Abstract
Actin is one of the most conserved proteins in nature. Its assembly and disassembly are regulated by many proteins, including the family of actin-depolymerizing factor homology (ADF-H) domains. ADF-H domains can be divided into five classes: ADF/cofilin, glia maturation factor (GMF), coactosin, twinfilin, and Abp1/drebrin. The best-characterized class is ADF/cofilin. The other four classes have drawn much less attention and very few structures have been reported. This study presents the solution NMR structure of the ADF-H domain of human HIP-55-drebrin-like protein, the first published structure of a drebrin-like domain (mammalian), and the first published structure of GMF β (mouse). We also determined the structures of mouse GMF γ, the mouse coactosin-like domain and the C-terminal ADF-H domain of mouse twinfilin 1. Although the overall fold of the five domains is similar, some significant differences provide valuable insights into filamentous actin (F-actin) and globular actin (G-actin) binding, including the identification of binding residues on the long central helix. This long helix is stabilized by three or four residues. Notably, the F-actin binding sites of mouse GMF β and GMF γ contain two additional β-strands not seen in other ADF-H structures. The G-actin binding site of the ADF-H domain of human HIP-55-drebrin-like protein is absent and distorted in mouse GMF β and GMF γ.
Keywords: actin-depolymerizing factor homology domain, G-actin binding, F-actin binding, structure stabilization
Introduction
Actin is one of the most conserved proteins in nature, differing by no more than 5% in species as diverse as algae and humans.1 The individual subunits of actin are known as globular actin (G-actin). G-actin spontaneously assembles to form a filamentous helical polymer, called F-actin. F-actin provides mechanical support for the cell, determines the cell shape, forms the cytoskeleton, and enables cell movement, cytokinesis, morphogenesis, endocytosis, and cell division.1 The relative concentrations of F-actin and G-actin are regulated by a variety of intricate signaling pathways involving many proteins.2,3 These include monomer sequestering proteins, end-capping proteins of actin filaments, actin filament crosslinking proteins, filament-severing and nucleating proteins, and numerous small actin-binding proteins. Despite the diverse biochemical activities of these domains, there are relatively few actin-binding motifs. One of these motifs is presented by the 15–20 kDa (∼150 amino acids) actin-depolymerization factor homology (ADF-H) domain.4
ADF-H domains are widely distributed: they are found in yeast, bacteria, and plants, and all eukaryotic cells appear to have at least one.5 ADF and cofilin are the founding members of what is now called the family of ADF-H domains. They were initially identified and named according to their ability to either depolymerize F-actin (ADF) or form cofilamentous structures with actin (cofilin). They are much alike (e.g., actin dynamics)6 and lack clearly distinctive biochemical properties. Thus, they are often considered as a single entity: ADF/cofilin.1 However, they are the products of different genes: Most vertebrates have one gene encoding ADF and two genes encoding cofilin (muscle and non-muscle cofilins). Many lower eukaryotes, such as D. melanogaster and S. cerevisiae, have only one gene. The genes commonly have introns and generate several different protein products; those without introns are likely pseudogenes.5 The eukaryotic ADF/cofilins share about 40% sequence identity.2 Mutations that inactivate cofilin/ADF are lethal in C. elegans, D. melanogaster, and S. cerevisiae.
With the advent of cloning and cDNA sequencing there have been many additions to the family of ADF-H domains based on sequence homology (Figs. 1 and 2). Phylogenetically (Fig. 3),7 the family members can be divided into five functionally distinct classes: I-cofilin/ADF, II-glia maturation factor (GMF), III-coactosin, IV-twinfilin, and V-Abp1/drebrin. They have diverse sequences (Figs. 1 and 2) and share about 20% sequence identity at the amino acid level.1,2 Very little is known about the ADF-H domains other than ADF/cofilin. They differ in their ability to bind G- and F-actin (Table I). All five classes can be found in species as diverse as H. sapiens, M. musculus, D. melanogaster, and D. discoideum. Two types of GMFs (Class II) have been identified in mouse and humans: GMF β was identified as a nerve growth factor, implicated in nervous system development,8 whereas GMF γ was initially identified as a molecule with high similarity to GMF β,9 although it is not expressed in brain, neuronal, or glia cells, but in microvascular endothelial and inflammatory cells.10 In Drosophila, twinfilin (Class IV) is required for actin-dependent developmental processes and may be involved in the localization of actin monomers in cells.11 Drebrins (Class V) contain an N-terminal ADF-H domain and a dynamin-binding C-terminal SH3 domain.12 The human HIP-55-drebrin-like protein is important for the function of the immune system and it is regulated by T cell antigen receptor (TCR) signaling.13 Few structures of ADF-H domains from the classes other than Class I (ADF/cofilin) have been reported (Table I).14–18 We now report the solution NMR structures of five domains from these four underrepresented classes: mouse GMF β and γ (Class II), mouse coactosin-like (Class III), the C-terminal domain (residues 161-313) of mouse twinfilin 1 (Class IV), and the ADF-H domain of human HIP-55 drebrin-like (Class V). The latter domain is the first published structure of a mammalian drebrin-like domain (the structure of the Abp1 ADF-H domain of S. cerevisiae was reported previously).18 The goal is to deduce the differences in F- and G-actin binding from the structures and to identify the common characteristics of each ADF-H class.
Figure 1.
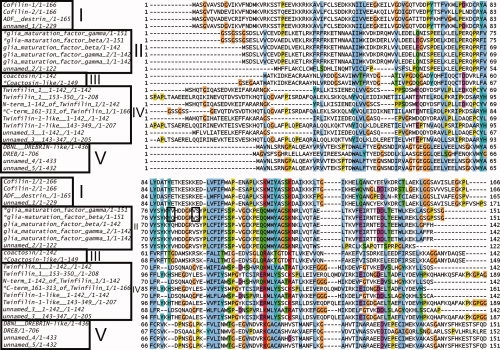
Sequence alignments of ADF-H domains from mouse. *: Proteins analyzed in this study. Sources: see Materials and Methods. The alignment was generated with the use of the CLUSTAL W2 program31 using ClustalX colors. The labels I, II, III, IV, and V refer to the classification of the proteins according to Figure 3. The residues in the box form β-strands.
Figure 2.
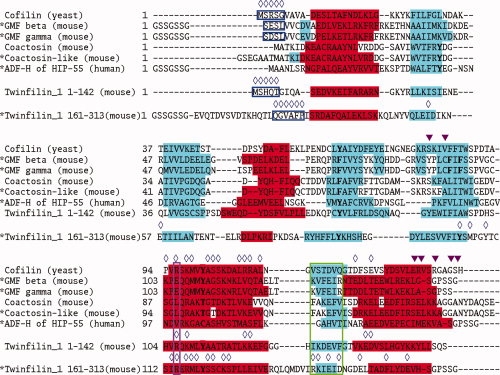
Sequence alignments of selected ADF-H domains across species. *: Proteins analyzed in this study. Sources: see Materials and Methods. Red indicates helices and cyan represents β-strands. Residues involved in structure stabilization are highlighted in bold type. Important residues for F- and G- actin binding are marked with blue open diamonds (⋄) and magenta closed triangles (▾), respectively. The data are derived from site-directed mutagenesis experiments for yeast cofilin (PDB, id. 1CFY)17 and the N-terminal domain of mouse twinfilin 1,14 and from the crystal structure of the C-terminal ADF-H domain of twinfilin 1 (176-316) bound to ATP-G-actin.16 The regions encircled in blue, purple, and green are relevant to G-actin binding.
Figure 3.
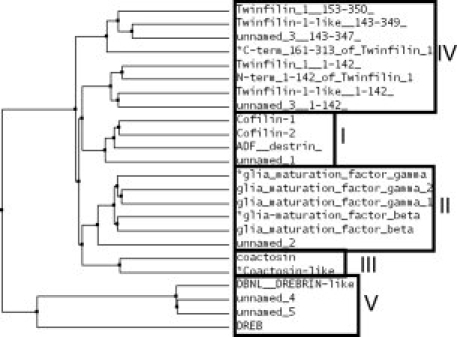
Phylogenetic tree of mouse ADF-H proteins reveals five classes. *: Proteins analyzed in this study. The figure is based upon data from Figure 1. The tree was generated with the use of CLUSTAL W2 and Jalview 2.4 (average distance tree using BLOSOM62).31
Table I.
Structural Statistics of the 5 ADF-H Domain Structures
| Glia maturation factor (GMF) |
|||||
|---|---|---|---|---|---|
| Name | β | γ | Coactosin-like | Twinfilin 1 (161–313) | ADF-H of HIP-55 (drebrin-like) |
| Species | Mouse |
Human |
|||
| ADF-H class | II-GMF | III-coactosin | IV-twinfilin | V-Abp1/drebrin-like | |
| General characteristics of class: | |||||
| (Number of ADF-H in protein) | 1 | 1 | 2 | 1 | |
| (Number of domains in protein) | 1 | 1 | 2 | Multidomain | |
| Expected to bind F-actin | Yes | Yes | Nob | Yes | |
| Expected to bind G-actin | ? | No | Yesb | No | |
| Members in PDBa | 3 (2) | 8 (1) | 4 (1) | 1 (1) | |
| Input data | |||||
| Dihedral angle constraints (φ,ψ) | 152 | 134 | 204 | 173 | 141 |
| Total number of NOEs | |||||
| From 15N-NOESY-HSQC | 1779 | 1984 | 2089 | 1777 | 1759 |
| From 3C-NOESY-HSQC | 5289 | 4869 | 4413 | 5116 | 4223 |
| Cyana-version used | 2.0.17 | 2.0.26 | 2.0.13 | 2.2.1 | 2 |
| Output data | |||||
| CYANA target function value (Å2) | 0.29 | 0.24 | 1.37 | 2.35 | 0.04 |
| NOE upper distance constraints: | |||||
| Intraresidual (|i − j|= 0) | 886 | 911 | 891 | 603 | 711 |
| Medium-range (1≤|i − j|≤4) | 1692 | 1653 | 1652 | 1370 | 1408 |
| Long-range (|i − j|> 4) | 1439 | 1255 | 1193 | 1112 | 1205 |
| RMS deviation from ideal geometry | |||||
| Bond lengths (Å) | 0.004 | 0.005 | 0.005 | 0.005 | 0.004 |
| Bond angles (degrees) | 0.7 | 0.7 | 0.6 | 0.7 | 0.7 |
| Statistics for structured region (residues): | 15–146 | 15–146 | 11–72,82–138 | 27–160 | 15–140 |
| RMS deviation from averaged coordinates (Å) | |||||
| Backbone | 0.31 | 0.29 | 0.31 | 0.31 | 0.30 |
| Heavy atoms | 0.72 | 0.71 | 0.66 | 0.68 | 0.64 |
| Ramachandran analysis (%) | |||||
| Most favored | 86.0 | 77.8 | 81.4 | 77.2 | 81.3 |
| Additionally allowed | 13.6 | 20.7 | 18.1 | 22.6 | 18.3 |
| Generously allowed | 0.4 | 1.4 | 0.5 | 0.2 | 0.3 |
| Disallowed | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 |
The number in parentheses represents structures determined in this study.
For the whole twinfilin protein, not for individual ADF-H constituents (see text).
Results
Structure determination of the five ADF-H domains
Experimental restraints and structural statistics are summarized in Table I. The assignments were almost complete: Among the backbone and side chain assignments, excluding the non-native expression-tag derived sequences, only the amide atom of G42 of the C-terminal ADF-H domain of twinfilin 1 could not be assigned.
Description of the structures and comparison with published structures
The resulting structures of the five domains share a common fold that is typical of the ADF domains (Fig. 4). There are five internal β-strands, four of them are antiparallel and the fifth runs parallel to the fourth. The β-strands are surrounded by at least four distinct helices. However, there are some significant differences, as described below.
Figure 4.
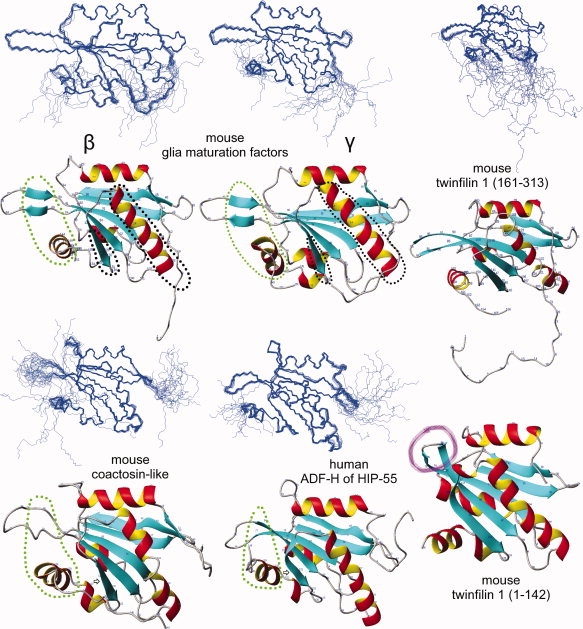
NMR structures of the five determined ADF-H domains and the twinfilin 1 (1-142) crystal structure [PDB id. 1M4J]. Each NMR structure is represented by a diagram showing the 20 structures superimposed on one another and the ribbon diagram of one of those structures. Areas relevant to F-actin and G-actin binding are circled in green and black, respectively (see Discussion). The β-strand (pointed out by arrows) has a different shape in the human ADF-H structure of HIP-55. The presence of the “tilted” loop (purple) in the N-terminal twinfilin 1 domain (1-142) prevents the binding of this domain to F-actin.15 However, the C-terminal twinfilin 1 domain (161-313) lacks this “tilted” domain, and thus can bind F-actin. The whole twinfilin protein, consisting of both N- and C- terminal ADF-H domains connected by a short linker, does not bind F-actin, likely probably because of steric hindrance (due to the large size of F-actin).
Class II
Our GMF structures are essentially identical to each other (they share ∼79% sequence identity) and to the crystal structure of mouse GMF γ [PDB, id. 1VKK; Joint Center for Structural Genomics (JCSG), http://www.topsan.org/Proteins/JCSG/1vkk] differing by RMSDs ∼1.0 Å. They share ∼16% sequence identity with yeast cofilin (class I) [PDB, id. 1CFY], with an RMSD ∼1.4 Å. All three GMF structures display two additional β-strands in the loop, which is highlighted by the green circle in Figure 4 (residues 81–82, 88–89). This was confirmed by the dihedral angles and the NOE data. These β-strands have not been seen in the protein structures of other ADF-H classes. Therefore, they may be a class-defining feature, since the other ADF-H classes have fewer amino acids with favorable β-strand propensities in this loop, that is large amino acids (Y, F, W) or β-branched amino acids (T, V, I) (Figs. 1 and 2). Another unusual β-strand is present at the N-terminus.
Class III
The mouse coactosin-like structure is very similar to that of mouse coactosin (PDB id, 1WM4)19 and to the six other coactosin and coactosin-like structures in the PDB (all from human). The most striking difference is the appearance of an additional β-strand at the N-terminus (Figs. 2 and 4), as in our GMF structures.
Class IV
There are very few differences (RMSD ∼0.80 Å) between our C-terminal mouse twinfilin 1 domain structure (161-313), PDB id, 2D8B, and the NMR structure of twinfilin 1 (176-316), PDB id, 2HD7. Their individual RMSDs are ∼0.31 Å and ∼0.41 Å, respectively. Our construct is longer at the N-terminus, and the proteins share 100% sequence identity over 139 residues. Incidentally, our structure was the first that became available to the general public, via the PDB.
Class V
The structures of the ADF-H domain of human HIP-55 drebrin-like and the Abp1 ADF-H domain of S. cerevisiae18 are quite similar: Both structures contain a loop, circled in green in Figure 4, that slightly bends towards the C-terminal helix. This loop seems to be a defining feature of this class of ADF-H domains. Other peculiarities can be found in the different shape of the β-strand, as pointed out by an arrow, and in the rotated long central helix (Fig. 5, bottom right).
Figure 5.
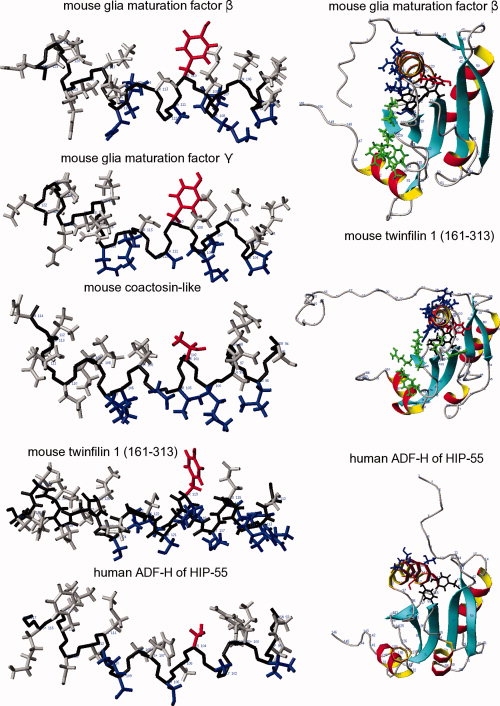
Actin binding sites on the long helix (left) and putative G-actin binding interfaces (right). Left: The red-colored sidechains are important for the stabilization of the structures: Y110 (mouse GMF β and γ); T101 (mouse coactosin-like); Y119 (human twinfilin 1 (161-313)); C104 (human ADF-H of HIP-55). The blue-colored sidechains are likely probably important for actin binding: P104, Q107, M108, A111, K114, and N115 for mouse GMF β and γ; L96, K100, T103, D104, T106, and L107 for mouse coactosin-like; I113, R114, R116, M117, L118, S120, S121, K123, and S124 for human twinfilin 1 (161-313); and V99, A103, S106, S109, and T110 for human ADF-H of HIP-55. Right: G-actin binding interface of twinfilin 1 (161-313), based on the crystal structure of twinfilin 1 (176-316) complexed with G-actin.16 This binding interface is compared with the putative G-actin binding interfaces of GMF β and human ADF-H of HIP-55. Binding residues are colored green in this figure and are labeled with blue open diamonds (⋄) in Figure 2: M106, Y109, R140, K141, E143, D145, E149, F154, and E158 for twinfilin 1 (161-313); and S98, K125, V126, E128, R130, D134, W139, and K143 for GMF β, based on the alignment of Figure 2. The red and black sidechains stabilize the structures, and are shown in bold type in Figure 2. The red sidechains on the right side correspond to the red sidechains on the left side. Notice the rotated long central helix of the human ADF-H in the HIP-55 structure.
Different stabilization mechanisms in the structures
The long central helix is crucial for the stabilization of the structures (Fig. 5). This helix is stabilized by four residues in GMF β and γ (Class II): F74, F94, F96, Y110, and by three residues in the other determined structures: Y38, F66, T101 (mouse coactosin-like; Class III); Y83, Y104, T119 (C-terminal domain (161-313) of mouse twinfilin 1; Class IV); and Y40, Y71, C104 (ADF-H domain of human HIP-55 drebrin-like; Class V). Other members of the respective classes are stabilized in similar manners. Interestingly, the N-terminal domain of mouse twinfilin 1 is stabilized by only two residues, Y78 and Y111,15 as residue 97 is an alanine.
Discussion
Implications for F- and G-actin binding
Figures 2 and 4 display the residues and the structural features that have been shown to be involved in F-actin and G-actin binding.14–18 The central helix seems to be crucial for both F- and G-actin binding. For F-actin to bind, the C-terminal helix and a loop region connecting some internal β-strands are apparently important. The three major sites of interaction with G-actin are as follows: (1) The long central helix; (2) The area before the C-terminal helix, that is the β-strand in between the long central helix and the C-terminal helix (pointed out by arrows in Fig. 4) and the adjacent loop; and (3) The N-terminal extension preceding the first helix, which is initially disordered but becomes ordered upon binding. For yeast cofilin, the N-terminal MSRSG sequence is essential, as its deletion was lethal.17 However, twinfilin 1 (176-316) binds G-actin with a QGVAFP sequence. The N-terminal extension involved in G-actin binding is either truncated or absent in the case of the coactosins (Fig. 2). In our mouse coactosin-like structure, the additional β-strand preceding the first helix probably interferes with the ordering of the N-terminal residues when G-actin approaches, and thus inhibits docking. Both reasons can serve as explanations for the lack of G-actin binding to coactosins. In Figure 5 (left), the residues that are likely to be important for actin binding to the long central helix are colored blue, and those involved in structural stabilization are red. It is apparent that the binding residues are not conserved in the ADF-H domains across the classes (Figs. 1 and 2). On the right side of Figure 5 (middle), the side chains of twinfilin 1 (161-313) involved in the G-actin binding interface are colored blue and green.16 This interface is missing in the ADF-H domain of HIP-55 (Fig. 5, bottom right), and not surprisingly, G-actin binding has not been observed. There are many reasons for the absence of this interface. The most important one may be that a different structural stabilization mechanism rotates the central helix. The total number of binding residues and the number of long-chained actin-binding amino acids (Fig. 5, bottom left) is also lower. Other reasons may include the different orientation of the C-terminal β-strand, pointed out by an arrow in Figure 4, and the different residues in the green-circled area in Figure 2 (the area around the C-terminal β-strand). The putative G-actin binding interface (Fig. 5, right) of mouse GMF is less prominent than that of twinfilin 1 (residues 161-313). In Figure 2, the structure-based alignment revealed several amino acid substitutions, including the replacement of the strongly basic arginine (R) with an acidic glutamic acid (E) or aspartic acid (D) within the putative G-actin binding interface, as highlighted in the blue-circled area of the N-terminal residues (SEDL and SDSL cf. SRSG of cofilin (yeast)) and in the purple-circled area of the long central helix. The long central helix is very basic in other ADF-H domains. Furthermore, the GMF structures also feature a β-strand preceding the first helix, reminiscent of our coactosin-like structure, which probably interferes with the ordering of the N-terminal residues when G-actin approaches, and thus inhibits docking. Therefore, the binding of G-actin to GMF may involve a different interface, if it occurs at all. So far, no binding has been demonstrated, and it appears that G-actin binding is not the main physiological role of GMF γ.10 Differences also exist in the putative F-actin binding interface, with the appearance of two additional β-strands in the green-circled loop in Figure 4 (residues 81, 82, 88–89). They may partially account for some results10 that suggested that GMF γ has a different function in actin dynamics from cofilin, as it lacks significant actin-depolymerizing activity. It may define a novel pathway in the regulation of actin-based cellular functions, and may be involved in the pathophysiology of cardiovascular diseases.
The determination of additional ADF-H structures will certainly clarify the characteristics of each ADF-H domain class.
Materials and Methods
Expression and purification of the five ADF-H proteins
Five ADF-H proteins were selected: mouse glia maturation factor β8 (SWISS-PROT: Q9CQ13) and γ (SWISS-PROT: Q9ERL7); mouse coactosin-like (SWISS-PROT: Q9CQ16); C-terminal domain (residues 161-313) of mouse twinfilin 1 (SWISS-PROT: Q91YR1); and ADF-H domain of human HIP-55 (SWISS-PROT: Q9UJU6). The cDNA fragments encoding these domains were obtained from the RIKEN full-length enriched mouse cDNA library20: clone IDs 3110001H22, 2310057N07, and 2010004C08, and the Ultimate ORF Clones (Invitrogen, USA): clone IDs IOM20752 and IOH21501, respectively. The DNA-fragments were cloned (separately) into the expression vector pCR2.1 (Invitrogen) as a fusion with an N-terminal 6-His affinity tag and a TEV protease cleavage site. The 13C/15N-labeled fusion proteins were synthesized by the cell-free protein expression system.21,22 The proteins were purified from the synthesis solution as follows: The solution was adsorbed to a HiTrap Chelating column (GE Healthcare), which was washed with buffer A (50 mM Tris-HCl buffer [pH 8.0] containing 500 mM sodium chloride and 10 mM imidazole) and eluted with buffer B (50 mM Tris-HCl buffer [pH 8.0] containing 500 mM sodium chloride and 500 mM imidazole). To remove the histidine tag, the eluted protein was incubated at 30°C for 1 h with TEV protease. After dialysis against buffer A without imidazole, the dialysate was mixed with imidazole, to a 10 mM final concentration, and then was applied to a HiTrap Chelating column, which was washed with buffer A. The flow-through fraction was loaded onto a HiTrap Desalting column (GE Healthcare) with buffer C (20 mM Tris-HCl buffer [pH 8.0]). The ADF-H-containing fractions were applied to a HiTrap Q column with a concentration gradient of buffer C and buffer D (20 mM Tris-HCl buffer [pH 8.0] containing 1 M sodium chloride). The ADF-H-containing fractions were collected, and a protease inhibitor cocktail (Complete [EDTA-free], Roche Applied Science) and DTT (final concentration, 1 mM) were added. All ADF-H constructs, except for the mouse coactosin-like, have linker sequences at the N-terminus (GSSGSSG) and the C-terminus (SGPSSG), which are both derived from the expression vector.
NMR spectroscopy, structure determination and analysis
For NMR measurements, the purified proteins were concentrated to around 1.0 mM (1.16 mM, 1.21 mM, 0.67 mM, 1.44 mM, 1.33 mM, respectively) in 1H2O/2H2O (9:1) 20 mM Tris d11-HCl buffer (pH 7.0) containing 100 mM NaCl, 1 mM 1,4-dl-dithiothreitol-d10 (d-DTT), and 0.02% NaN3. All NMR measurements were performed at 25°C on Bruker AVANCE 600, AVANCE 800, and AVANCE 900 (for the twinfilin 1 sample) spectrometers. Sequence-specific backbone assignments were accomplished with the 13C/15N-labeled sample using standard HNCO, HNCOCA, HNCACO, CBCACONNH, and HNCOCA triple-resonance experiments.23 Assignments of side chains were obtained from HBHACONH, HCCONNH, CCCONNH, HCCH-TOCSY, and CCH-TOCSY spectra. 3D 15N- and 13C-edited NOESY-HSQC spectra with 80 ms mixing times were used to obtain distance restraints. The spectra were processed with the program NMRPipe24 and were analyzed with the programs Kujira25 and NMRView.26 Automated NOE cross-peak assignments and structure calculations with torsion angle dynamics were performed using CYANA.27 Dihedral angle restraints (φ,ψ) were derived using TALOS.28 No hydrogen bond constraints were used. A total of 100 structures were independently calculated and the 20 best ones (according to the target function) were selected for further analysis. Several different versions of CYANA were used for each data set. Each version of CYANA calculates a slightly different structure. The quality of the structure, quantified for example by the number of unassigned NOE cross peaks and the number of residues in favorable areas of the Ramachandran plot, depends to a small degree on which CYANA version is used for a particular data set, and we chose to present our results using the most favorable CYANA version in that regard. However, it should be emphasized that the variations between CYANA-versions 2.x.x are so minor that the conclusions made in this paper are the same, regardless of which version was actually used. To overcome some minor problems associated with CYANA, Pro-132 of the C-terminal ADF-H domain of twinfilin 1 was placed in the up-form (default: indifferent) [Peter Güntert, pers. comm.]. The structures were validated using PROCHECK-NMR29 and were analyzed using MOLMOL,30 which was also used to produce the figures. The atomic coordinates have been deposited into the Protein Data Bank (PDB) under the accession codes 1V6F, 1WFS, 1UDM, 2D8B, and 1X67, respectively.
The sequences used in Figure 1 were obtained from the NCBI Reference sequences: Cofilin-1/1-166 (NP031713.1), Cofilin-2/1-166 (NP031714.1), ADF destrin/1-165 (NP062745.1), glia maturation factor beta/1-142 (NP071306.1), glia maturation factor gamma 2/1-142 (NP001034281.1), glia maturation factor gamma 1/1-142 (NP071307.1), twinfilin 1/1-350 (NP032997.3), twinfilin 1-like/1-349 (NP036006.1); the DNA Database of Japan (DDBJ): unnamed 1/1-229 (BAB32114.1), unnamed 2/1-122 (BAE31257.1), unnamed 3/1-347 (BAB22293.1); SWISS-PROT: DBNL DREBRIN-like/1-436 (Q62418); GenBank: DREB/1-706 (AAF25189.1| AF187147_1), unnamed 4/1-433 (BAE31953.1), unnamed 5/1-432 (BAC32592.1); and the PDB IDs: 1CFY (cofilin (yeast)), 1WM4 (coactosin (mouse)), and 1M4J (twinfilin 1-142 (mouse)).
Acknowledgments
The authors would like to thank the following persons: Yasuko Tomo, Takayoshi Matsuda, Eiko Seki, Masaomi Ikari, Kazuharu Hanada, Yukiko Fujikura, Takashi Yabuki, Masaaki Aoki, Yoshiko Ishizuka-Katsura, Yuri Tomabechi, Masato Aoshima, Satoshi Morita, and Ryoko Ushikoshi-Nakayama.
Glossary
Abbreviations:
- ADF-H domain
actin depolymerizing factor homology domain
- NOE
Nuclear Overhauser Effect
- GMF
glia maturation factor
- PDB
Protein Data Bank, Research Collaboratory for Structural Bioinformatics (http://www.rcsb.org)
References
- 1.Gungabissoon RA, Bamburg JR. Regulation of growth cone dynamics by ADF/cofilin. J Histochem Cytochem. 1999;51:411–420. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- 2.Sun HQ, Kwiatkowski K, Yin HL. Actin monomer binding proteins. Curr Opin Cell Biol. 1995;7:102–110. doi: 10.1016/0955-0674(95)80051-4. [DOI] [PubMed] [Google Scholar]
- 3.Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 4.Hartwig JH, Kwiatkowski DJ. Actin-binding proteins. Curr Opini Cell Biol. 1991;3:87–97. doi: 10.1016/0955-0674(91)90170-4. [DOI] [PubMed] [Google Scholar]
- 5.Maciver SM, Hussey PK. The ADF/cofilin family: actin-remodeling proteins. Genome Biology. 2002;3:3007.1–3007.12. doi: 10.1186/gb-2002-3-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeoh S, Pope B, Mannherz HG, Weeds A. Determining the differences in actin binding by human ADF and cofilin. J Mol Biol. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- 7.Nevalainen EM, Paavilainen VO, Lappalainen P. Twinfilin family of actin monomer-binding proteins. In: Lappalainen P, editor. Actin-monomer-binding proteins. Landes Bioscience and Springer Science + Business Media. 2007. pp. 53–60. [Google Scholar]
- 8.Lim R, Miller JF, Zaheer A. Purification and characterization of glia maturation factor beta: a growth regulator for neurons and glia. Pro Natl Acad Sci USA. 1989;86:3901–3905. doi: 10.1073/pnas.86.10.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asai K, Fujita K, Yamamoto M, Hotta T, Moriyama K, Kokubo M, Moriyama A, Kato T. Isolation of novel cDNA (hGMF-gamma) homologous to glia maturation factor-beta gene. Biochim Biophys Acta. 1998;1396:242–244. doi: 10.1016/s0167-4781(97)00222-4. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda K, Kundu RK, Ikeda S, Kobara M, Matsubara H, Quertermous T. Glia maturation factor-{gamma} is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ Res. 2006;99:424–433. doi: 10.1161/01.RES.0000237662.23539.0b. [DOI] [PubMed] [Google Scholar]
- 11.Wahlström G, Vartiainen M, Yamamoto L, Mattila PK, Lappalainen P, Heino TI. Twinfilin is required for actin-dependent developmental processes in drosophila. J Cell Biol. 2001;155:787–796. doi: 10.1083/jcb.200108022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessels MM, Engqvist-Goldstein AEY, Drubin DG, Qualmann B. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J Cell Biol. 2001;153:351–366. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Kori R, Shui J-W, Chen U-R, Yao Z, Tan T-H. The SH3 domain-containing adaptor HIP-55 mediates c-Jun N-terminal kinase activation in T cell receptor signaling. J Biol Chem. 2003;278:52195–52202. doi: 10.1074/jbc.M305026200. [DOI] [PubMed] [Google Scholar]
- 14.Paavilainen VO, Merckel MC, Falck S, Ojala PJ, Pohl E, Wilmanns M, Lappalainen P. Structural conservation between the actin monomer-binding sites of twinfilin and actin-depolymerizing factor (ADF)/Cofilin. J Biol Chem. 2002;277:43089–43095. doi: 10.1074/jbc.M208225200. [DOI] [PubMed] [Google Scholar]
- 15.Paavilainen VO, Hellman M, Helfer E, Bovellan M, Annila A, Carlier M-F, Permi P, Lappalaninen P. Structural basis and evolutionary orgin of actin filament capping by twinfilin. Natl Acad Sci USA. 2007;104:3113–3118. doi: 10.1073/pnas.0608725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the actin-depolymerizing factor homology domain in complex with actin. J Cell Biol. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lappalainen P, Federov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintero-Monzon O, Rodal AA, Strokopytov B, Almo SC, Goode BL. Structural and functional dissection of the Abp1 ADFH actin-binding domain reveals versatile in vivo adapter functions. Mol Biol Cell. 2005;16:3128–3139. doi: 10.1091/mbc.E05-01-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellman M, Paavilainen VO, Naumanen P, Lappalainen P, Annila A, Permi P. Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins. FEBS Lett. 2004;576:91–96. doi: 10.1016/j.febslet.2004.08.068. [DOI] [PubMed] [Google Scholar]
- 20.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schönbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyo-oka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y, RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium Functional annotation of a full-length cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 21.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genom. 2004;5(1–2):63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J Biomol NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- 23.Bax A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr Opin Struct Biol. 1994;4:738–744. [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 26.Johnson B, Blevins R. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 27.Güntert P. Automated NMR structure calculation. Prog Nucl Magn Reson Spectrosc. 2003;43:105–125. [Google Scholar]
- 28.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 30.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]


