved D162 did not greatly interfere in the ability of σA to bind [Fig. 4(D,E)]. In the wild-type model, D162 forms a hydrogen bond with β′ R264 [Fig. 5(D)]. Mutations in this position appear to be tolerated because the interaction is towards the end of the interface. Hence, the lysine in the D162K mutant may be able to bend out of the way, with binding still possible across the rest of the interface (Fig. 5, compare E to D). However, there would still be a loss of a hydrogen bond, which may translate to slightly weaker binding using more sensitive assays. Similarly, the M172A mutation does not show dramatically decreased interaction, possibly because it is also positioned at the end of the interface [Fig. 3(B)].
Discussion
We detected a mutation in the published sequence of B. subtilis σA, and this was recently confirmed by other authors.17 The correct amino acid at position 165, in region 2.2 of σA is glutamine (Q), not histidine (H), as previously thought.16 This is significant for the current work, because the major interaction between β′ and σA in the RNAP holoenzyme involves σA region 2.2.
Previously, a homology model of B. subtilis HE was published15 which included the incorrect amino acid in the σA sequence. This model has now been corrected and extensively refined and validated. A model of the core enzyme has also been built and compared to the HE. The differences between core and HE models showed that the β and β′ subunits move as rigid bodies (as has previously been suggested11), rather than the subunits being flexible within themselves. The comparison between the β′ and σA interface in the models and the crystal structures used as templates has yielded interesting results. The crystal structures are both of RNAP from thermophiles, where a greater number of hydrogen bonds seem to be involved in the σA-β′ interaction than in the mesophilic B. subtilis, where more hydrophobic interactions appear to be involved.
Our models propose that the corrected amino acid Q165 is crucial for the σA-β′ interaction. We designed several mutants to test our model and show that targeted disruption of residues at the σA-β′ interface could lead to decreased interaction.
σA mutants were prepared and used to demonstrate that mutation of Q165 dramatically reduces the ability of σA to bind to β′. Binding was significantly decreased for the Q165A mutant, and was undetectable in the Q165K mutant. Previous studies have also shown that mutation of the equivalent residue in E. coli σ32 (Q80; an alternate σ factor) to R or N, reduces core association markedly and greatly lowers the level of transcription.29 Similar results were seen with an E. coli σ70 Q406A mutant (equivalent to B. subtilis Q165A).30 Interestingly, this study also had a D403N mutation (equivalent to B. subtilis σA D162) which showed that 9 times the amount of mutant protein was required to achieve the same level of binding as wild-type σ70. Our results did not show a significant effect on binding when mutating this residue, but this could be due to the lack of sensitivity of our assay. Importantly, mutation of this residue in E. coli did not abolish the interaction completely, which fits well with our modeling of mutant D162K. Additionally, mutation of hydrophobic residues along the interface in E. coli σ70 had little or no effect on core binding. This was also observed in this study with the M172A mutation.30
Combining computer-modeling and in vitro binding assays, we have thus been able to take the first steps towards validating the σA-β′ interaction in bacterial RNAP holoenzymes as a potential new target for the development of antibacterial agents with a novel mode of action.
Materials and Methods
Sequence and template acquisition
All sequences of B. subtilis proteins were obtained from the SubtiList web server (genolist.pasteur.fr/SubtiList). Gene names were as follows: α-rpoA, β-rpoB, β′-rpoC, ω-yloH, σA-sigA. Protein sequences from all other organisms were obtained from NCBI (www.ncbi.nih.gov). These represent RNAP subunits from a range of Gram positive and Gram negative bacteria.
Crystal structure templates were obtained from the Protein Data Bank (PDB)31 (www.rcsb.org/pdb). The template for core RNAP was of T. aquaticus core,8 PDB code 1HQM, chains A–E; and the template for the HE was of T. thermophilus HE,3 PDB code 1IW7, chains A–F.
DNA sequencing
sigA from three different strains of B. subtilis was amplified by PCR and then sequenced (Australian Genome Research Facility, Brisbane, Australia). Two of the strains used were “wild-type” 168 laboratory strains from different research groups; this is important as the provenance of strains is often hard to determine. Strain 168, a highly transformable tryptophan auxotroph, was originally obtained by X-ray mutagenesis in the 1940s32 and has since been passaged through many laboratories, making unambiguous establishment of strain provenance problematic. This accounts for slight differences between strains/stocks between labs. The strains used in this study included: 168 (laboratory stock, P.J. Lewis), 168CA,33 the type of strain used in the B. subtilis genome sequencing project and SB19 (R.G. Wake, The University of Sydney). Refer Supporting Information, Table SI for strain details.
Homology modeling
Multiple sequence alignments were completed for each subunit using the ClustalW2 program34 (www.ebi.ac.uk/Tools/clustalw2) and then submitted to the SWISS-MODEL server35 (http://swissmodel.expasy.org) using the “alignment interface.” In the case of β′ (rpoC) and ω (yloH), the position of gaps would not allow a model to be created, so the gaps in the alignment had to be manipulated manually using Swiss-Pdb Viewer35 and then submitted to SWISS-MODEL using the project mode. Alignments for these subunits are provided in the Supporting Information. Core β′ had to be split into two parts to obtain the homology model.
Structure assembly and clean-up
To create the entire enzyme, each subunit of B. subtilis core and HE was imported on top of the crystal structure template to check for correct positioning. The template was then deleted, leaving behind B. subtilis RNAP models only.
Swiss-Pdb Viewer35 was used to clean-up models before further refinement. Amino acids with steric clashes were selected and then “fixed” (repaired) using the “quick and dirty” method. This was repeated until there was no improvement in the number of amino acids with clashes.
Structure minimization
Energy minimization of the structures was carried out using Discovery Studio 2.0 (Accelrys). Conditions were set by the CHARMM forcefield36 as implemented in the Accelrys software. First, hydrogens were added to the model or crystal structure and the remainder of the structure was constrained (fixed position in space) to refine the positioning of the hydrogens. Next, the backbone was constrained and minimization carried out to refine the positioning of the sidechains; and finally, the entire structure was minimized with no constraints. For mutation analysis, single amino acids were substituted and minimized alone (i.e., with the rest of the enzyme constrained) followed by minimization of sidechains within 10 Å of the σA-β′ interface (backbone constrained).
Each set of refinements involved first minimizing with the Steepest Descent algorithm until the maximum derivative was less than 0.1 kJ/(mol Å); and then minimizing with the Adopted-Basis Newton Raphson (ABNR) algorithm until the maximum derivative was less than 0.01 kJ/(mol Å). All other parameters were kept at default values.
Structure evaluation
Quality of models was checked using online programs, including MolProbity18 (http://kinemage.biochem.duke.edu) and PROCHECK19 (Protein Data Bank). These packages make use of Ramachandran plots20 and clash-scores, and determine outliers in areas such as bond lengths, angles, and spatial arrangement. To assess subunit flexibility, the ProFlex2.0 program21 was used to obtain distance difference matrix (DDM) plots. The β and β′ subunits were split into two parts because the program cannot evaluate chains longer than 999 amino acids.
Strain construction
A full strain and plasmid list can be found in the Supporting Information, Table SI.
E. coli DH5α was used for all cloning procedures. B. subtilis sigA was amplified from 168 genomic DNA and inserted into pETMCSIII via the XbaI and Acc65I sites to create pNG590 (wild-type sigA with an N-terminal 6xHis tag to enable protein purification). PCR site-directed mutagenesis37 was used to create mutants of sigA that were similarly inserted into pETMCSIII. The plasmids pNG570, 571, 572, 573, and 600 code for σA mutations M172A, D162G, D162K, Q165A, and Q165K, respectively.
Protein overproduction, purification, and evaluation
E. coli strain C41(DE3) (Table SI) was transformed with each sigA-containing plasmid and resulting colonies placed into 1L of Luria-Bertani broth (LB) containing 100 μg/mL ampicillin and grown shaking at 37°C. At A600 0.6–1.0 isopropyl-β-d-thiogalactoside (IPTG) was added to 0.5 mM and shaking continued at room temperature for further 5 h. Cells were harvested by centrifugation and stored at −80°C.
Cells were resuspended in 20–40 mL buffer A (20 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, pH 7.5) and lysed using an EmulsiFlex-C5 homogenizer (Avestin). Proteins were purified by affinity chromatography using a 1 mL HisTrap HP column (GE Healthcare) and an AKTA FPLC system (GE Healthcare). The cell lysate (soluble fraction) was applied to the column then washed with 6.6% buffer B (50 mM NaH2PO4, 500 mM NaCl, 400 mM imidazole, pH 7.5) and eluted with a step to 100% buffer B. The protein peak was collected and dialyzed into storage buffer by two step dialysis (step 1: 50 mM NaH2PO4, 150 mM NaCl, 1 mM EDTA, 0.1 mM diothiothreitol, pH 7.5; step 2: 50 mM NaH2PO4, 150 mM NaCl, 0.1 mM diothiothreitol, 30% glycerol, pH 7.5) and stored at −80°C.
To check for correct protein folding, CD spectra were recorded from 300 nm to 190 nm on a JASCO J-810 spectropolarimeter, with proteins diluted to ∼0.25 mg/mL in 10 mM NaH2PO4.
Far-Western blotting
pNG482 and pNG48327 containing overlapping β′ fragments (both with the coiled-coil region of interest) were transformed into BL21(DE3)pLysS. Colonies were added to 10 mL LB containing 100 μg/mL ampicillin and grown at 37°C until A600 reached 0.5–0.8, where IPTG was then added (to 0.5 mM) and the culture grown for a further 3 h at 37°C; 1 mL aliquots were centrifuged and cells resuspended to an A600 of 10 in TES [200 mM Tris-HCl (pH 7.5), 5 mM EDTA, 100 mM NaCl]; 5–10 μL of each cell lysate was subjected to SDS-PAGE and then transferred to a nitrocellulose membrane. Following transfer, the membrane was washed three times (5 min each) in PBS and three times in FWB (20 mM Tris, 100 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20). The membranes were blocked and proteins refolded by immersing in FWB + 5% skim milk (w/v) for 1 h while shaking. The membrane was then probed with a σA protein (∼10 μg protein added to FWB + 5% skim milk) for 1 h and then washed three times in FWB. Rabbit polyclonal anti-σA antibody (M. Yudkin, University of Oxford) diluted 1:3000 in FWB + 5% milk was added to the membrane and left to incubate for 1 h, followed by another three washes and incubation with 1:3000 goat-anti-rabbit horsereadish peroxidase (BioRad). After the three final washes, the antibody was detected using an Opti-4CN™ substrate kit (BioRad) following the manufacturer's instructions.
Acknowledgments
The authors thank M. Yudkin, University of Oxford, for kindly supplying polyclonal anti-σA anti serum. E.J. was supported by a Ph.D. scholarship from the ARC and the University of Newcastle.
Glossary
Abbreviations
- core
core RNA polymerase
- DDM
distance difference matrix
- HE
RNA polymerase holoenzyme
- HtH
helix-turn-helix
- RNAP
RNA polymerase.
References
- 1.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 2.Burgess RR, Travers AA, Dunn JJ, Bautz EKF. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 3.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 4.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 5.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 8.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 10.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 11.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 12.Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 13.Andre E, Bastide L, Michaux-Charachon S, Gouby A, Villain-Guillot P, Latouche J, Bouchet A, Gualtieri M, Leonetti JP. Novel synthetic molecules targeting the bacterial RNA polymerase assembly. J Antimicrob Chemother. 2006;57:245–251. doi: 10.1093/jac/dki426. [DOI] [PubMed] [Google Scholar]
- 14.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDougall IJ, Lewis PJ, Griffith R. Homology modeling of RNA polymerase and associated transcription factors from Bacillus subtilis. J Mol Graph Model. 2005;23:297–303. doi: 10.1016/j.jmgm.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 17.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Medigue C, et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology. 2009;155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 21.Keller PA, Leach SP, Luu TT, Titmuss SJ, Griffith R. Development of computational and graphical tools for analysis of movement and flexibility in large molecules. J Mol Graph Model. 2000;18:235–241. 299. doi: 10.1016/s1093-3263(00)00028-0. [DOI] [PubMed] [Google Scholar]
- 22.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Tsai C-J, Nussinov R. Thermodynamic differences among homologous thermophilic and mesophilic proteins. Biochemistry. 2001;40:14152–14165. doi: 10.1021/bi0106383. [DOI] [PubMed] [Google Scholar]
- 24.Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 25.Helmann JD. Purification of Bacillus subtilis RNA polymerase and associated factors. Methods in enzymology. In: SAaS Garges., editor. RNA polymerases and associated factors, Part C. Vol. 370. San Diego, CA: Academic Press; 2003. pp. 10–24. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Lewis PJ. Overproduction and purification of recombinant Bacillus subtilis RNA polymerase. Protein Expr Purif. 2008;59:86–93. doi: 10.1016/j.pep.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, Hankamer B, Lewis RJ, Lewis PJ. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 10:997–1002. doi: 10.1038/embor.2009.155. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthur TM, Burgess RR. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta′ subunit. J Biol Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- 29.Joo DM, Ng N, Calendar R. A σ32 mutant with a single amino acid change in the highly conserved region 2.2 exhibits reduced core RNA polymerase affinity. Proc Natl Acad Sci USA. 1997;94:4907–4912. doi: 10.1073/pnas.94.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. The interface of σ, with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burkholder PR, Giles NH. Induced biochemical mutations in Bacillus subtilis. Am J Bot. 1947;34:345–348. Jr. [PubMed] [Google Scholar]
- 33.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 36.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 37.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]


