Abstract
The homopentameric ρ1 GABAC receptor is a ligand-gated ion channel with a binding pocket for γ-aminobutyric acid (GABA) at the interfaces of N-terminal extracellular domains. We combined evolutionary analysis, structural modeling, and experimental testing to study determinants of GABAC receptor assembly and channel gating. We estimated the posterior probability of selection pressure at amino acid residue sites measured as ω-values and built a comparative structural model, which identified several polar residues under strong selection pressure at the subunit interfaces that may form intersubunit hydrogen bonds or salt bridges. At three selected sites (R111, T151, and E55), mutations disrupting intersubunit interactions had strong effects on receptor folding, assembly, and function. We next examined the role of a predicted intersubunit salt bridge for residue pair R158–D204. The mutant R158D, where the positively charged residue is replaced by a negatively charged aspartate, yielded a partially degraded receptor and lacked membrane surface expression. The membrane surface expression was rescued by the double mutant R158D–D204R, where positive and negative charges are switched, although the mutant receptor was inactive. The single mutants R158A, D204R, and D204A exhibited diminished activities and altered kinetic profiles with fast recovery kinetics, suggesting that R158–D204 salt bridge perhaps stabilizes the open state of the GABAC receptor. Our results emphasize the functional importance of highly conserved polar residues at the protein–protein interfaces in GABAC ρ1 receptors and demonstrate how the integration of computational and experimental approaches can aid discovery of functionally important interactions.
Keywords: GABAC receptor, protein–protein interactions, ω-value, evolutionary analysis
Introduction
γ-Aminobutyric acid (GABA), the main inhibitory neurotransmitter of the central nervous system, is the native ligand of three classes of postsynaptic receptors, GABAA, GABAB, and GABAC, where GABAA and GABAC are Cys-loop ligand-gated ion channels (LGICs).1 The Cys-loop superfamily is named after a 13-residue signature loop formed by two conserved cysteine residues found in the N-terminal extracellular domain and comprises a variety of channels, including nicotinic acetylcholine receptors (nAChR), GABAA, GABAC, glycine receptors (GlyR), and serotonin (5-HT3) receptors. The subunits of these receptors assemble into functional homo- or hetero-pentameric structures with ligand-binding pockets at the subunit interfaces of the N-terminal extracellular domains, and a central ion channel.
X-ray structures have been determined for nAChR2,3 and a number of soluble proteins4,5 and prokaryotic membrane proteins (pLGIC)6–8 distantly related to LGIC family. Soluble acetylcholine binding proteins (AChBP)4,5 modulate the synaptic transmission of acetylcholine9 and are distantly related to the N-terminal ligand-binding domain of LGICs. High-resolution X-ray structures of pentameric AChBPs are widely used as templates for comparative modeling of LGICs.10–13
Native GABAC receptors are heteropentamers consisting of ρ1, ρ2, and/or ρ3 subunits and are found in retina, thalamus, hippocampus, pituitary, and gut.14 GABAC receptors are involved in visual processing, regulation of sleep-waking rhythms, pain perception, memory, learning, regulation of hormones, and neuroendocrine gastrointestinal secretion. However, no structural information is available for the GABAC receptor. It is important to understand the molecular mechanisms of GABAC receptor activation and inhibition by agonists and antagonists, respectively. Mutational studies of the GABAC receptor, including those of the N-terminal domain, have identified residues that rendered GABAC inactive, altered the sensitivity of the receptor to the agonists, or kept the protein constitutively active.15–19 These studies have relied primarily on information about known locations of protein loops that contain binding residues in other homologous well-studied LGICs, such as nAChR.20
The present study was undertaken to develop a homology model of the ρ1 GABAC receptor, in which detailed evolutionary analysis is used specifically to guide structure generation and to predict protein sites under strong evolutionary selection pressure. Such an approach differs from previously published work17,18,21,22 in that it combines Markovian evolutionary analysis with homology modeling and is not limited to determinations of residues specifically in the ligand-binding pocket. To develop the homology model, we first carried out a comprehensive maximum likelihood evolutionary analysis of GABAC ρ1 subunits at the cDNA level with the purpose of identifying a set of ρ1-specific residues under a strong evolutionary selection pressure. We then used evolutionary information as a guide for the sequence alignment with a template AChBP sequence from Lymnaea stagnalis, for which a high-resolution X-ray structure is available.23 According to our model, several residues are predicted to be at the protein–protein interfaces and under strong evolutionary purifying selection pressure. Using site-directed mutagenesis, Western blot study, and electrophysiological analysis, we have tested the effects of these residues on receptor assembly and channel activation.
Results
Evolutionary analysis
We first assessed the evolutionary selection pressure at each amino acid residue position of the GABAC extracellular domain. Here, the selection pressure was measured as the ratio of synonymous versus nonsynonymous substitutions, which is usually termed as the ω-ratio.24,25 We constructed explicitly a phylogenetic tree using only orthologous cDNA sequences from different species and modeled the evolution as a Markovian process. Figure 1 shows the estimated posterior probability based on an empirical Bayes approach24,26 of different ω-ratios for amino acid residues 53–259 of the 11 investigated DNA sequences of the ρ1 subunits. Here, darkly shaded, medium shaded, and lightly shaded vertical segments at a given amino acid position represent, respectively, the predicted posterior probability Pi (where i = 1–3) of belonging to the class of ωi-values of relatively high (ω1 ≤ 0.0078), medium (0.0078 < ω2 ≤ 0.026), or low purifying selection pressure (ω3 ≥ 0.0258). The amplitudes of the probabilities of these three regions sum to unity. The expected ω-value at each amino acid residue position used in this study is calculated as
Figure 1.
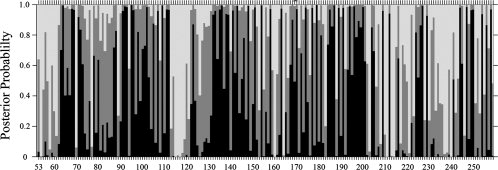
Selection pressure as measured by ω-ratio of nonsynonymous versus synonymous codon substitution rates at each amino acid residue of the N-terminal domain of GABAC receptor (residues 53–259). The x-axis shows the residue numbers of the protein. The y-axis shows the posterior probabilities of ω belonging to one of the three classes, where all probabilities sum up to unity at each residue position. The ω-ratios are grouped as follows: ω1 ≤ 0.0078 (darkly shaded vertical segment), 0.0078 < ω2 ≤ 0.026 (gray segment), and ω3 ≥ 0.0258 (light segment). Residues with large probability for ω1 (dark) are experiencing strong purifying selection and are highly conserved.
Several sequence regions exhibit alternating high and low or medium conservation patterns, which correspond well with the solvent-exposed β-strand secondary structure, revealing evolutionary constraints existing at conserved buried residues.27 For example, the stretch of residues between R104 and R109 displays this type of alternating graphic pattern. That is, within this sequence of residues, medium or weak purifying selection is observed for even-numbered, solvent-accessible residues, for example, R104 (ω = 0.017), Y106 (0.021), and K108 (0.030). By contrast, the odd-numbered residues are under significantly stronger purifying selection pressure; they exhibit low ω-values [H105 (ω = 0.005), W107 (0.003), D109 (0.001)], consistent with the notion that these residues are buried in the protein core. This sequence stretch includes residues that form loop D, which is a part of the ligand-binding pocket.28,29 A similar conservation pattern is observed between residues S168 and A175. Comparison with the template AChBP structure shows that this stretch of residues correlates well with the respective secondary structure, indicating that our method provides accurate estimations and useful predictions. Because of the insufficient diversity of the available GABAC sequences, a rather low mean ω-value of 0.023 was obtained for residues 53–259. The highly conserved positions with the lowest ω-value (ω = 0.001) were identified as D64, M95, D109, D136, N154, M179, and D187. The following residues were estimated to have ω = 0.002: R68, P69, E92, V93, Y102, R111, L112, P135, K143, H148, T151, R158, G163, R170, T174, C177, P185, Q189, C191, E194, E196, Y200, F227, T244, Y247, L250, and R258. Supporting Information Table 1 contains calculated ω-values for ρ1 residues 53–259.
Table I.
Amino Acid Residues Identified by Geometric Calculations as Forming GABA-Binding Pockets in the Pentameric Model of the ρ1 GABAC Receptor
| Residue | ω-value | ASA | Residue | ω-value | ASA | Residue | ω-value | ASA |
|---|---|---|---|---|---|---|---|---|
| Q83 | 0.027 | 0.09 | Q16017 | 0.089 | 0.21 | T21817,37,38 | 0.025 | 0.13 |
| E85 | 0.020 | 0.20 | L16617 | 0.034 | 0.28 | I22217 | 0.017 | 0.45 |
| L87 | 0.080 | 0.08 | Y16717 | 0.010 | 0.07 | S22317 | 0.042 | 0.27 |
| Y10215,37 | 0.002 | 0.03 | S16817,37,39 | 0.005 | 0.25 | L22417 | 0.006 | 0.05 |
| R10415,39 | 0.017 | 0.08 | L16917 | 0.011 | 0.04 | S22517 | 0.011 | 0.05 |
| H10515,39,40 | 0.005 | 0.17 | R17017,39 | 0.002 | 0.08 | Q22617 | 0.005 | 0.73 |
| M122 | 0.014 | 0.24 | C177 | 0.002 | 0.01 | F22717 | 0.002 | 0.20 |
| F13817,37 | 0.004 | 0.03 | S192 | 0.031 | 0.21 | L238 | 0.070 | 0.38 |
| V14017 | 0.027 | 0.12 | E194 | 0.002 | 0.09 | F24037 | 0.089 | 0.29 |
| H14117,40 | 0.004 | 0.22 | E196 | 0.002 | 0.03 | Y24137 | 0.017 | 0.56 |
| T152 | 0.021 | 0.04 | S197 | 0.007 | 0.02 | S24237,39 | 0.041 | 0.65 |
| D153 | 0.041 | 0.23 | Y19815 | 0.005 | 0.20 | S24315,39 | 0.033 | 0.24 |
| M15617 | 0.014 | 0.09 | A199 | 0.006 | 0.05 | T244 | 0.002 | 0.90 |
| L15717 | 0.023 | 0.02 | K21138,40 | 0.090 | 0.19 | Y247 | 0.002 | 0.08 |
| R15817,39 | 0.002 | 0.16 | N21338 | 0.080 | 0.23 | R24915,39 | 0.003 | 0.02 |
Accessible solvent area (ASA) of amino acid residues is calculated as a fraction of the extended standard state for a residue in tripeptide Gly-X-Gly.41
Construction and analysis of homology model
A structure of AChBP from L. stagnalis23 (PDB ID: 1I9B) served as the template for homology modeling. This L. stagnalis structure was chosen over the structures of AChBPs from A. californica30 and from B. trancatus31 because it has the longest N-terminal α-helix, similar to that of nAChR subunits, and hence was anticipated to provide a better template for the GABAC receptor. Because the sequence identity between AChBP and N-terminal domain of GABAC receptor is low (∼16%), an accurate sequence alignment between the target and template proteins is challenging. To obtain a suitable sequence alignment, we combined evolutionary data with an additionally predicted secondary structure of the GABAC ligand-binding domain. Consensus results obtained from JPRED32 strongly indicated that the N-terminal α-helix begins near residues E55 or Q56. A similar conclusion was reached by comparing GABAA and GABAC receptor sequences with sequences and structures of α, β, γ, and δ subunits of nAChR2 (PDB ID: 2BG9) (Fig. 2).
Figure 2.
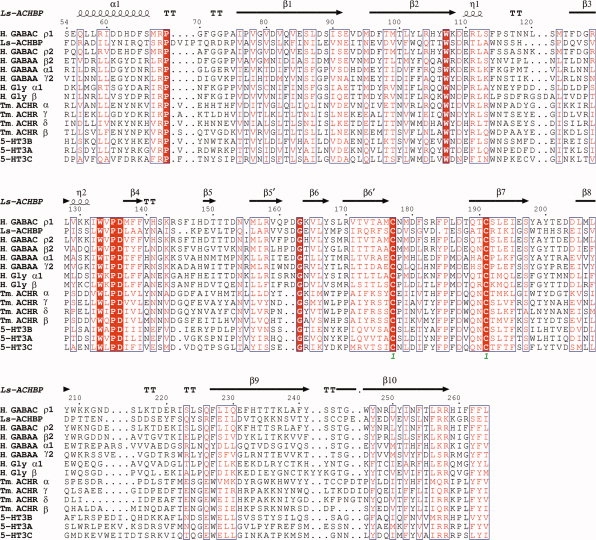
Multiple sequence alignment of L. stagnalis AChBP (Ls-ACHBP), H. sapiens ligand-gated ion channels GABAC ρ1, ρ2 (H. GABAC ρ1, H. GABAC ρ2), GABAA α1, β2, γ2 (H. GABAA α1, H. GABAA β2, H. GABAA γ2), glycine receptor α1, β (H. Gly α1, H. Gly β), 5-HT3 receptor A, B, C (5-HT3A, 5-HT3B, 5-HT3C), and T. marmorata nAChR α, β, γ, δ (Tm. ACHR α, Tm. ACHR β, Tm. ACHR γ, Tm. ACHR δ).
Computational geometry calculations33–36 were performed on the homology model obtained by the above approach, to identify residues that form the GABA-binding pocket at the subunit interfaces of the modeled structure. These calculations identified a total of 185 pockets and voids in the modeled GABAC ρ1 homopentamer. There are five large symmetric pockets found at the subunit interfaces, which contain residues that previously have been proposed to be involved in GABA binding.15,17,18 Figure 3 illustrates details of one of these intersubunit pockets, and Table I lists 45 amino acid residues that are predicted to form the pocket and their expected ω-values and the accessible surface area (ASA). These values of ASA are presented as a fraction of the solvent-accessible surface calculated using the VOLBL program33 and the respective residue's accessible solvent area in the extended standard state in tripeptide Gly-X-Gly.41 A literature search revealed that mutation data on 30 out of 45 pocket residues are available,15,17,37–40 as listed in Table I.
Figure 3.
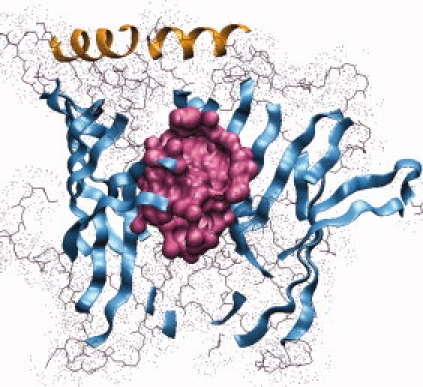
A side view, from outside the receptor, of a GABA-binding pocket (shown in magenta) at the interface of two N-terminal subunits of the GABAC receptor model.
General locations and properties of residues predicted to be under strong evolutionary selection pressure
Analysis of the ω-values listed in Table I showed that none of the residues with the strongest selection pressure (lowest calculated ω-value = 0.001) is found in the GABA-binding pocket. We mapped all residues with the lowest ω-value = 0.001 to the homology model and combined them into three groups by their locations in the model structure: (i) residues M95, M179, and D187, which are found at the protein–membrane interface; (ii) residues D64, D136, and N154, which are located at the receptor vestibule side; and (iii) D109, which is within the N-terminal α-helix region. Relatively little experimental information is available for these highly conserved residues, although, within this group, mutations at D136 and D187 positions are known to have a marked effect on GABAC function.15,17
Residues with ω = 0.002 in the present model were relatively widely distributed throughout the N-terminal domain and also found at locations such as the protein–membrane interface (E92, V93, P185, Q189, R258), the N-terminal α-helix and nearby loops (R68, P69, R111, L112), the vestibule side of the receptor (T174), the protein–protein interface (K143, H148, T151, Y200), and buried within the monomeric core (P135, G163, L250). Several studies of ρ1 GABAC mutants15,17,42 at the positions that we found to be under strong purifying selection pressure have shown that mutations of these residues in many cases substantially affect receptor function or expression. For example, the K143C mutant exhibited a low level of activity when expressed in oocytes, precluding electrophysiological investigation.17 The mutant receptor with the conservative Y200F substitution exhibited a GABA EC50 value about 10-fold greater than that of the wild-type receptor, while the mutant with the more drastic substitution Y200S was inactive.15 Furthermore, substitutions of P135C and G163C yielded nonfunctional mutants.17 Both of these residues have lowest ω-values and are conserved throughout the LGIC and AChBP families. Experimental measurements also showed that cysteine mutants of loop E residues L166, S168, and R170 significantly altered GABA binding.17 Mutational studies by Wang et al.42 identified an intrasubunit salt bridge between residues E92 and R258, which are at the protein–membrane interface. These authors proposed that this interaction contributes to the gating pathway and stabilizes the receptor in the resting state. Three additional residues (V93, P185, and Q189) located at the putative protein–membrane interface are under strong purifying selection pressure and may play important roles in channel gating.
Mutations of highly conserved nonpocket residues near protein–protein subunit interfaces
Mapping of the residues under strong purifying selection to the GABAC model revealed a number of gaps in the sites of the studied mutations. To the best of our knowledge, previous studies have not addressed the roles of the residues in the α-helical/loop region of the protein–protein interface of ρ1 GABAC, or at the protein–protein interface formed by the β-strands. To elucidate the functional roles of the protein sites under strong purifying selection at the protein–protein interfaces, we chose two residues to generate alanine and cysteine mutants for such study.
The first residue, R111, is located in the α-helical region of the protein, distant from both the GABA-binding pocket and the protein–membrane interface [Fig. 4(A)]. The second, T151, is located on the vestibule side of the modeled protein–protein interface, in the vicinity of the binding pocket [Fig. 4(B)]. Western blot analysis [Fig. 4(C)] showed that the R111C, T151C, and T151A mutant receptors were expressed at the oocyte membrane, although the expression level of R111C [Fig. 4(C), Lane 4] and T151C [Fig. 4(C), Lane 6] was low and multiple protein degradation products were evident. Mutant T151A [Fig. 4(C), Lane 5] showed an expression level similar to that of a wild-type receptor, while the R111A mutant [Fig. 4(C), Lane 3] produced no detectable bands. Figure 4(D–G) shows electrophysiological recordings obtained from Xenopus oocytes expressing these mutant receptors and compares these results with responses of wild-type receptors to both GABA and muscimol. As shown by these data, there was no significant responsiveness to GABAC agonist GABA, or to the GABAC agonist muscimol, by the R111C, R111A, and T151C mutants. The T151A mutant receptor showed responsiveness to GABA and muscimol, with EC50 = 0.8 μM for GABA and EC50 = 0.6 μM for muscimol, which are in the same range as those obtained with wild-type receptor.
Figure 4.
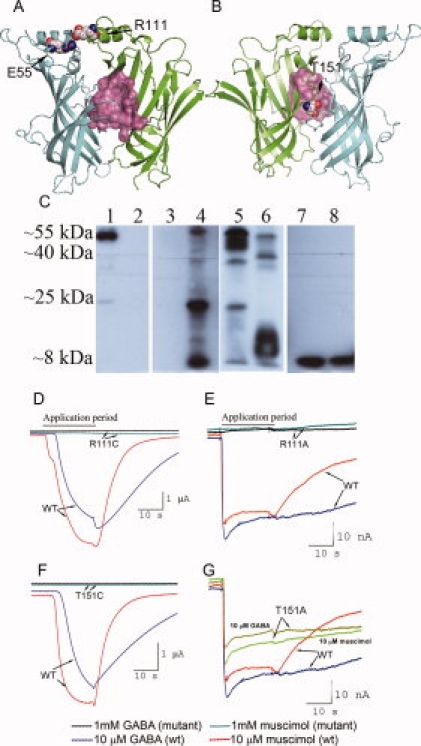
Novel mutants of GABAC receptor at protein–protein interfaces. A: Salt bridge E55–R111 from neighboring subunits in side view. This salt bridge may provide important lateral connections between the N-terminal α-helices and the first 310 helices from the adjacent subunits, and stabilize intersubunit interactions. Atoms forming the GABA-binding pocket are shown in pink. B: Proposed intersubunit location of T151, as viewed from the vestibule side of the ρ1 GABAC pentamer. T151 is buried in the protein–protein interface in our model and is likely located in the second layer of residues immediately behind the binding pocket. Atoms forming a GABA-binding pocket are shown in pink. C: Western blot of oocyte membrane protein preparations assayed with the anti GABAC ρ1 antibody. The native ρ1 polypeptide shows a band at ∼55 kDa, while all other bands are the products of protein degradation. Membrane preparations were obtained from oocytes expressing the following ρ1 GABAC receptors: Lane 1: wild type; Lane 2: nonexpressing control oocytes; Lane 3: R111A mutant; Lane 4: R111C mutant; Lane 5: T151A mutant; Lane 6: T151C mutant; Lane 7: E55A mutant; Lane 8: E55C mutant. D–G: Membrane currents recorded from oocytes expressing wild-type ρ1 GABAC (wt) receptors and the following mutant receptors R111C (Panel D), R111A (E), T151C (F), and T151A (G). Response currents to GABA and muscimol were recorded for the T151A mutant only, while no currents were recorded for the R111A, R111C, and T151C mutants. These results show that mutations of polar residues under strong evolutionary selection pressure at the protein–protein interfaces affect the ability of the GABAC receptors to bind agonists and induce recordable currents.
For the protein–protein interface in the α-helical/loop region, our model predicts that an intersubunit salt bridge may form between R111 and E55 in the ρ1 GABAC receptor [Fig. 4(A)]. E55 is located at the beginning of the N-terminal α-helix, close to the protein–protein interface, and has a medium conservation with ω-value = 0.035. We obtained alanine and cysteine mutants at this position. Similar to R111A mutant [Fig. 4(C), Lane 3], the mutant receptors E55A [Fig. 4(C), Lane 7] and E55C [Fig. 4(C), Lane 8] were not expressed as full-length receptors. Only a low molecular weight degradation product was detected by Western Blot for both mutants.
Predicted intersubunit salt bridge R158–D204
The evolutionary analysis (Fig. 2) indicates that residues R158 and D204 experience strong purifying selection pressure. The AChBP structure reveals an intersubunit salt bridge between R104 and E14731 that align with R158 and D204 in ρ1 GABAC, respectively. The role of this intersubunit salt bridge has not been previously studied in either L. stagnalis AChBP, or in ρ1 GABAC. This prompted us to investigate the effects of mutations at R158 and D204 sites [Fig. 5(A)]. To this end, we prepared and tested a total of five mutants at R158 and D204 positions of the ρ1 subunit, namely R158A, R158D, D204A, D204R, and the double mutant R158D–D204R. The mutant receptors were tested with GABA and muscimol at concentrations of up to 1 mM, that is, up to several orders of magnitude above the EC50 for wild-type GABAC receptors for these two ligands.
Figure 5.
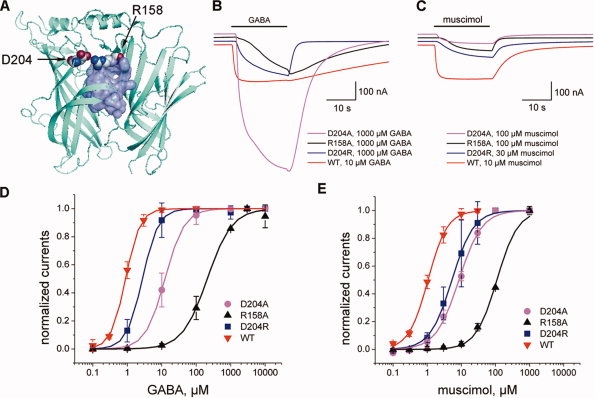
Modeling of the predicted R158–D204 intersubunit salt bridge and electrophysiological properties of the R158 and D204 mutants. A: Modeled intersubunit salt bridge R158–D204 where the atoms forming a binding pocket are shown in lilac. In this model, several side-chain atoms of R158 are located in the binding pocket. B,C: Representative membrane current responses to GABA (B) and muscimol (C) for mutants D204A, R158A, and D204R and for the native ρ1 receptor. D204 mutant induces peak currents that are approximately five-fold higher than those for the native receptor in response to GABA, but not in response to muscimol. D,E: Dose-response functions for GABA (Panel D) and muscimol (E) obtained with the native ρ1 GABAC and mutant receptors. Peak amplitudes of membrane current responses to GABA and muscimol for each mutant receptor are normalized to the saturating current response obtained at the highest concentration of GABA and muscimol (mean ± SD for four to eight determinations in two to three experiments). Smooth curves show fitting of the Hill equation. The single mutants R158A, D204R, and D204A exhibited diminished activities with higher EC50 values and altered kinetic profiles with fast recovery kinetics suggesting that R158–D204 salt bridge perhaps stabilizes the open state of the GABAC receptor.
The results of electrophysiological measurements are presented in Table II and in Figure 5(B–E). Here, the wild-type GABAC receptor shows responses to muscimol and GABA that are similar in peak amplitudes. The measured wild-type receptor EC50 values are 1.0 and 0.89 μM with Hill coefficients of 1.53 and 2.04, and with mean recovery time constant τ values of ∼6 and 38 s for muscimol and GABA, respectively. No agonist-induced currents were detected from oocytes injected with cRNA coding for the single mutant R158D or for the double mutant R158D–D204R. Mutant R158A exhibited an increase of ∼100- and ∼200-fold in EC50 values for muscimol and GABA, respectively. The D204A mutant exhibited 8- and 14-fold increase in EC50 values for muscimol and GABA. Although the amplitude of GABA-elicited responses from D204A mutant was larger than those observed for the wild-type receptor, the currents induced by muscimol in D204A mutant were smaller by an order of magnitude than those induced by GABA (51–28 nA vs. 1003–520 nA, respectively). Additionally, while the substitution D204R did not significantly affect EC50 values for either of the tested agonists, the recovery time constant τ was significantly altered in all functional mutants (R158A, D204A, D204R) to much smaller values in the range of ∼1–2 s for muscimol and ∼1–7 s for GABA responses. The largest change was observed for the GABA response, where τ values decreased from ∼38 s for the wild-type receptor to ∼1–7 s for all active R158 and D204 mutants (Table II).
Table II.
Functional Properties of the ρ1 GABAC Receptor Mutants at Positions R158 and D204
| Peak currents (nA) |
EC50 (μM) |
Hill coefficient, n |
τ (s) |
|||||
|---|---|---|---|---|---|---|---|---|
| Receptor | GABA | Muscimol | GABA | Muscimol | GABA | Muscimol | GABA | Muscimol |
| WT ρ1 | 237–169 | 298–161 | 0.89 ± 0.01 | 1.05 ± 0.02 | 2.04 ± 0.06 | 1.53 ± 0.05 | 37.80 ± 10.06 | 5.97 ± 2.24 |
| R158D | No response | No response | N/A | N/A | N/A | N/A | N/A | N/A |
| R158A | 333–157 | 348–61 | 208.9 ± 19.66 | 110.2 ± 6.65 | 1.20 ± 0.10 | 1.41 ± 0.12 | 1.04 ± 0.36 | 1.17 ± 0.44 |
| D204R | 484–201 | 266–127 | 2.75 ± 0.13 | 5.3 ± 0.19 | 1.87 ± 0.08 | 1.36 ± 0.05 | 7.25 ± 1.83 | 1.81 ± 0.29 |
| D204A | 1003–530 | 51–28 | 12.4 ± 0.28 | 8.5 ± 0.52 | 1.53 ± 0.08 | 1.29 ± 0.09 | 1.89 ± 0.22 | 2.34 ± 0.54 |
| R158D– D204R | No response | No response | N/A | N/A | N/A | N/A | N/A | N/A |
The above data are the averages obtained from five or more different oocytes from two or more different batches. Dose-response data (mean ± SD for four to eight determinations in two to three experiments) for the wild-type (WT) human ρ1 GABAC receptor as well as the mutant receptors are obtained by fitting of the Hill equation. Peak amplitudes of responses to GABA and muscimol for each mutant receptor are normalized to the saturating response obtained at the highest concentration of the respective agonist. τ values obtained by fitting a simple exponential to the recovery phase of the membrane current responses to the agonist concentrations in the range EC30–E70.
Figure 6 shows Western blot data obtained from membrane protein preparations of nonexpressing oocytes, oocytes expressing the wild-type GABAC ρ1 receptor, and oocytes expressing the R158A, R158D, D204A, D204R, and R158D–D204R mutant receptors. A polyclonal antihuman ρ1 GABAC antibody directed against a 14-mer segment of the extracellular domain (GABAC Ab N-14) was used for detecting expressed protein.43 A 55-kDa band is evident in the samples expressing wild-type ρ1 subunit, and in those expressing mutant receptors except for the R158D mutant. In the R158D mutant, an intense band corresponding with a degradation product of ∼25 kDa was observed. Additionally, a faint degradation product band of ∼40 kDa was observed for all mutants except R158D. Notably, the oocyte membrane preparation of the double mutant R158D–D204R contained only 55 kDa and ∼40 kDa products and no 25-kDa product, implying that receptor assembly was rescued by the additional mutation at Position 204 (Fig. 6, Lane 7).
Figure 6.
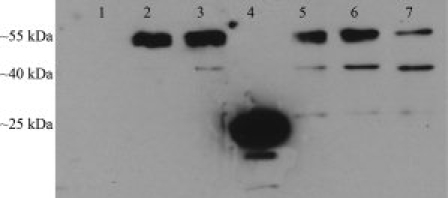
Western blot of oocyte membrane protein preparations assayed with anti-GABAC ρ1 antibody. Membrane preparations were obtained from oocytes expressing the following ρ1 GABAC receptors: Lane 1: nonexpressing control oocytes. Lane 2: wild-type ρ1 GABAC. Lane 3: R158A mutant. Lane 4: R158D mutant. Lane 5: D204A mutant. Lane 6: D204R mutant. Lane 7: R158D–D204R double mutant. Mutations at D204 result in the receptors with molecular weights similar to the native ρ1 subunits, while R158D mutation yields partially degraded receptors (band at ∼24 kDa, Lane 4), possibly as a result of misfolding. Double mutant R158D–D204R (Lane 7) contains only ∼55 kDa and ∼40 kDa products, implying that receptor subunit folding is rescued by the additional mutation at Position 204.
To probe surface expression of the wild-type and mutant ρ1 subunit, we performed immunolabeling using the GABAC Ab N-14 antibody.43 Figure 7 shows the fluorescence images of oocytes obtained following labeling of oocytes expressing the wild-type GABAC ρ1 receptor (top row), the R158D mutant (middle row), and the R158D–D204R double mutant. When GABAC Ab N-14 was applied extracellularly to the intact oocytes, fluorescence was observed at the surface membrane of the oocyte expressing the wild-type receptor and the double mutant receptors, but not at the surface of the oocyte expressing the R158D mutant. These results, together with those from the Western blot (Fig. 6), strongly suggest that the R158D mutation disrupts receptor assembly and thus prevents receptor expression at the surface membrane. Importantly, the double mutation R158D–D204R is able to restore receptor assembly and surface membrane expression, but not receptor function.
Figure 7.
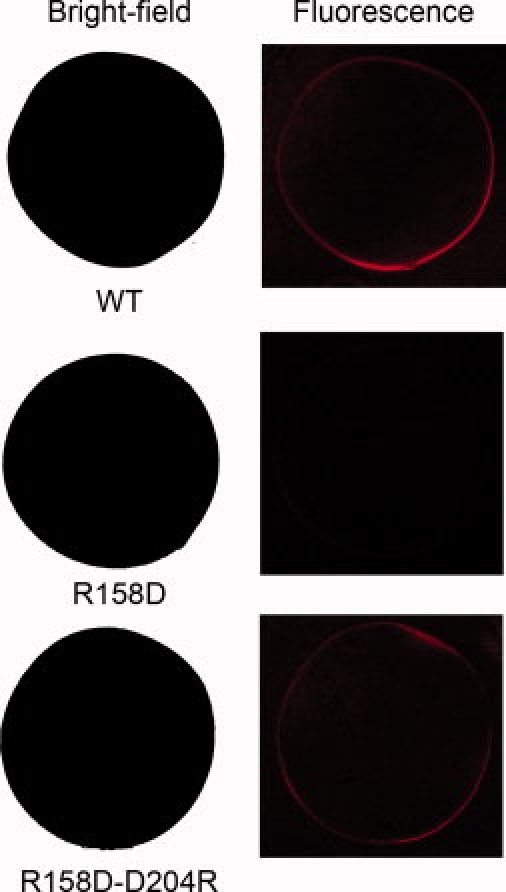
Bright-field and fluorescence images obtained with oocytes labeled with anti-GABAC ρ1 antibody. Top row: Oocyte expressing the wild-type GABAC ρ1 receptor. Middle row: Oocyte expressing the R158D mutant ρ1 GABAC. Bottom row: Oocyte expressing the double mutant R158D–D204R ρ1 GABAC. These results show that the partially degraded R158D mutant did not express on the surface of the oocyte, while the surface expression was rescued by the additional mutation at Position 204.
Discussion
In this study, we have combined computational modeling and experimental measurements to examine the subunit interactions in the homomeric ρ1 GABAC receptor. We have used posterior probability analysis of evolutionary selection pressure measured as ω-ratio to identify residues under strong purifying selection pressure in ρ1 receptors. Based on additional information from the structural model, we have obtained and tested several mutants of highly conserved polar residues located at or near the protein–protein interfaces, which can potentially form intersubunit salt bridges or hydrogen bonds and have shown that they play important roles in the folding, assembly, and function of GABAC receptors.
Probability analysis of evolutionary selection pressure
To identify residues in the GABAC receptor experiencing purifying selection pressure, only orthologous sequences of the same type of subunit (e.g., ρ1) from different species should be analyzed. A challenge is that frequently the number of orthologous sequences is limited. In addition, the protein sequences may not have diverged sufficiently to be informative enough for detailed analysis, as was found for ρ1 subunits.44 In this case, the pairwise amino acid sequence identity reaches a mean value of ∼88%, making it difficult to distinguish residues that are experiencing purifying selection pressure from residues that have not had sufficient time to diverge. By contrast, ω-ratio analysis based on DNA sequences that are different but may encode the same or very similar protein sequences can detect subtle evolutionary signals. In the present work, we examined the evolution of corresponding coding cDNA sequences at the codon level. We used an explicit codon evolutionary model based on a continuous-time Markov process,45 which yields deeper insights about the mechanisms of molecular evolution. This approach distinguishes mutations fixed by evolution from mutations fixed by chance. It takes into account bias in codon frequency, bias-favoring transition over transversion, as well as explicit phylogenetic information.45 This approach has the advantage over other approaches such as entropy-based calculations, as it is more accurate in accounting for bias due to different evolutionary distances between species. Although only a limited number of sequences were available for this study, our probabilistic approach allowed identification of residue positions that had the highest probabilities of being under strong purifying selection pressure. Remarkably, experimental data obtained from the literature demonstrated that mutations at positions with the lowest ω-values usually had significant effects on the receptor function or expression.15–17,39,40,42
Additionally, we have assessed the advantage of using evolutionary information in improving the quality of the homology model of GABAC receptor. We have compared our model with the model obtained using standard programs with default settings and default search results (as described in “Methods”). We found that in the model generated by default program settings, the α-helix is shifted ∼20 residues upstream, and begins at K34 and ends at S49. By contrast, the α-helix in our model begins near S54 and ends near M67. The predicted polar interactions between side-chains of E55 and R111, between R158 and D204, as well as the salt bridge E92–R258 experimentally confirmed by Wang et al.42 are absent in the model obtained using default parameters.
Polar residues at subunit interfaces are critical for GABAC receptor assembly and function
Combined study of evolutionary analysis and structural modeling suggested that several highly conserved polar amino acid residues play important roles in the assembly and/or function of the GABAC receptor. Guided by computational predictions, we tested the effects of mutation at Positions R111, T151, R158, and D204, which were hypothesized to be involved in protein–protein interactions, with their side chains contributing to polar interactions at the subunit interfaces.
Residue R111
The α-helical region (α-helix with the adjacent loops) has not been studied in GABAC receptors, and its functional role is unknown. Using alanine and cysteine mutations, we investigated the functional role of R111 (ω-value = 0.002), which belongs to the 310 helix η1 between beta strands β2 and β3 (Fig. 2) and is spatially close to the N-terminal α-helix [Fig. 4(A)]. We found that both mutations had a strong effect on the membrane surface expression of the mutant receptor, where R111C exhibited a diminished surface expression with multiple protein degradation products, while R111A was not expressed on the membrane surface as shown by Western blot [Fig. 4(C)]. The weak band corresponding to the native ρ1 receptor in R111C mutant [Fig. 4(C), Lane 4] may be due to a weak hydrogen bond formed by cysteine with another residue from the neighboring subunit. Neither the R111C nor the R111A mutant showed a significant membrane current response to GABA or muscimol [Fig. 4(D,E)]. Based on our model, we hypothesize that R111 plays an important role in receptor oligomerization and in maintaining its stability by forming an intersubunit salt bridge with the residue from the N-terminal α-helix of the adjacent subunit. Support for this hypothesis comes from the results obtained with the E55 mutants. Our structural model suggests residue E55 at the beginning of the N-terminal α-helix as the mostly likely candidate for interaction with R111 [Fig. 4(A)]. We tested this possibility by constructing alanine and cysteine mutants of E55. Both mutations had strong impact on folding and/or assembly of the receptor, as only degradation products were recovered for both of these E55 mutants [Fig. 4(C), Lanes 7 and 8].
Residue T151
We also investigated the role of T151 in GABAC ρ1 receptors. Position T151 has a low ω-value (0.002) and is predicted to be located at the protein–protein interface. T151 is not highly conserved in LGICs, although other GABA-binding receptor subunits, for example, ρ2, β2, α1, and γ2, as well as α1 and β subunits of the glycine receptor, contain threonine at the same position (Fig. 2). Previously, mutation T125C in the GABAA γ2 subunit, which is homologous to T151, was shown to significantly affect surface expression of the mutant receptor (∼40% of the wild-type level).46 Additionally, reduced coprecipitation of γ2T125C with α1A108C mutant subunits was observed, and it was concluded that this position is important for assembly of the GABAA receptor.46 In the case of the GABAC receptor, T151C mutation also led to reduced surface expression, with the majority of the protein degraded [Fig. 4(C), Lane 6]. However, the T151A mutant exhibited a normal level of surface expression and an activity comparable to wild-type, although the induced currents were smaller than those of wild-type receptors [Fig. 4(G)]. These data suggest that T151C substitution may result in nonspecific intra- or intersubunit interactions that promote protein degradation, while T151A substitution permits assembly of a receptor with a near-wild-type activity. Alternatively, the observed effect of T151C substitution may derive from a reduced ability of the cysteine side-chain to pack as efficiently as the threonine side-chain against the neighboring protein subunit (due to differences in size and shape of the cysteine vs. the threonine residue), impairing receptor oligomerization.
Salt bridge R158–D204
The present model of the GABAC receptor also indicates the possible occurrence of an intersubunit salt bridge between highly conserved R158 and moderately conserved D204. Prior studies have shown that R158, a position located in the GABA-binding pocket, is crucial for GABAC function. Sedelnikova et al.17 found that R158C mutant has low expression, while Harrison and Lummis39 demonstrated that alanine, glutamic acid, or lysine substitutions at R158 result in fully assembled but nonfunctional GABAC receptors. Studying the mutant receptor by Western blot assays, immunofluorescence, and by electrophysiological recordings enabled us to distinguish the effects of mutations on receptor folding versus effects on receptor function. Western blot results indicated that introduction of the negative charge at R158 position, for example, R158D substitution, leads to degradation of the protein, while the R158A mutant is assembled and expressed at a level similar to that of the wild-type receptor (Fig. 6). Furthermore, immunofluorescence data showed that mutant R158D is not expressed at the oocyte surface (Fig. 7). The present model predicts that in the R158D mutant, negatively charged residues E202, D203, and D204 of the subunit that forms a principal face of the GABA-binding site are close to the introduced aspartic acid at position 158 of the complementary face of the binding pocket, and this proximity to the introduced aspartic acid may strongly destabilize the native protein fold. It is thus likely that R158D mutant ρ1 subunits misfold or misassemble, and the N-terminal domain becomes susceptible to protease activity.
Preparation of the double mutant R158D–D204R, that is, a mutant designed to restore the putative native salt bridge between R158 and D204, yielded a receptor that is fully assembled, as indicated by the Western blot analysis (Fig. 6), and is properly expressed on the oocyte surface membrane, as shown by the immunofluorescent experiment (Fig. 7). However, the double mutant did not respond either to GABA or to muscimol (Table II), even at concentrations as high as 1 mM, which is ∼1000 times the EC50 of the wild-type receptor. This absence of responsiveness is perhaps not surprising, as both GABA and muscimol are zwitterionic at neutral pH, and the charged residue R158 presumably plays an important role in agonist binding and positioning.39 The findings that removing arginine in the R158A mutant yields a 200-fold increase in EC50 and that the double mutant R158D–D204R is inactive together suggest that the absence of a positively charged residue specifically at Position 158 strongly diminishes the binding affinity of the GABA and muscimol.
The electrophysiological data obtained with functional mutants R158A, D204R, and D204A, where the putative ion pair R158–D204 is disrupted, suggest that this intersubunit interaction stabilizes the open channel state. Table II shows that the recovery times τ are significantly smaller in all three active mutants than in the wild-type receptor, particularly in response to GABA. The decrease in recovery time is strongly pronounced in case of alanine substitutions at both R158 and D204 (decreases of up to 36- and 20-fold, respectively). Recent comparative structural studies of pLGICs in closed and open conformations indicate that the rotation of individual N-terminal domains occurs during channel activation.7,8 In the wild-type receptor, the salt bridge between R158 and D204 may form in response to ligand binding and persist while the channel is in the conducting state. Once the channel closes, the ion pair no longer exists. By contrast, in the mutant receptors, this ion pair is not formed, the conducting conformation is not stabilized, and, as a result, the receptor returns to a closed state within significantly shorter time. Overall, the electrophysiological, immunofluorescence, and Western blot data indicate that the positively charged R158 side chain plays a crucial role in the binding of GABA and muscimol to the GABAC receptor, while the R158–D204 salt bridge may stabilize the channel in the open state.
Methods
Determination of evolutionary selection pressure
To identify residues under strong evolutionary selection pressure in the GABAC extracellular domain, we carried out a posterior probability analysis of evolutionary selection pressure at the individual amino acid residue sites using a maximum likelihood estimation.47 The evolutionary selection pressure was calculated as the ratio of synonymous versus nonsynonymous substitutions, here termed the ω-ratio, which measures selection at each amino acid residue position. To avoid over- and under-representation of sequences from a specific part of the phylogeny, we selected and aligned 11 orthologous DNA sequences of ρ1 subunits retrieved from the National Center for Biotechnology Information (NCBI) NR database using the alignment of protein sequences as a guide. The synonymous versus nonsynonymous ratios were calculated at the DNA level using the PAML package, which is based on a continuous-time Markov model of codon substitution.45,48,49
Homology modeling
LGIC and AChBP sequences were retrieved from the NCBI (http://www.ncbi.nlm.nih.gov/) databases and from the Protein Data Bank (PDB), using the L. stagnalis model of AChBP (PDB ID: 1I9B) as template. To increase confidence in the sequence alignment, we incorporated additional results from the analysis of ω-values together with the predicted secondary structure from JPRED server (http://www.compbio.dundee.ac.uk/www-jpred). We generated 30 homology models using MODELLER 9v2.50 All five subunits of the pentamer were modeled simultaneously. The positions of Cys-loop cysteine residues C177 and C191 were constrained. A structure with the lowest MODELLER's target energy function was chosen as the final model. A multiple alignment of the L. stagnalis AChBP protein sequence with sequences of H. sapiens GABAC (ρ1, ρ2), GABAA (α1, β2, γ2), glycine receptor (α1, β), and T. marmorata nAChR (α, β, γ, δ) was obtained using CLUSTALW,51 followed by a manual adjustment with PFAAT program52 (Fig. 2). Pockets and voids in the homology model were calculated using the CASTp program (http://sts.bioengr.uic.edu/castp/), which computes geometric constructs (Voronoi diagram, Delaunay triangulation, and the alpha complex) derived from the protein structure coordinates.33–35,53
For comparison purposes, we built a homology model using default program settings. We chose the first AChBP structure returned with the BLAST searches of PDB database as a template, which was AChBP from Aplysia californica (PDB ID: 2BYN). We next aligned the A. californica sequence with the sequence of the extracellular GABAC domain in CLUSTALX54 using default parameters. The MODELLER program50 was run locally using input files generated with the obtained alignment.
Mutagenesis and receptor expression
Site-directed mutagenesis was performed on the pGEMHE plasmid containing the human ρ1 sequence (gift from Dr. David S. Weiss University of Texas at San Antonio) using Pfu polymerase (Stratagene, La Jolla, CA) and primers containing the desired mutation (Integrated DNA Technologies, Coralville, IA). PCR products were introduced into XL-1 competent cells (Stratagene), and the presence of desired mutation was confirmed by nucleotide sequencing. Expression of the mutated GABAC receptors in Xenopus laevis oocytes employed procedures described previously.55,56 Briefly, mRNA, obtained for each selected sequence from in vitro transcription (mMessage mMachine Ambion, Austin, TX) from linearized cDNAs, was injected into the oocytes (Drummond Nanoject II; Drummond Scientific, Broomall, PA). Oocytes were analyzed for GABAC receptor expression after 24–72 h incubation in physiological saline (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, and 10 mM glucose, at pH 7.2–7.4) containing 0.1 mg/mL gentamycin at 16–19°C.
Electrophysiology
Membrane current responses to the presentation of defined concentrations of GABA, and of the GABAC agonist muscimol, were recorded using a two-microelectrode voltage clamp apparatus (GeneClamp 500B amplifier; Axon Instruments, Foster City, CA) and Clampex 8.2 software (Axon Instruments). Procedures used to record the responses were similar to those described.56 The oocyte, positioned in a perfusion chamber (Bioscience Tools, San Diego, CA), was superfused with physiological saline (Ringer's solution) at a rate of ∼1 mL/min. The Ringer's solution consisted of 100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES, at pH 7.2–7.4. Test solutions used during the experiments were delivered to the oocyte chamber using ValveLink 16 (AutoMate Scientific, Berkeley, CA) from separate reservoirs under gravity flow, controlled by pinch valves operating under computer command. Signals were filtered at 10 Hz and sampled at 100 Hz by a DIGIDATA 1322A A/D board (Axon Instruments) and analyzed using Clampfit 8.2 (Axon Instruments) software, SigmaPlot Versions 8.0 and 10.0 (SYSTAT Software, Point Richmond, CA) and OriginPro Version 7.5 (OriginLab Corporation, Northampton, MA). Dose-response functions were obtained by averaging the normalized peak amplitude of the response (normalized to the saturating peak amplitude obtained with the highest concentration of agonist) and plotting these normalized amplitudes as a function of agonist concentration. The data were analyzed in relation to the Hill equation
R/Rmax = Cn/(EC50n + Cn) where C is the ligand concentration, R is the peak amplitude of the resulting response at concentration C, Rmax is the saturating peak amplitude, EC50 is the ligand concentration producing 50% saturation current, and n is the Hill coefficient.
Western blotting
The membrane protein fraction of the control nonexpressing oocytes and oocytes transfected to express the wild-type or mutant human ρ1 GABAC receptor was prepared using the method described by Wible et al.57 Fifty micrograms of total membrane proteins was loaded in each lane and separated on SDS-PAGE. Preparations were probed with a 1/10,000 dilution of GABAC Ab N-14, a polyclonal antibody targeting the human ρ1 GABAC receptor,43 followed by HRP-conjugated, goat-anti-guinea pig secondary antibody at a 1/5000 dilution (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunofluorescence
Oocytes expressing either the wild-type, the R158D mutant, or the R158D–D204R double mutant GABAC ρ1 receptor were fixed for 1 h in 4% paraformaldehyde, washed with PBS three times, and incubated in blocking buffer (10% normal goat serum + 1% BSA in PBS) for 1 h. They were incubated in the presence of 1/1000 dilution of GABAC Ab N-14 (diluted in 1% normal goat serum + 0.1% BSA in PBS) for 2 h at RT and washed three times (15 min each) in PBS. They were then incubated with Cy5-conjugated goat anti-guinea-pig IgG diluted 1/400 in 1% normal goat serum + 0.1% BSA in PBS. Following three washes (15 min each) in PBS, images of the oocytes were obtained on a Leica DM-IRE2 confocal microscope, at 10× magnification.
Summary
In this study, we have combined computational and experimental approaches to gain insight into the determinants of the mechanism of GABAC ρ1 receptor assembly and ion channel gating. We have identified and experimentally studied a set of polar residues under strong selection pressure predicted to be at the protein–protein interfaces of the ρ1 GABAC pentamers. We show that these residues play critical roles in the receptor assembly, as the majority of the alanine and cysteine mutants at the modeled protein–protein interface positions do not express at the cell membrane, while those that do express either show diminished functional activity or are inactive. The results also suggest a functional role for the intersubunit salt bridge R158–D204, perhaps based on stabilization of the conducting state of the receptor.
Acknowledgments
We thank Dr. D.S. Weiss and Dr. A. Sedelnikova for providing the pGEMHE-hρ1 construct, and Mr. Zheng Ouyang and Mr. Yun Xu for technical assistance.
References
- 1.Johnston GAR. GABAc receptors: relatively simple transmitter-gated ion channels? Trends Pharmacol Sci. 1996;17:319–323. [PubMed] [Google Scholar]
- 2.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Dellisanti CD, Yao Y, Stroud JC, Wang Z-Z, Chen L. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94 Å resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 4.Celie PHN, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 5.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 8.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux J-P, Delarue M, Corringer P-J. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 9.Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, Lodder H, van der Schors RC, van Elk R, Sorgedrager B, Brejc K, Sixma TK, Geraerts WPM. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 10.Speranskiy K, Cascio M, Kurnikova M. Homology modeling and molecular dynamics simulations of the glycine receptor ligand binding domain. Proteins. 2007;67:950–960. doi: 10.1002/prot.21251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves DC, Lummis SCR. The molecular basis of the structure and function of the 5-HT3 receptor: a model ligand-gated ion channel. Mol Membr Biol. 2002;19:11–26. doi: 10.1080/09687680110110048. [DOI] [PubMed] [Google Scholar]
- 12.Henchman RH, Wang H-L, Sine SM, Taylor P, McCammon JA. Asymmetric structural motions of the homomeric α7 nicotinic receptor ligand binding domain revealed by molecular dynamics simulation. Biophys J. 2003;85:3007–3018. doi: 10.1016/S0006-3495(03)74720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cromer BA, Morton CJ, Parker MW. Anxiety over GABAA receptor structure relieved by AChBP. Trends Biochem Sci. 2002;27:280–287. doi: 10.1016/s0968-0004(02)02092-3. [DOI] [PubMed] [Google Scholar]
- 14.Chebib M. GABAC receptor ion channels. Clin Exp Pharmacol Physiol. 2004;31:800–804. doi: 10.1111/j.1440-1681.2004.04083.x. [DOI] [PubMed] [Google Scholar]
- 15.Amin J, Weiss DS. Homomeric ρ1 GABA channels: activation properties and domains. Receptors Channels. 1994;2:227–236. [PubMed] [Google Scholar]
- 16.Torres VI, Weiss DS. Identification of a tyrosine in the agonist binding site of the homomeric ρ1 γ-aminobutyric acid (GABA) receptor that, when mutated, produces spontaneous opening. J Biol Chem. 2002;277:43741–43748. doi: 10.1074/jbc.M202007200. [DOI] [PubMed] [Google Scholar]
- 17.Sedelnikova A, Smith CD, Zakharkin SO, Davis D, Weiss DS, Chang Y. Mapping the ρ1 GABAC receptor agonist binding pocket: constructing a complete model. J Biol Chem. 2005;280:1535–1542. doi: 10.1074/jbc.M409908200. [DOI] [PubMed] [Google Scholar]
- 18.Harrison NJ, Lummis SCR. Molecular modeling of the GABAC receptor ligand-binding domain. J Mol Model. 2006;12:317–324. doi: 10.1007/s00894-005-0034-6. [DOI] [PubMed] [Google Scholar]
- 19.Carland JE, Moorhouse AJ, Barry PH, Johnston GAR, Chebib M. Charged residues at the 2′ position of human GABAC ρ1 receptors invert ion selectivity and influence open state probability. J Biol Chem. 2004;279:54153–54160. doi: 10.1074/jbc.M410625200. [DOI] [PubMed] [Google Scholar]
- 20.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Halim H, Hanrahan JR, Hibbs DE, Johnston GAR, Chebib M. A molecular basis for agonist and antagonist actions at GABAC receptors. Chem Biol Drug Des. 2008;71:306–327. doi: 10.1111/j.1747-0285.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- 22.Osolodkin DI, Chupakhin VI, Palyulin VA, Zefirov NS. Molecular modeling of ligand–receptor interactions in GABAC receptor. J Mol Graph Model. 2009;27:813–821. doi: 10.1016/j.jmgm.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z. Handbook of statistical genetics. New York: Wiley; 2001. [Google Scholar]
- 25.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 26.Tseng YY, Liang J. Are residues in a protein folding nucleus evolutionarily conserved? J Mol Biol. 2004;335:869–880. doi: 10.1016/j.jmb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Lecompte O, Thompson JD, Plewniak F, Thierry JC, Poch O. Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene. 2001;270:17–30. doi: 10.1016/s0378-1119(01)00461-9. [DOI] [PubMed] [Google Scholar]
- 28.Corringer PJ, Le Novère N, Changeux JP. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 29.Karlin A, Akabas MH. Toward a structural basis for the function of nicotinic acetylcholine receptors and their cousins. Neuron. 1995;15:1231–1244. doi: 10.1016/0896-6273(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 30.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celie PHN, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, Smit AB, Sixma TK. Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem. 2005;280:26457–26466. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- 32.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelsbrunner H. The union of balls and its dual shape. Discrete Comput Geom. 1995;13:415–440. [Google Scholar]
- 34.Edelsbrunner H, Facello M, Liang J. On the definition and the construction of pockets in macromolecules. Discrete Appl Math. 1998;88:83–102. [PubMed] [Google Scholar]
- 35.Edelsbrunner H, Mucke EP. Three-dimensional alpha shapes. ACM Trans Graph. 1994;13:43–72. [Google Scholar]
- 36.Edelsbrunner H, Shah NR. Incremental topological flipping works for regular triangulations. Algorithmica. 1996;15:223–241. [Google Scholar]
- 37.Zhang J, Xue F, Chang Y. Structural determinants for antagonist pharmacology that distinguish the ρ1 GABAC receptor from GABAA receptors. Mol Pharmacol. 2008;74:941–951. doi: 10.1124/mol.108.048710. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Xue F, Chang Y. Agonist- and antagonist-induced conformational changes of loop F and their contributions to the ρ1 GABA receptor function. J Physiol. 2009;587:139–153. doi: 10.1113/jphysiol.2008.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison NJ, Lummis SCR. Locating the carboxylate group of GABA in the homomeric rho GABAA receptor ligand-binding pocket. J Biol Chem. 2006;281:24455–24461. doi: 10.1074/jbc.M601775200. [DOI] [PubMed] [Google Scholar]
- 40.Kusama T, Wang JB, Spivak CE, Uhl GR. Mutagenesis of the GABA ρ1 receptor alters agonist affinity and channel gating. NeuroReport. 1994;5:1209–1212. doi: 10.1097/00001756-199406020-00012. [DOI] [PubMed] [Google Scholar]
- 41.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Lester HA, Dougherty DA. Establishing an ion pair interaction in the homomeric ρ1 γ-aminobutyric acid type A receptor that contributes to the gating pathway. J Biol Chem. 2007;282:26210–26216. doi: 10.1074/jbc.M702314200. [DOI] [PubMed] [Google Scholar]
- 43.Gussin HA, Khasawneh FT, Xie A, Qian H, Le Breton GC, Pepperberg DR. Characterization of a novel polyclonal anti human ρ1 GABAC antibody. Invest Ophthalmol Vis Sci. 2008;49 e-abstract 1288. [Google Scholar]
- 44.Zhang D, Pan Z-H, Awobuluyi M, Lipton SA. Structure and function of GABAC receptors: a comparison of native versus recombinant receptors. Trends Pharmacol Sci. 2001;22:121–132. doi: 10.1016/s0165-6147(00)01625-4. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol. 1993;10:1396–1401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- 46.Sarto-Jackson I, Ramerstorfer J, Ernst M, Sieghart W. Identification of amino acid residues important for assembly of GABAA receptor α1 and γ2 subunits. J Neurochem. 2006;96:983–995. doi: 10.1111/j.1471-4159.2005.03626.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 49.Tseng YY, Liang J. Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: a Bayesian Monte Carlo approach. Mol Biol Evol. 2006;23:421–436. doi: 10.1093/molbev/msj048. [DOI] [PubMed] [Google Scholar]
- 50.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caffrey DR, Dana PH, Mathur V, Ocano M, Hong E-J, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinformatics. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules II. Inaccessible cavities in proteins. Proteins. 1998;33:18–29. [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcqettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Vu TQ, Chowdhury S, Muni NJ, Qian H, Standaert RF, Pepperberg DR. Activation of membrane receptors by a neurotransmitter conjugate designed for surface attachment. Biomaterials. 2005;26:1895–1903. doi: 10.1016/j.biomaterials.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Qian H, Hyatt G, Schanzer A, Hazra R, Hackam AS, Cutting GR, Dowling JE. A comparison of GABAC and ρ subunit receptors from the white perch retina. Vis Neurosci. 1997;14:843–851. doi: 10.1017/s0952523800011585. [DOI] [PubMed] [Google Scholar]
- 57.Wible BA, Yang Q, Kuryshev YA, Accili EA, Brown AM. Cloning and expression of a novel K+ channel regulatory protein, KChAP. J Biol Chem. 1998;273:11745–11751. doi: 10.1074/jbc.273.19.11745. [DOI] [PubMed] [Google Scholar]


