Abstract
Cell division is commonly thought to involve the equal distribution of cellular components into the two daughter cells. During many cell divisions, however, proteins, membrane compartments, organelles, or even DNA are asymmetrically distributed between the two daughter cells. Here, we review the various types of asymmetries that have been described in yeast and in animal cells. Asymmetric segregation of protein determinants is particularly relevant for stem cell biology. We summarize the relevance of asymmetric cell divisions in various stem cell systems and discuss why defects in asymmetric cell division can lead to the formation of tumors.
Keywords: Asymmetric cell division, cell polarity, neurogenesis, stem cell, tumorigenesis
When we think of cell division, we usually have a process in mind where one cell gives rise to two identical daughter cells. In many cases, however, cell divisions are asymmetric and generate two daughter cells that are different in protein content, cell size, or developmental potential (Chia et al. 2008; Doe 2008; Gonczy 2008; Knoblich 2008). In fact, many secrets of the cell cycle were resolved in the yeast Saccharomyces cerevisiae, an organism that divides in a highly asymmetric fashion (Chant 1999; Thorpe et al. 2008). But even in cultured cells, cytoplasmic structures like the midbody are often inherited by only one of the two daughter cells (Gromley et al. 2005). The two centrosomes that are inherited by the two daughter cells can be different in protein composition (Piel et al. 2000; Spradling and Zheng 2007). And even the chromatin in the two daughter cells could be different, as several results have indicated that the two DNA strands generated during S phase might be unequal in their segregation behavior (Rando 2007).
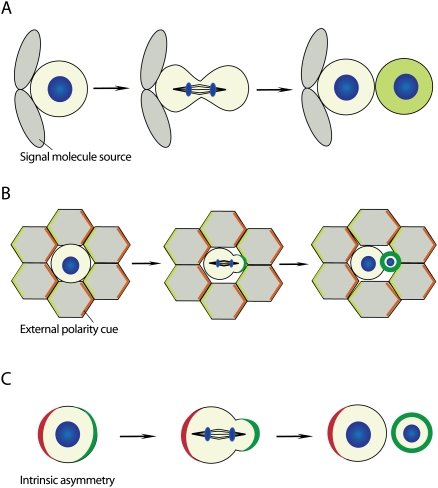
While the significance of these mitotic asymmetries in cultured cells remains to be demonstrated, asymmetric cell division in a developing organism is known to play a major role (Horvitz and Herskowitz 1992). Within a whole tissue, a cell division can become asymmetric in several ways (Fig. 1): In the first example, the dividing cell is within a polar environment. In this case, the two daughter cells are initially identical, but their exposure to different environments induces alternative fates. Alternatively, external polarity can induce the asymmetric distribution of cellular components during mitosis so that their unequal inheritance induces different cell fates. Finally, an asymmetric division can be purely intrinsic when a pre-existing cellular polarity is used to polarize cell fate determinants in a cell-autonomous fashion. In this review, we focus on these intrinsic mechanisms, and we refer to excellent recent reviews for extracellular pathways (Spradling et al. 2001; Lin 2002; Fuller and Spradling 2007; Kirilly and Xie 2007; Morrison and Spradling 2008).
Figure 1.
Modes of asymmetric cell division. (A) Asymmetric cell fate specification is regulated by a niche-derived signal. Cells that contact the niche retain their identity, whereas cells that become detached from the niche after division adopt a different cell fate. (B) External polarity induces the asymmetric localization of cell fate determinants (green). (C) Intrinsic asymmetry localizes polarity proteins (red), which instruct cell fate determinants (green) to segregate asymmetrically during mitosis in the absence of extracellular cues (DNA, blue).
Early theories of development postulated that all cellular identities are assigned through the asymmetric inheritance of nuclear determinants. And as early as 1904, Conklin (1905) could demonstrate that a segregating cytoplasmic determinant conveniently colored in yellow is responsible for the induction of muscle development in the ascidian Styela partita. But it was not before 1994 that a general regulator of asymmetric cell division was found that acts in organisms as different as fruit flies and mice (Rhyu et al. 1994). This regulator is called Numb, and its discovery has sparked research efforts that led to the identification of what is now thought to be a fundamental mechanism for intrinsically asymmetric cell division. We discuss this mechanism with a focus on more recent insights, and discuss the exciting implications of intrinsically asymmetric cell divisions for stem cell biology. In particular, we outline the interesting connections that have been made between defects in asymmetric cell division and the generation of a stem cell pool that loses control over growth and proliferation to form eternally proliferating deadly tumors.
Mitotic asymmetries
A cell division is considered asymmetric when the two daughter cells have different sizes, when one or more cellular constituents are preferentially segregated into only one of the two daughter cells, or when the two daughter cells are endowed with different potentials to differentiate into a particular cell type (Horvitz and Herskowitz 1992). Below, we explain how different daughter cell sizes are generated, and how daughter cells can inherit different fate determinants to enter distinct differentiation pathways. We also summarize asymmetrically segregating cellular components where the functional implications of the asymmetry are still obscure. We focus on animal cells and occasionally include yeast to illustrate functional principles. Asymmetric cell division also plays a major role in plants, but since the mechanisms are quite distinct, we refer to several excellent recent reviews on this topic (Abrash and Bergmann 2009; Menke and Scheres 2009).
Generating different daughter cell sizes
The size of the two daughter cells is determined by the cleavage furrow, which in turn is specified by the position of the mitotic spindle (Glotzer 2004). A centrally located mitotic spindle will result in two daughter cells of the same size, whereas any displacement of the spindle toward one pole will generate one larger and one smaller daughter cell. In some cases, like polar body extrusion, this can lead to extreme asymmetry, where one of the daughter cells is barely large enough to hold one copy of the genetic material. In somatic divisions, however, cell size asymmetry is mild and, only rarely, one daughter cell is more than double the size of the other.
The Par protein complex
One of the best-understood model systems for generating cell size asymmetry is the zygote of Caenorhabditis elegans. After fertilization, C. elegans embryos divide into a larger anterior AB and a smaller posterior P1 daughter cell (Cowan and Hyman 2004a). Sperm entry triggers a series of events that result in a subdivision of the cell cortex into an anterior and a posterior domain (Cowan and Hyman 2004a). This results in a stronger capacity of the posterior cortex to exert force on the mitotic spindle. The spindle is displaced toward the posterior end, and, therefore, the division plane forms in an asymmetric manner. It turns out that the proteins controlling these events are part of a fundamental mechanism for cell polarity and asymmetric cell division that is conserved in worms, flies, and vertebrates.
Core components of this machinery were discovered in a landmark genetic screen for “par” (partitioning defect) mutants in which AB and P1 have the same size (Kemphues et al. 1988). Based on their localization pattern, three classes of Par proteins can be distinguished: The serine/threonine kinase PAR-1 (Guo and Kemphues 1995) and the RING finger protein PAR-2 (Boyd et al. 1996) accumulate on the posterior cell cortex, whereas the anterior cell cortex is occupied by the PSD95/Dlg/ZO1 (PDZ) domain proteins PAR-3 and PAR-6, and by an atypical protein kinase C (aPKC, called PKC-3 in C. elegans). In addition, the serine–threonine kinase PAR-4 (Watts et al. 2000) and the 14–3–3ɛ protein PAR-5 (Morton et al. 2002) are necessary for generating asymmetry but are not asymmetrically distributed themselves. The individual Par proteins show distinct levels of functional conservation: While par-2 is found only in C. elegans, the other Par proteins are conserved in animals but not in fungi or plants (Ohno 2001; Suzuki and Ohno 2006). In animals, the conserved Par proteins regulate epithelial apical–basal polarity and many other aspects of cell polarity (Ohno 2001; Suzuki and Ohno 2006). Par-3, Par-6, and aPKC regulate asymmetric cell division in diverse organisms including Drosophila, mouse, and chicken. More recently, a function for the Drosophila homologs of PAR-5 and PAR-1 in this process has been described as well (Krahn et al. 2009). Par-3, Par-6, and aPKC form a complex that localizes asymmetrically, controls the activity of aPKC in space and time, and acts as a scaffold for the assembly of further asymmetry factors. Par-6 has been reported to bind and regulate aPKC kinase activity, and the two proteins are strictly codependent for their asymmetric localization. Par-6 also binds to the small GTPase Cdc42 through an N-terminal Cdc42/Rac interactive binding (CRIB) domain, and Cdc42 is another important regulator of aPKC activity (Joberty et al. 2000; Johansson et al. 2000; Lin et al. 2000; Qiu et al. 2000). Par-3 is less tightly bound and localizes asymmetrically without the other partners in certain mutant conditions (Beers and Kemphues 2006). It competes with other partners for binding to Par-6 (Yamanaka et al. 2006), and might act as an adaptor that regulates substrate specificity by simultaneously binding both aPKC and its substrates.
Establishing polarity
In C. elegans, subdivision of the cell cortex into an anterior and a posterior domain starts with fertilization (Cowan and Hyman 2004a; Gonczy 2008). Before fertilization, PKC-3, PAR-3, and PAR-6 are distributed along the entire cell cortex (Fig. 2A). Upon sperm entry, however, PAR-3 and PAR-6 disappear from the cortical area overlying the sperm centrosome, and this allows PAR-2 to be recruited to the cortex. Subsequently, the PAR-2 area expands until the entire cell cortex is divided equally into an anterior PAR-3/6 and a posterior PAR-2 domain. Elegant centrosome ablation experiments have demonstrated that an interaction between the sperm centrosome and the cell cortex is the initial symmetry-breaking event. This interaction was initially thought to be microtubule-independent (Cowan and Hyman 2004b), but subsequent RNAi experiments have suggested that microtubules play an important role (Tsai and Ahringer 2007). Polarization of the cell cortex is accompanied by characteristic asymmetries in the actin cytoskeleton (Munro et al. 2004). Around the time of PAR-2 recruitment, a cortical meshwork consisting of actin and nonmuscle myosin II translocates toward the anterior. This generates a cortical flow of cytoplasm toward the anterior that is balanced by a corresponding posterior flow in the center of the cell. At the same time, cortical ruffling movements, which occur throughout the embryo before polarization, cease in the area that is occupied by PAR-2. It is thought that the cortical actin network is under tension, and a local weakening by the sperm centrosome causes it to collapse toward the anterior much like a mesh stocking stretched over a ball (Bray and White 1988). This local weakening is mediated by the Rho-GAP CYK-4 that is carried in the sperm and is localized in organelles around the sperm derived centrosome (Jenkins et al. 2006; Motegi and Sugimoto 2006; Schonegg and Hyman 2006). CYK-4 inactivates Rho by stimulating GTP hydrolysis. Since Rho controls the phosphorylation levels of the myosin regulatory light chain MLC-4, this could inhibit myosin contractility and thereby weaken the actin cytoskeleton.
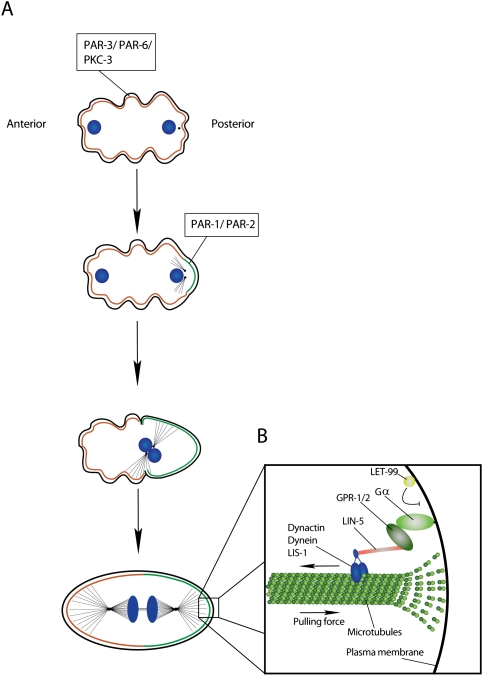
Figure 2.
Asymmetric division of the C. elegans one-cell embryo. (A) Separation of the PAR-3/PAR-6/PKC-3 and PAR-1/PAR-2 occupied cortical domains. (B) The mitotic spindle is anchored along the anterior–posterior axis. LIN-5 and GPR1-/2 link the spindle via simultaneous binding to the microtubule-associated Dynactin/Dynein/LIS-1 complex and membrane-anchored Gα (DNA, blue).
Although the centrosome is essential for polarity establishment, it is no longer required once the cortical domains have formed (Cowan and Hyman 2004b). At later stages, inhibitory interactions between the anterior and posterior Par proteins maintain cortical polarity. Anterior PKC-3 phosphorylates PAR-2 within its cortical localization domain, thereby preventing its recruitment to this part of the cell cortex (Hao et al. 2006). On the posterior side, PAR-2 prevents the cortical localization of PAR-3, and thereby restricts the PAR-3/6 complex to the anterior side. This inhibition is likely to be indirect, since it also requires PAR-1 and PAR-5 (Hao et al. 2006). Work in Drosophila epithelial cells has demonstrated that Par-1 can directly phosphorylate Par-3 (Benton and Johnston 2003). Phosphorylation generates binding sites for 14–3–3ɛ (PAR-5 in C. elegans), and thereby prevents binding to aPKC and assembly into the Par complex. Thus, cortical polarity in C. elegans is established by interaction between the sperm centrosome and the actin cytoskeleton. Later, it is maintained by mutual inhibition of the anterior and posterior cortical domains through reciprocal phosphorylation.
Asymmetric spindle positioning and heterotrimeric G proteins
How do cortical domains translate into posterior spindle displacement? The position of the mitotic spindle is determined by the extent of pulling forces exerted on the anterior and posterior poles (Hyman and White 1987; Hyman 1989; Grill et al. 2001; Colombo et al. 2003; Labbe et al. 2003). These forces can be calculated from the speed with which the spindle poles move toward the cell cortex after cutting the central part of the mitotic spindle with a laser beam (Grill et al. 2001). The forces are higher on the posterior pole, and this depends on the anterior–posterior polarity established by the Par proteins. Experiments in which the spindle poles are disintegrated by a laser beam (Grill et al. 2003) demonstrate that this is not because more microtubules connect the posterior pole to the cell cortex, but because of a larger number of pulling force generators acting on astral microtubules on the posterior cortex. Thus, different interactions of the anterior and posterior cell cortex with astral microtubules are ultimately responsible for the differences in daughter cell sizes.
A membrane-anchored protein complex containing the minus end-directed microtubule motor Dynein and its binding partner LIS-1 generates pulling forces on the astral microtubules (Gonczy 2008). Microtubules emanating from the centrosome contact the cell cortex via their plus end (Fig. 2B). Dynein binds to this end and generates a cortex-directed pulling force on the microtubule by trying to move toward the minus end (Gonczy et al. 1999). On the cortex, microtubule ends depolymerise, and this depolymerization is essential for pulling forces to emerge (Kozlowski et al. 2007; Nguyen-Ngoc et al. 2007). The interaction of Dynein with the cell cortex is regulated by a protein complex containing the C. elegans NuMA homolog LIN-5 (Gotta et al. 2003; Srinivasan et al. 2003) and one of the two heterotrimeric G-protein α subunits, GOA-1 or GPA-16 (Gotta and Ahringer 2001; Nguyen-Ngoc et al. 2007). LIN-5 is directly associated with Dynein, while G proteins bind via one of two highly related adaptor proteins: GPR-1 or GPR-2. Since Gα subunits are myristoylated, they might act as the membrane anchor of the spindle-positioning complex.
Why the forces generated by those protein complexes are higher on the posterior side is not precisely understood. The levels of Gα subunits as well as the levels of cortical Dynein and LIS-1 are not significantly different. The difference may lie in GPR-1/2, which are enriched on the posterior side (Colombo et al. 2003; Gotta et al. 2003). GPR-1 and GPR-2 contain GoLoco domains. GoLoco domains can bind Gα subunits specifically in the GDP-bound state (Cismowski et al. 2001). During signal transduction downstream from seven-transmembrane receptors, ligand binding switches heterotrimeric G proteins from their inactive, GDP-bound state into an active, GTP-bound state where both Gα-GTP as well as the free Gβγ subunit can interact with downstream components. In the spindle pathway, only Gα-GDP is active, whereas Gβγ plays an inhibitory role. Nevertheless, cycling between GDP- and GTP-bound forms seems to be important, since efficient control of pulling forces requires both the GOA-1 exchange factor RIC-8 and the Gα GAP RGS-7 (Afshar et al. 2004; Hess et al. 2004). In the absence of RGS-7, which increases the rate of GTP hydrolysis, posterior pulling forces are actually increased, whereas ric-8 mutants have phenotypes that resemble those of GPR-16/GOA-1 or GPR-1/2. The ric-8 mutant phenotype has been explained by an alternative G-protein signaling cycle in which Gα has to go through one round of GTP hydrolysis in order for the Gα/GPR-1/2 complex to form (Hampoelz and Knoblich 2004). Although this is an interesting model, it may have to be reconsidered, since further mutant analysis has suggested a role for RIC-8 in delivering G proteins to the plasma membrane rather than controlling their hydrolysis cycle (Afshar et al. 2005; David et al. 2005; Hampoelz et al. 2005; Wang et al. 2005). Thus, although Dynein and Lis-1 are clearly the business end of the spindle-positioning machinery, the way they are differentially regulated is still unclear.
Asymmetric daughter cell size outside C. elegans
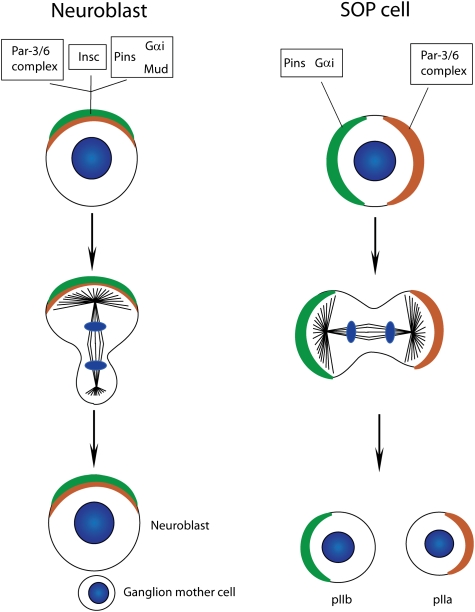
The molecular machinery that generates cellular asymmetry in C. elegans is almost entirely conserved in Drosophila (Betschinger and Knoblich 2004; Doe 2008; Knoblich 2008). In Drosophila, asymmetric cell division is mostly studied in two cell types: neuroblasts, which are the precursors of the CNS, and sensory organ precursor (SOP) cells, which are the founder cells of external sensory (ES) organs (see below for more details). In both cell types, the Drosophila homologs of PAR-3 (called Bazooka) (Schober et al. 1999; Wodarz et al. 1999), PAR-6 (Petronczki and Knoblich 2001), and PKC-3 (called aPKC) (Wodarz et al. 2000; Rolls et al. 2003) set up polarity for asymmetric cell division. Like in C. elegans, a complex consisting of a heterotrimeric G-protein α subunit (called Gαi) (Schaefer et al. 2001; Yu et al. 2003; Izumi et al. 2004), a GoLoco domain protein (either Pins or Loco) (Schaefer et al. 2000; Yu et al. 2000, 2005), and a NuMA-related Dynein-binding protein (called Mud) (Bowman et al. 2006; Izumi et al. 2006; Siller et al. 2006) controls the position of the mitotic spindle during mitosis (Fig. 3). How these complexes regulate daughter cell size asymmetry in Drosophila, however, is slightly different from C. elegans. In SOP cells, the Par-3/6/aPKC complex localizes to the posterior cell cortex, whereas Pins and Gαi are on the anterior side (Bellaiche et al. 2001b). Unlike in C. elegans, this results in daughter cells that are similar in size. In neuroblasts, both complexes are apical, and the spindle is displaced to generate a smaller basal daughter cell. An adaptor protein called Inscuteable (Insc) can bind to Pins and Par-3 simultaneously and connects the two complexes (Fig. 3). When Insc is ectopically expressed in epithelial cells, it is sufficient for reorienting the spindle along an apical–basal axis and for creating a smaller basal and a larger apical daughter cell (Kraut et al. 1996). Taken together, these results suggest that both Pins/Gαi and Par-3/6/aPKC can exert pulling forces in Drosophila: When the two complexes are on the opposite side, this results in equal daughter cells, but when they are combined by the expression of Insc, their concerted action displaces the spindle, resulting in unequal daughter cell size. While Pins/Gαi act through Dynein and Lis-1 (Siller et al. 2005), the mechanism through which Par-3/6/aPKC act on the mitotic spindle remains to be resolved.
Figure 3.
Differences in protein localization between Drosophila neuroblasts and SOP cells. The Par-6/Par-3 and the Pins/Gαi/Mud complex colocalize at the apical cortex of Drosophila neuroblasts. Pins/Gαi/Mud are recruited to the apical cortex through the presence of Insc, which simultaneously binds members of both complexes. These complexes localize to opposite cortical domains in SOP cells, as the linker molecule Insc is not expressed in this cell type (DNA, blue).
Asymmetric inheritance of cell fate determinants
Asymmetric protein segregation in C. elegans
A hallmark of any intrinsically asymmetric cell division is the differential inheritance of cell fate determinants by one of the two daughter cells. In C. elegans, the CCCH-Zn finger proteins PIE-1, MEX-5, and MEX-6 are among these segregating determinants (Cowan and Hyman 2004a; Gonczy 2008). PIE-1 is a germline determinant that segregates into the posterior P1 cell, where it represses general transcription and promotes the expression of germline-specific genes (Seydoux et al. 1996; Tenenhaus et al. 2001). MEX-5 and MEX-6 segregate into the opposite, anterior AB cell. They are highly homologous and act redundantly in correctly specifying muscle lineages (Schubert et al. 2000). At the time of fertilization and during most of interphase, all three proteins are distributed uniformly in the cytoplasm of the zygote. During mitosis, however, PIE-1 concentrates in the posterior and MEX-5/6 in the anterior half of the cytoplasm, so that the proteins are differentially segregated into the two daughter cells.
The asymmetric localization of cytoplasmic determinants in C. elegans involves a combination of cytoplasmic anchoring and regulated protein degradation (Spike and Strome 2003). Before mitosis, a reaction-diffusion mechanism seems to be responsible for the initial concentration of PIE-1 in the posterior cytoplasm that is destined for germline incorporation (Daniels et al. 2009). PIE-1 exists in a rapidly diffusing form that can distribute through the embryo and a slowly diffusing form whose long-range movement is hindered by the association with some cytoplasmic structure. Association of PIE-1 with the P granules could be responsible for changes in mobility. P granules are ribonucleoprotein particles that accumulate in the posterior half of the cell. They mark the C. elegans germline and cosegregate with the PIE-1 protein during each division. Mathematical modeling demonstrates that this reaction-diffusion mechanism can explain the graded distribution of PIE-1 that is seen before and during the early stages of mitosis. Later, however, the regulated degradation of PIE-1 protein seems to play an important role (Reese et al. 2000).
The degradation of PIE-1 is controlled by MEX-5. MEX-5 is phosphorylated by PAR-1 on a C-terminal serine residue (Tenlen et al. 2008). Through an unknown mechanism, this selectively increases the mobility of MEX-5 in the posterior cytoplasm, causing the protein to accumulate in the anterior domain. In the anterior, MEX-5 (and MEX-6) stimulate PIE-1 degradation. MEX-5 activates a protein called ZIF-1 that recruits PIE-1 and other CCCH-Zn finger proteins into an E3 ubiquitin ligase complex. This complex is also called an ECS-type ubiquitin ligase and contains the proteins Elongin B and C, a Cullin, a SOCS box protein, and a ring finger protein (Kile et al. 2002). It is thought that after activation by ZIF-1, the complex targets PIE-1 for degradation through the proteasome. Interestingly, however, local degradation has been proposed as a mechanism for P-granule localization as well, suggesting that various interconnections and feedback loops that exist between the various localization mechanisms ensure the formation of a sharp cytoplasmic boundary (Spike and Strome 2003). Thus, the asymmetric segregation of cytoplasmic determinants in C. elegans involves three steps: First, the Par proteins establish differences in cytoplasmic diffusion rates to generate an initial asymmetric distribution. (Formally, this has only been demonstrated for Mex-5 so far, and future experiments are needed to demonstrate the general use of this attractive mechanism.) Next, the initial asymmetries are refined by differences in protein degradation rates between the anterior and posterior cytoplasm. Finally, feedback loops between anterior and localized determinants result in the formation of sharp boundaries.
Drosophila as a model system for asymmetric cell division
Besides C. elegans, the fruitfly Drosophila melanogaster is the model organism of choice for the analysis of asymmetric cell division. Unlike C. elegans, the proteins that act as segregating determinants in Drosophila are highly conserved and regulate asymmetric cell division in vertebrates as well.
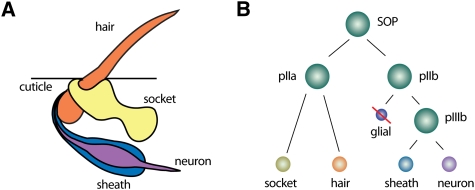
Our knowledge about asymmetric cell division in Drosophila derives mostly from the analysis of two tissues: SOP cells (Fig. 4) in the Drosophila peripheral nervous system, and neuroblasts in the CNS (Fig. 5). SOP cells undergo three rounds of asymmetric cell division to form the different cell types in ES organs (Bardin et al. 2004). They first divide into an anterior pIIa and a posterior pIIb cell. Next, the pIIb cell gives rise to an apical pIIIb cell and a basal glia cell that undergoes apoptosis. Finally, pIIa and pIIIb undergo a terminal division to form the two outer (hair and socket) and the two inner (neuron and sheath) cells of the organ (Fig. 4). ES organs are not essential for viability, and cell fate transformations generate externally visible morphological changes. For these reasons, ES organs have been used recently to analyze asymmetric cell division on a genome-wide level using transgenic RNAi (Mummery-Widmer et al. 2009).
Figure 4.
The Drosophila ES organ as a model system for asymmetric cell division. (A) The Drosophila ES organs consist of two outer (hair and socket) and two inner (neuron and sheath) cells. (B) SOP cells divide asymmetrically in a stem cell-like fashion to generate the various cells of the ES organ. Note that the glial cell undergoes programmed cell death.
Figure 5.
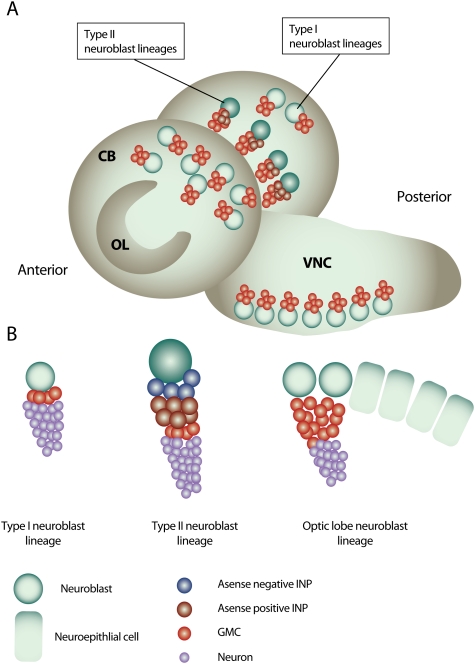
Neurogenesis in the Drosophila larval brain. (A) The Drosophila third instar larval brain contains three main neurogenic regions: the OL neurogenic center, located at the lateral surface of the two brain hemispheres, and the CB neurogenic center, which is located medially of the OL and descends to the VNC on the anterior side of the brain. (B) Type I neuroblast lineages constitute the majority of neuroblast lineages in the CB and VNC. A type I neuroblast gives rise to another neuroblast and a GMC that terminally divides to produce two neurons. Type II neuroblasts are situated on the medial, posterior surface of the brain lobes and give rise to transit-amplifying INP cells. The neuroblasts of the OL originate from OL neuroepithelial cells.
Neuroblasts are stem cell-like progenitors that generate all neurons present in the Drosophila CNS (Doe 2008). During embryogenesis, they undergo a limited number of divisions and do not grow significantly, and therefore become smaller with each round of mitosis. During larval development, they grow in volume between each division, and therefore maintain a constant size that allows them to divide many more times. Based on their position in the brain, larval neuroblasts are subdivided in ventral nerve chord (VNC), central brain (CB), and optic lobe (OL) neuroblasts (Fig. 5A).
All neuroblasts divide asymmetrically into a larger and a smaller daughter cell. While the larger daughter cell maintains stem cell identity, the smaller so-called ganglion mother cell (GMC) divides only once more to generate two differentiating neurons. Although most neuroblasts obey this lineage, eight CB neuroblasts located in the dorso–posterior and medioposterior part of each brain hemisphere proliferate in a more complex manner (Bello et al. 2008; Boone and Doe 2008; Bowman et al. 2008). These so-called type II neuroblasts (also called PAN neuroblasts) divide into one neuroblast and one intermediate neural progenitor (INP). The INP continues to divide asymmetrically and generates one daughter INP and one GMC, which undergoes a terminal division. (Fig. 5B).
CB and VNC neuroblasts arise from embryonic neuroblasts, which become quiescent during embryogenesis but re-enter the cell cycle during larval development (Truman and Bate 1988). In the OL, however, neuroblasts are generated during larval stages. The OL originates from the embryonic optic placode and starts to proliferate and expand in size during larval development (White and Kankel 1978). It is composed of a columnar neuroepithelium that does not express the neuroblast identity genes asense (ase) or deadpan (dpn) (Egger et al. 2007). During larval development, a synchronized wave of proneural gene expression spreads through the epithelium and triggers the transition of epithelial cells into neuroblasts (Yasugi et al. 2008). Before this transition, epithelial cells divide symmetrically, but afterward, they lose their epithelial junctions and divide asymmetrically following a typical neuroblast lineage. Since vertebrate neurogenesis passes through a similar neuroepithelial stage of expansion by symmetric division (Gotz and Huttner 2005), the OL might be a particularly suitable model system for brain development in higher organisms.
Segregating determinants in Drosophila
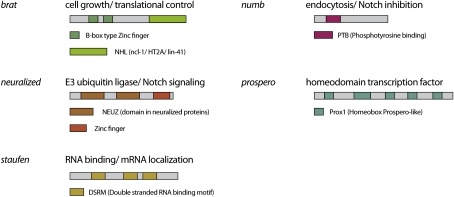
The proteins Numb (Rhyu et al. 1994; Spana et al. 1995), Neuralized (Le Borgne and Schweisguth 2003), Prospero (Pros) (Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995), and Brat (Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c) are known to act as segregating determinants in the Drosophila nervous system (Fig. 6). During late prophase, the proteins concentrate in the plasma membrane area overlying one of the two spindle poles, and upon cytokinesis they segregate into one of the two daughter cells. Numb contains an N-terminal phosphotyrosine-binding (PTB) domain (Li et al. 1998) and a C-terminal DPF/NPF motif that is known to bind components of the endocytic machinery (Berdnik et al. 2002). Numb is a membrane-associated protein that contains an N-myristoylation signal (Benetka et al. 2008) and several N-terminal positively charged amino acids that might interact with membrane phospholipids. During interphase, Numb is distributed uniformly around the plasma membrane. During mitosis, it concentrates in the plasma membrane area overlying one of the two spindle poles so that it segregates into one of the two daughter cells during cytokinesis (Rhyu et al. 1994). In this cell, it inhibits signal transduction through the Notch/Delta pathway. In numb mutants, SOP cells divide symmetrically into two pIIa cells, leading to the formation of abnormal ES organs with only outer, but no inner, cells (Uemura et al. 1989; Rhyu et al. 1994). In numb mutant larval neuroblasts, defects in asymmetric cell division result in the formation of a stem cell-derived tumor (Lee et al. 2006a; Wang et al. 2006). This tumor phenotype is characteristic for many regulators of asymmetric cell division (see below), and results from the massive generation of neuroblasts at the expense of GMCs. Numb also acts in the malphighian tubules, in the gut, and in muscles (Carmena et al. 1998), indicating that it is the most general regulator of asymmetric cell division in flies.
Figure 6.
Domain architecture and main function of Drosophila cell fate determinants. Asymmetric localization and segregation of these proteins upon cell division requires the action of Par proteins.
Numb is a regulator of endocytosis that binds to the endocytic adaptor protein α-adaptin (Santolini et al. 2000; Berdnik et al. 2002). Through its N-terminal PTB domain, Numb also interacts with the Notch receptor (Guo et al. 1996), and it could regulate Notch trafficking at an early endocytic step. Numb also binds to the four-transmembrane protein Sanpodo (O'Connor-Giles and Skeath 2003; Hutterer and Knoblich 2005). Sanpodo is required for Notch signaling only in those tissues where Numb acts (Skeath and Doe 1998). It is localized on intracellular vesicles, but relocalizes to the plasma membrane in the absence of Numb (O'Connor-Giles and Skeath 2003; Hutterer and Knoblich 2005). Thus, Numb could inhibit Notch by facilitating the translocation of Sanpodo into an endocytic compartment where it cannot fulfil its role in the Notch pathway.
Like Numb, the E3 ubiquitin ligase Neuralized segregates into the pIIb cell during SOP division (Le Borgne and Schweisguth 2003). In neuralized mutants, SOP cells generate two pIIb cells, a cell fate transformation opposite to that observed in numb mutants. Neuralized acts as a ubiquitin ligase for the Notch ligand Delta (Lai 2002), and this modification is essential for Delta to activate Notch in the neighboring cell (Fig. 6). Thus, Neuralized is an activator of Notch signaling while Numb is an inhibitor, and this explains the opposite phenotypes. Whether Neuralized has a function in neuroblasts as well is currently unclear.
Pros is a homeodomain transcription factor that transiently associates with the coiled-coil protein Miranda (Mira) during mitosis (Fig. 6; Ikeshima-Kataoka et al. 1997; Shen et al. 1997). Together with Mira, Pros segregates into one of the two daughter cells, where it dissociates from Mira and enters the nucleus (Hirata et al. 1995; Knoblich et al. 1995; Spana and Doe 1995). In SOP cells, Pros plays only a minor role (Reddy and Rodrigues 1999). In the neuroblast lineage, however, it is a major determinant of the GMC fate. During embryogenesis, mutations in pros lead to the loss of differentiated neurons (Doe et al. 1991) and to the transformation of GMCs into ectopic neuroblasts (Choksi et al. 2006). In the larval CNS, pros mutant neuroblasts give rise to tumors consisting of neuroblasts that have lost their ability to generate differentiating neurons (Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c). A technique called DamID (DNA adenine methyltransferase identification) has been used to determine the nuclear-binding sites on a genome-wide level (Choksi et al. 2006). This technique involves the expression of a fusion protein between Pros and the Escherichia coli adenine methyltransferase Dam in transgenic flies. Pros will target Dam to its endogenous binding sites, which can then be identified upon DNA digestion with a methylation-sensitive restriction endonuclease. Among the identified Pros targets are key cell cycle regulators and genes required for neuroblast self-renewal, but also genes involved in neuronal differentiation. Microarray analysis reveals that Pros can act as both a transcriptional activator of cell cycle genes and an inhibitor of differentiation genes (Choksi et al. 2006). Thus, Pros modulates the transcription pattern of the small neuroblast daughter to exit the cell cycle and enter a differentiation pathway.
The most recently identified segregating determinant is called Brat (Fig. 6; Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c). Like Pros, Brat binds to Mira in mitosis and cosegregates into the GMC during neuroblast and SOP division. In SOP cells, Brat is not required for asymmetric division, but in embryonic neuroblasts, it acts redundantly with Pros to induce neuronal differentiation (Betschinger et al. 2006). In larval neuroblasts, Brat acts as a tumor suppressor, and in brat mutants, neuroblasts overproliferate at the expense of differentiating neurons. In contrast to pros, however, brat is required only in type II CB neuroblasts. These neuroblasts do not express pros, and this may be why they are particularly sensitive to the loss of other determinants like brat. Brat is particularly interesting because it inhibits proliferation also in symmetrically dividing cells. When overexpressed in epithelial cells, brat reduces the size of the nucleolus and inhibits mitotic proliferation (Frank et al. 2002). Since the nucleolus is the site of ribosomal RNA biogenesis, a reduction of overall protein biosynthesis rates may explain the growth inhibitory function of Brat.
Brat is a member of a conserved protein family whose common function may be the control of stem cell proliferation. One or more N-terminal B boxes, a coiled-coil region, and a C-terminal NHL (NCL-1, HT1A, and LIN-41) domain characterize these proteins (Fig. 6; Reymond et al. 2001). In Drosophila, at least one other member of this family besides brat acts as a tumor suppressor. This protein is called Mei-P26, and it controls differentiation and cell growth in the Drosophila ovary (Page et al. 2000; Neumuller et al. 2008). Drosophila ovarian stem cells normally generate one daughter cell that retains stem cell identity and one so-called cystoblast that switches to a transit-amplifying division mode with incomplete cytokinesis (Wong et al. 2005). While stem cells maintain their size over many cell divisions, cystocytes (the daughter cells of the cystoblast) become smaller because cell growth is no longer coupled to cell division. Mei-P26 is specifically expressed in cystocytes and reaches a peak at the end of mitotic proliferation. In mei-P26 mutants, cystocytes maintain their size during the transit-amplifying divisions and continue to divide mitotically, leading to the formation of an ovarian tumor. Like brat, mei-P26 reduces the size of the nucleolus, suggesting that inhibition of protein biosynthesis may be a common function of the Brat/Mei-P26 protein family. This function seems to be conserved as a role in proliferation control, and regulation of the nucleolus has also been described for the mouse homolog TRIM32 (Schwamborn et al. 2009) and the C. elegans homolog Ncl-1 (Frank and Roth 1998) as well.
The molecular function of brat in neuroblasts is unclear. Brat has been suggested to regulate the activity of Pros (Bello et al. 2006), but this is unlikely since brat tumors arise in type II CB neuroblasts where pros is not expressed (Bowman et al. 2008). During early embryogenesis, brat forms an RNA-binding complex with Nanos and Pumilio. This complex inhibits the translation of a protein called Hunchback and is involved in establishing the anterior–posterior body axis (Sonoda and Wharton 2001). So far, however, a role for this complex in neuroblasts has not been demonstrated. Some hints as to how brat may act come from the analysis of its relatives, Mei-P26 and TRIM32. Both proteins act as inhibitors of the transcription factor Myc. They contain an N-terminal RING finger domain that has ubiquitin ligase activity and targets Myc for proteasomal degradation. Brat does not have a RING finger domain, but it might regulate Myc in another way. Brat (Neumuller et al. 2008), Mei-P26 (Neumuller et al. 2008), TRIM32 (Schwamborn et al. 2009), and their C. elegans homolog NHL-2 (Hammell et al. 2009) can bind to the RNase Argonaute-1 (Ago1) via their C-terminal NHL domain. Ago1 is a key component of the microRNA (miRNA) pathway, and, indeed, NHL-2 and TRIM32 were shown to activate certain miRNAs. Mei-P26 regulates miRNAs as well, but in this case, the effect is inhibitory. As we do not know the mechanistic basis for these regulatory effects, the reasons for those differences are currently unclear. In any case, determining whether Brat inhibits proliferation through miRNAs as well will be an interesting question to answer.
Asymmetric localization of Drosophila cell fate determinants
As in C. elegans, the asymmetric segregation of cell fate determinants in Drosophila involves the Par-3/6/aPKC complex. Par-3/6 and aPKC localize apically in neuroblasts (Schober et al. 1999; Wodarz et al. 1999, 2000; Petronczki and Knoblich 2001) and posteriorly in SOP cells (Bellaiche et al. 2001b). In SOP cells, Par protein polarity follows planar polarity of the overlying epithelium. Embryonic neuroblasts arise from a polarized epithelium, and the apical localization of Par-3/6 and aPKC is simply maintained during the delamination process and the first mitotic division. This directs the first division along the apical–basal axis. During subsequent divisions, Par-3/6/aPKC localization is maintained through an extracellular signal that is received via a cadherin-mediated contact with the overlying epidermis (Siegrist and Doe 2006). The molecular nature of this signal is unknown. In larval neuroblasts, the mechanisms that set up and maintain polarity are less clear. One of the two centrosomes occupies an invariant position between individual division cycles (Rebollo et al. 2007), and this may serve as a positional cue for orienting asymmetric cell divisions along the same polarity axis in this cell type. Thus, the Par proteins direct asymmetric cell division in Drosophila as well, but in contrast to C. elegans, their initial polarization is determined by pre-existing polarity in the surrounding tissues.
How the Par protein complex directs the asymmetric localization of cell fate determinants has only recently become clear. Brat and Pros localize by binding to the adaptor protein Mira (Ikeshima-Kataoka et al. 1997; Shen et al. 1997; Matsuzaki et al. 1998; Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c). Numb binds to the adaptor protein Pon (Partner of Numb), but, unlike Mira, Pon is not essential for the asymmetric localization of its binding partner (Lu et al. 1998). Thus, Numb, Mira, and, to some extent, Pon are the key targets of Par proteins during asymmetric cell division.
Numb and Mira localization is microtubule-independent (Knoblich et al. 1995) but requires the actin cytoskeleton (Knoblich et al. 1997; Shen et al. 1998). Genetic experiments have suggested that two myosin motors might be responsible for directional transport of the two proteins along the cell cortex: Inhibition of myosin II activity by a Rho kinase inhibitor prevents Mira localization (Barros et al. 2003). In addition, myosin VI is required for localization of Mira (Petritsch et al. 2003) but not Pon (Erben et al. 2008). However, photobleaching experiments did not reveal any evidence for directional transport along the cell cortex (Lu et al. 1999; Mayer et al. 2005). Instead, these experiments show that Numb and Pon exchange rapidly between plasma membrane and cytoplasm, and suggest that local differences in cortical affinity are responsible for the apparent asymmetric localization of these proteins. As it turns out, cell cycle-dependent phosphorylation of Numb, Pon, and Mira regulates those differences in plasma membrane affinity.
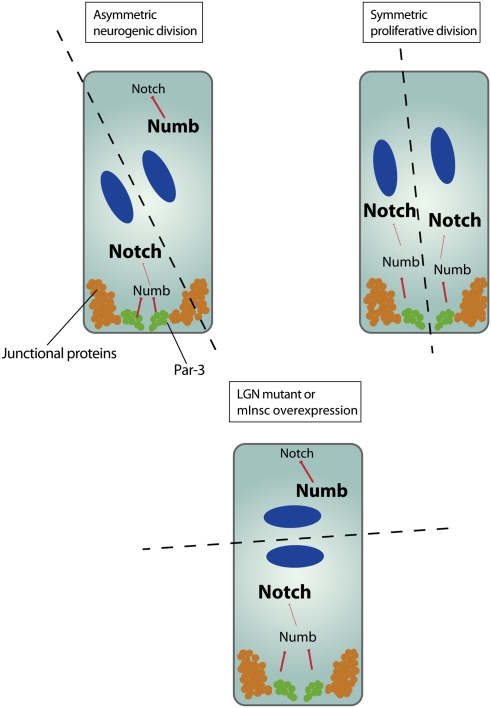
The plasma membrane affinity of Numb is regulated by phosphorylation. Numb is attached to membranes through its N terminus (Knoblich et al. 1997). The positively charged N-terminal region of Numb that is responsible for membrane association contains three aPKC phosphorylation sites. Phosphorylation of those sites neutralizes the charges and prevents membrane localization of Numb (Smith et al. 2007). Interestingly, a nonphosphorylatable form of Numb is cortical but fails to localize asymmetrically, indicating that aPKC phosphorylation might be responsible for the asymmetric localization of Numb. In a simple model, Numb is phosphorylated by aPKC on one side of the cell, and therefore concentrates on the opposite side of the plasma membrane (Fig. 7A; Smith et al. 2007; Wirtz-Peitz et al. 2008).
Figure 7.
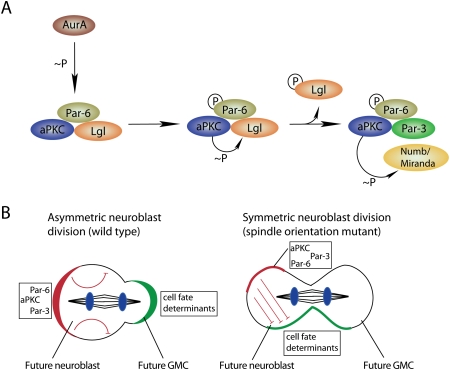
The apical domain regulates cell fate determinant localization and activity. (A) Aurora-A (AurA)-induced complex remodeling leads to the phosphorylation-mediated exclusion of the Lgl form and entry of Par-3 into the Par complex. This complex remodelling is associated with an alteration of the aPKC substrate specificity toward Numb and Miranda, which are hence excluded from the cortical domain occupied by aPKC. (B) The ratio of apical/basal determinants specifies cellular identity. Asymmetric neuroblast divisions can occur, although both daughter cells inherit cell fate determinants (green) that are inhibited by aPKC (red) in one daughter cell (DNA, blue).
Why is Numb only asymmetric in mitosis, although aPKC is asymmetric already in interphase? Recent experiments have shown that aPKC substrate specificity is regulated in a cell cycle-dependent manner by the mitotic kinase Aurora A (Fig. 7A). aPKC is part of two protein complexes (Yamanaka et al. 2006): In mitosis, it binds to Par-6 and Par-3. In interphase, Par-6 is still in the complex, but Par-3 is replaced by Lgl [Lethal (2) giant larvae], a WD40 repeat protein (Betschinger et al. 2005) that was originally identified as a tumor suppressor in Drosophila (Gateff 1978; Mechler et al. 1985). Biochemical experiments show that only the Par-3-containing form of the complex can phosphorylate Numb, while the Lgl complex has almost no activity (Wirtz-Peitz et al. 2008). This can be explained because Par-3 can bind to Numb and might act as an adaptor, allowing aPKC to recognize Numb as a substrate. Transition between the two complexes is triggered by Aurora A. At the onset of mitosis, when Aurora A becomes active, it phosphorylates Par-6 on a residue involved in aPKC binding. Through some conformational change, this increases the activity of aPKC, and the first protein that is phosphorylated is Lgl. Phosphorylated Lgl no longer binds aPKC, and this allows Par-3 to bind and Numb to be phosphorylated. Thus, a cascade of phosphorylation events initiated by the mitotic kinase Aurora A is responsible for the asymmetric localization of Numb.
Besides clarifying Numb localization, this mechanism could also explain why the segregation of aPKC into the daughter neuroblast is important for cell fate specification (Rolls et al. 2003; Lee et al. 2006b; Cabernard and Doe 2009). Since Numb acts at the plasma membrane, phosphorylation should prevent its inhibitory action on the Notch pathway. Therefore, aPKC should not only localize but also inhibit the downstream functions of Numb. Immediately after mitosis, aPKC is bound to Par-3, and therefore it can phosphorylate any residual Numb protein that is accidentally inherited by the neuroblast daughter. This hypothesis is supported by several genetic interactions between Numb and components of the apical Par-3/6/aPKC complex (Wirtz-Peitz et al. 2008). It could explain why neuroblast divisions are completely normal even when aPKC is asymmetric, but basal determinants are inherited by both daughter cells in mutants affecting spindle orientation (Fig. 7B; Cabernard and Doe 2009). Thus, the asymmetric inheritance of aPKC provides a backup mechanism to ensure asymmetric cell division even when determinant localization fails.
How are the other determinants localized? Like Numb, Mira is a substrate for aPKC (Wirtz-Peitz et al. 2008), and aPKC phosphorylation is important for Mira localization as well (Fig. 7A; Atwood and Prehoda 2009). Neuralized carries an N-terminal myristoylation signal followed by a positively charged domain that binds to membrane phospholipids (Skwarek et al. 2007). Consensus sites for aPKC phosphorylation are located near this domain, and it is quite conceivable that its localization mechanism is highly similar to that of Numb. Pon can also be phosphorylated by aPKC (Wirtz-Peitz et al. 2008), but is also a substrate of the mitotic kinase Polo (Wang et al. 2007). Polo is required for Pon and Numb localization and it acts as a tumor suppressor. Therefore, phosphorylation by Polo might provide a secondary regulatory signal to connect determinant localization with cell cycle progression. Since Polo is asymmetrically localized in C. elegans (where it is called PLK-1) and is required for asymmetric cell division as well (Budirahardja and Gonczy 2008; Rivers et al. 2008), this role may well be conserved in evolution.
Asymmetric protein localization in Drosophila also requires the phosphatases PP2A and PP4 (Chabu and Doe 2009; Krahn et al. 2009; Sousa-Nunes et al. 2009; Wang et al. 2009). PP4 is essential for centrosomes to nucleate astral microtubules (Helps et al. 1998), and acts with its regulatory subunit, PP4R3 (called Falafel [Flfl] in flies), in the asymmetric localization of Mira (Sousa-Nunes et al. 2009). PP2A works together with its regulatory subunit, Twins, to regulate the asymmetric localization of aPKC and Numb and the orientation of the mitotic spindle (Chabu and Doe 2009; Wang et al. 2009). PP2A may revert the phosphorylation events catalyzed by Aurora-A or Polo. Interestingly, PP2A can also revert the phosphorylation of Par-3 by Par-1 (Krahn et al. 2009), and its absence can lead to a complete reversal of neuroblast polarity in Drosophila embryos. How this surprising phenotype fits with the regulatory networks described so far will need to be determined.
Telophase rescue
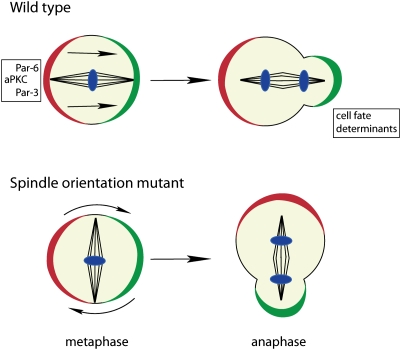
Although the initial asymmetric localization of cell fate determinants is microtubule-independent, a second, microtubule-dependent mechanism called “telophase rescue” ensures the asymmetric segregation of Numb or Mira even when they are not asymmetric in metaphase (Fig. 8; Schober et al. 1999; Wodarz et al. 1999; Peng et al. 2000). While cortical polarity normally instructs both spindle orientation and determinant localization, in this case, astral microtubules emanating from the centrosome to the plasma membrane instruct polarization of the cell cortex. Telophase rescue requires the membrane-bound guanylate kinase Dlg (discs large) and its interaction partner, Khc-73, a kinesin motor heavy chain (Siegrist and Doe 2005). Khc-73 localizes to microtubule plus ends, and its motor activity may act on the cortical actin cytoskeleton to focus cortical protein localization. The pathway is clearly required in mutants affecting cortical polarity (Siegrist and Doe 2005) or spindle orientation (Bowman et al. 2006; Izumi et al. 2006; Siller et al. 2006). The fact that mutations affecting astral microtubules cause an occasional missegregation of determinants indicates that the pathway is important during normal asymmetric cell division as well (Basto et al. 2006).
Figure 8.
Spindle orientation defects in metaphase are frequently corrected by the “telophase rescue” pathway in anaphase/telophase. This rescue pathway restores the correct cell fate determinant segregation in late cell cycle phases in a majority of divisions in spindle orientation mutants (DNA, blue).
Drosophila and C. elegans: variations on a common theme
The asymmetric localization of cell fate determinants during asymmetric cell division in C. elegans and Drosophila follows similar conserved principles. In both systems, initial asymmetric localization of cell fate determinants is established by differential regulation of mobility in different parts of the cell. In Drosophila, Numb is immobilized on the plasma membrane on one side of the cell but not the other. In C. elegans, diffusion of cell fate determinants is limited in one-half of the cytoplasm, possibly by differential association with ribonucleoprotein particles. It is intriguing to speculate that phosphorylation by an asymmetrically localized protein kinase (PKC-3 or PAR-1) might be responsible for the difference in cytoplasmic diffusion rates in C. elegans as well.
In both C. elegans and Drosophila, the initial asymmetry is enhanced by a secondary mechanism. In C. elegans, this involves differential regulation of protein stability in the anterior and posterior compartments. In Drosophila, interactions of astral microtubules with the cell cortex are responsible for refinement of cortical determinant localization. Thus, principally similar mechanisms are employed to achieve asymmetric localization in the cytoplasm or at the membrane in the two systems.
Asymmetric inheritance of damaged and misfolded proteins
Several cellular components are asymmetrically inherited, although they do not act in cell fate determination. Among these are damaged proteins that arise as by-products of cellular metabolism (Garcia-Mata et al. 2002). Oxygen radicals can lead to carbonylation of amino acids. These modifications are irreversible and accumulate over time. In addition, nonenzymatic Maillard reactions between reduced carbohydrates and proteins generate advanced glycation end products (AGE). Both of these metabolic by-products are increased in neurodegenerative diseases like Alzheimer's or Parkinson's, and, therefore, the mechanisms leading to their elimination are of high medical relevance.
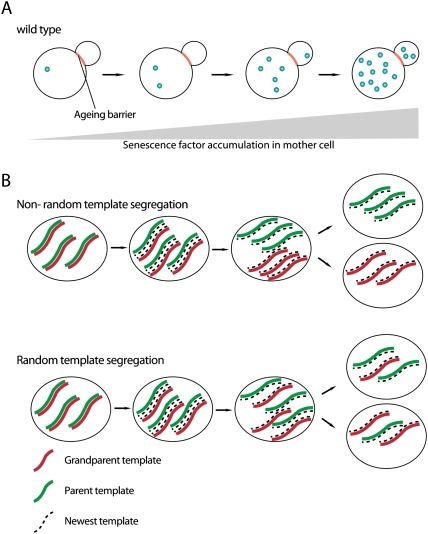
Specific antibodies exist to detect AGE products (Horiuchi et al. 1991). Carbonylated proteins can be visualized by a histochemical reaction generating a product that is recognized by a specific antibody (Aguilaniu et al. 2001). In yeast, carbonylated proteins are inherited by just one of the two daughter cells during mitosis (Aguilaniu et al. 2003). Yeast cells divide by budding. Daughter cells are generated by a protrusion growing from the plasma membrane and constriction of the plasma membrane, as in animal cell cytokinesis. Yeast used to be the leading model system for asymmetric cell division (Horvitz and Herskowitz 1992; Chant 1999), before it turned out that most of the mechanisms are not conserved in higher animals. However, yeast may provide highly useful insights into cellular aging, particularly in stem cells (Tissenbaum and Guarente 2002). Lineage analysis revealed that the two daughter cells generated during yeast division have unequal potential to survive (Fig. 9A). While the mother cell has a limited life span (Mortimer and Johnston 1959) and shows signs of cellular aging (Sinclair et al. 1998), the daughter cell is rejuvenated and will live much longer. Aging in yeast is due to the accumulation of cellular damage. Besides extrachromosomal DNA circles that are a by-product of replication of repetitive sequences, accumulation of damaged proteins is a major factor. Therefore, the asymmetric inheritance of carbonylated proteins may be one of the mechanisms through which yeast cells prevent colony extinction due to cellular senescence of mother and daughter cells. The ability to segregate damaged proteins diminishes in old yeast cells, and this may explain why daughter cells of very old mothers have a shorter replicative life span than daughter cells of young mother cells (Fig. 9A).
Figure 9.
Mitotic asymmetries of senescence factors and DNA. (A) Aging/senescence factors are selectively retained in the mother cell and accumulate during the replicative life span, leading to cellular senescence and death of the mother. Daughter cells born later inherit small amounts of senescence factors, leading to a shortened replicative life span compared with younger daughter cells. (B) Asymmetric versus symmetric segregation of DNA template strands during cell division.
The mechanism by which damaged proteins are asymmetrically inherited is unclear. The actin cytoskeleton is required and so is the histone deacetylase sir-2 (Aguilaniu et al. 2003). This is different for the asymmetric inheritance of extrachromosomal DNA circles. These circles arise from errors during replication of the highly repetitive ribosomal DNA clusters and are a major factor in determining yeast life span (Sinclair and Guarente 1997). During mitosis, they are inherited specifically by the mother cell and cleared from the daughter (Shcheprova et al. 2008). This is because they transiently associate with nuclear pore components during mitosis. In yeast, the nuclear envelope is maintained during mitosis, and elegant live-imaging and photobleaching experiments have demonstrated that a diffusion barrier exists within the nuclear envelope. This barrier allows the nuclear membrane to be extended into the daughter cell, but retains all nuclear pore complexes in the mother. In the daughter, nuclear pores are synthesized de novo. As a consequence, the associated DNA circles are retained in the mother cell, while the daughter is free of this burden.
To what extent is the asymmetric inheritance of cellular waste conserved? The amount of damaged, carbonylated proteins is high in mouse embryonic stem cells (Hernebring et al. 2006). In a developing mouse embryo, damaged proteins are enriched in the inner cell mass from where ES cells originate. Upon differentiation, the amount of carbonylated proteins is significantly reduced due to a proteasomal mechanism that allows animal cells to get rid of damaged proteins altogether. The mechanism involves the proteasome, but its precise molecular nature is unclear. Recent experiments have revealed that signaling intermediates targeted for degradation can be segregated asymmetrically in mitosis (Fuentealba et al. 2008), and it is quite possible that this applies to damaged proteins as well.
Asymmetric inheritance has also been demonstrated for another form of damaged proteins. Proteins that have not been properly folded during biosynthesis accumulate in particulate structures called the aggresomes (Johnston et al. 1998; Garcia-Mata et al. 2002). They typically form in the area surrounding one of the two centrosomes. They can be specifically induced by the expression of Huntingtin or the cystic fibrosis transmembrane conducting regulator CFTR, where particular mutant forms exist that are misfolded, aggregate, and contribute to disease formation. When expressed in various cell lines, those proteins accumulate in a single aggresome that is typically located next to the centrosome (Johnston et al. 1998). In Drosophila embryonic neuroblasts, aggresomes are asymmetrically inherited by the neuroblast daughter cell (Rujano et al. 2006). Although it needs to be demonstrated whether this asymmetry is observed in longer-lived larval neuroblasts as well, this mechanism might protect neurons from those potentially neurotoxic protein aggregates. As neuroblasts are a developmental cell type compared with neurons that persist throughout the entire life span of the fly, clearing damaged proteins from neurons into neuroblasts might have a role in preventing neurodegeneration. Asymmetric segregation of aggresomes is also seen in mouse gut stem cells, but in this case, non-stem cells inherit the structures (Rujano et al. 2006). While it makes a lot of sense to protect stem cells from cellular garbage, it is unclear why the polarity of segregation is inverted compared with Drosophila. In any case, the functional relevance of this potentially interesting phenomenon needs to be determined. Thus, the asymmetric inheritance of damaged or misfolded proteins may also occur in organisms other than yeast, but presently the mechanisms are entirely enigmatic.
Asymmetric inheritance of centrioles and centrosomes
Centrosomes each contain a pair of centrioles surrounded by pericentriolar material. Although the centrosomes at both spindle poles usually appear identical, the history of their individual centriole pairs is different. Centriole pairs split into individual centrioles early in the cell cycle and are then replicated semiconservatively, meaning that, in mitosis, each pair consists of one old and one new centriole. In the next cell cycle, one pair will consist entirely of recently synthesized centrioles, while the other will contain one centriole that can be many cell cycles old. As it turns out, many cell types can distinguish between the resulting old and new centrioles, and, in some cases, it seems like they are distinctly segregated into the two daughter cells.
Asymmetric segregation of microtubule-organizing centers (MTOCs) was first studied in yeast (Pereira et al. 2001). In yeast, the equivalent of the centrosome is called the spindle pole body (SPB). It does not contain centrioles and does not replicate semiconservatively. Instead, the entire SPB is duplicated during each cell cycle, resulting in one old and one new SPB. SPBs can be labeled by GFP fusions to Spc42p, a constitutive and stable component of the SPB. Upon photobleaching, the old SPB will remain unlabeled for several cell cycles, while the newly assembled SPB will recruit Spc42p from the cytoplasm and regain fluorescence after one cell cycle. This technology was used to demonstrate unequivocally that budding daughter cells almost always inherit the old SPB, while the newly synthesized spindle pole always remains in the mother cell (Pereira et al. 2001; Bornens and Piel 2002). What is the biological significance of this asymmetry in SPB behavior? The late stages of mitosis in yeast cells are regulated by two protein networks called the mitotic exit network (MEN) and the separation initiation network (SIN) (Bardin and Amon 2001). Key regulatory components of both networks are known to accumulate on the bud SPB but not the mother cell SPB. It would be attractive to speculate that centrosome asymmetry is responsible for this differential protein recruitment. However, when SPB inheritance is randomized by a transient treatment with microtubule inhibitors, key components of the MEN are still found exclusively on the bud SPB, although this can now be either the old or the new SPB (Pereira et al. 2001). Instead, it is the interaction between the SPB and the cell cortex that is different between the bud and the mother cell, and this difference is responsible for the differential recruitment of regulatory proteins (Pereira et al. 2001). Thus, yeast cells are able to segregate old and new SPBs differentially into the two daughter cells, but the physiological relevance of this asymmetry is unclear. Most likely, the old SPB can simply nucleate microtubules earlier and is therefore more likely to become the target of the microtubule-dependent machinery that moves one SPB into the budding daughter cell.
Asymmetry between the two centrosomes has also been demonstrated in the Drosophila germline (Yamashita et al. 2007). Like female germline stem cells (GSCs), GSCs in the Drosophila testes divide asymmetrically because one daughter cell receives an extracellular signal from the surrounding stem cell niche (Fuller and Spradling 2007). For this, the mitotic spindle in the stem cell needs to be oriented perpendicularly to the niche so that one daughter cell is positioned at maximum distance from the source of the signal. This is achieved by anchoring the centrosome to a DE-cadherin/β-catenin-rich (called armadillo in Drosophila) structure at the stem cell/niche interface throughout the cell cycle (Yamashita et al. 2003). After centriole duplication, it will always be the newly generated centrosome that migrates to the opposite pole, resulting in perpendicular spindle orientation (Yamashita et al. 2007). As a consequence, it is always the centrosome containing the old centriole that is inherited by the stem cell daughter (Yamashita et al. 2007). Although the functional relevance of this asymmetric centrosome inheritance is unclear, it is tempting to speculate that the permanent inheritance of a centriole contributes to the ability of stem cells to proliferate forever (Spradling and Zheng 2007).
A similar mode of centrosome inheritance might be used by Drosophila neuroblasts (Castellanos et al. 2008). Early reports demonstrated that, during the first division of embryonic neuroblasts, mitotic spindles rotate in metaphase to achieve their correct orientation (Kaltschmidt et al. 2000). During subsequent divisions and in larval neuroblasts, however, subsequent live-imaging studies revealed a different mode of orientation (Rebollo et al. 2007, 2009). Similar to male GSCs, one centriole remains anchored on a fixed position at the cell cortex. After centrosome duplication, one of the two centrioles loses its pericentriolar material and migrates to the opposite pole, where it recruits pericentrosomal material and sets up a mitotic spindle, which is already in its final orientation. Although this has not been formally demonstrated, it is quite likely that, in analogy to male GSCs, it is the older centriole that is anchored at a fixed position. In analogy to the germline, this would result in the asymmetric inheritance of the centrioles and the permanent retention of the oldest centriole in the stem cell daughter. Consistently, loss of centrioles in Drosophila DSas-4 mutants is associated with defects in asymmetric cell division of larval neuroblasts. Surprisingly, however, these mutant flies develop into adults, suggesting that centrioles are dispensable for a wide range of developmental processes (Basto et al. 2006).
However, asymmetric centrosome inheritance is not a general feature of all stem cell lineages. In the female Drosophila germline, stem cell division is oriented similarly to males. Despite this similarity, however, centrosomes segregate randomly in this lineage, and, in fact, centrosomes are not required for proper orientation of stem cell division (Stevens et al. 2007). Thus, the asymmetric behavior of centrosomes is not an essential feature of stem cell divisions.
So far, there is no evidence for asymmetric inheritance of centrosomes in mammalian stem cells, although the protein content of older and newer centrosomes can be quite distinct. For example, ɛ-tubulin is found specifically in the pericentriolar material of the old centrosome (Chang and Stearns 2000) while the poly (ADP-ribose) polymerase hPARP3 is found preferentially at the daughter centriole (Augustin et al. 2003). In cell lines expressing a centrin-GFP fusion, newly synthesized centrioles are less intensely labeled, and this allows the observation of mother and daughter centrosome behavior in real time (Piel et al. 2000). After cell division, it is always the mother (older) centrosome that remains near the cell center, while the newer centrosome migrates extensively throughout the cytoplasm. However, those differences disappear as cells go into mitosis and do not result in different behaviors of the two spindle poles. In addition, the mother centriole is known to move toward the cleavage plane during cytokinesis, where it contributes to proper abscission of the two daughter cells (Piel et al. 2001). Whatever the mechanism is by which cells distinguish older and younger centrioles, it will be interesting to determine whether stem cells show phenomena related to centrosome asymmetry in vivo as well.
Asymmetric inheritance of ribosomal components
The rate of cellular growth is highly correlated with overall protein biosynthesis. Rapidly proliferating cells are therefore characterized by extensive protein biosynthesis and a high rate of ribosome biogenesis. Recent live-imaging studies in Drosophila stem cell lineages have shown that ribosomal components can be distributed asymmetrically themselves, and this can contribute to different growth rates in the two daughter cells of an asymmetric division.
When Drosophila female GSCs divide asymmetrically, one daughter cell remains a stem cell and maintains its cell size over many cell divisions. The other daughter cell, the so-called cystoblast, will become smaller with each cell division. Since the nucleolus is much larger in the stem cell (Neumuller et al. 2008), a faster protein biosynthesis rate in the stem cell may contribute to this size difference. Although an extrinsic signal coming from the stem cell niche is primarily responsible for the different fates of the two daughter cells, recent experiments have demonstrated that the protein Wicked is distributed asymmetrically and is inherited preferentially by the daughter cell that retains GSC fate (Fichelson et al. 2009). Wicked encodes for a nucleolar protein required for correct processing of ribosomal RNA (rRNA). In wicked mutants, rRNA intermediates accumulate and, ultimately, GSCs are lost and undergo premature differentiation. It is tempting to speculate that the inability of GSCs to maintain high protein biosynthesis rates required for lifelong self-renewal is the cause of this phenotype. Interestingly, the asymmetric segregation of Wicked is not directed by the stem cell niche signal, but by a cell-intrinsic mechanism. Thus, the asymmetric segregation of core components of the protein biosynthesis machinery can contribute to self-renewal capacity in stem cell lineage. Asymmetric distribution of Wicked is observed in Drosophila neuroblasts as well (Fichelson et al. 2009), and it will be exciting to determine whether mammalian stem cells display asymmetries of this kind as well.
Asymmetric inheritance of DNA
Each round of DNA replication can potentially introduce mutations via incorporation of incorrect nucleotides. While sophisticated repair mechanisms ensure that the mutation rate during each individual S phase is minimal, the problem is more significant in stem cells, which proliferate throughout the lifetime of an animal. One way around this problem would be to retain the template DNA strand in the stem cell and continuously pass on the newly synthesized copy to the more short-lived non-stem cell daughter. This hypothesis was put forward by Cairns in the 1970s (Cairns 1975) and is called the “immortal strand hypothesis” (Fig. 9B). Indeed, several studies have provided evidence for asymmetric segregation of DNA strands in various stem cell lineages, and this has led to a vigorous debate on whether these results are simply artefacts or whether asymmetric DNA segregation is a widespread phenomenon in stem cell lineages (see Lansdorp [2007] and Rando [2007] for an excellent account of the pros and cons of the immortal strand hypothesis).
Currently, evidence for the immortal strand hypothesis relies on the same experimental principle. If stem cells always retain one DNA strand, DNA labeling with radioactive nucleotides or BrdU would always label only one of the two strands and, after two divisions, all labels should be lost. Conversely, when a label is administered before stem cells are generated, both strands are labeled, and the label should be retained by the stem cell forever. This method was first applied to tongue papilla and intestinal epithelia under stress or regenerative conditions where stem cells are thought to divide symmetrically and both strands would be labeled (Potten et al. 1978). While the label was rapidly diluted in most cells, some cells retain the label for very long times, and this was interpreted as evidence for asymmetric segregation of newly synthesized, unlabeled DNA. More recent experiments with muscle tissue cells revealed a similar label retention phenomenon in satellite cells, the stem cells of adult skeletal muscles (Shinin et al. 2006). Here, the use of BrdU allowed direct visualization of asymmetric DNA segregation in dividing cultured cells by immunofluorescence. Perhaps the best evidence for asymmetric DNA segregation comes from experiments in which use of two different DNA labels during two subsequent rounds of replication allowed staining artefacts to be excluded, and revealed that it is always the less differentiated daughter cell that retains the incorporated label (Conboy et al. 2007). Asymmetric DNA segregation also seems to occur in neural stem cells (Karpowicz et al. 2005), whereas use of similar labeling protocols in hair follicle stem cells (Sotiropoulou et al. 2008) or hematopoietic stem cells (Kiel et al. 2007) failed to reveal any evidence for differential segregation of DNA strands. In these cell types, the retention of labeled DNA is rather explained by long-term quiescence of the labeled stem cells (Tumbar et al. 2004; Wilson et al. 2008). Thus, the results described so far would suggest that asymmetric segregation of DNA strands occurs in some but not in other stem cell types.
So far, the molecular mechanisms leading to template strand retention in stem cells are entirely unclear. DNA strands could be distinguished by different labels that are attached during replication. Since DNA replication is bidirectional, however, this would require that the labeling machinery could distinguish replication forks going in opposite directions. Another potential mechanism could involve histone octamers that need to be newly assembled for one of the two sister chromatids. An interesting possibility has been suggested recently in yeast, where the old and the new kinetochore can segregate into different daughter cells under particular conditions (Thorpe et al. 2009). In yeast cells in which the two copies of a key kinetochore protein are labeled by different GFP variants, each of the haploid cells resulting from sporulation will express just one label. The other labeled form, however, is inherited from the diploid mother cell, and this can be used to track inheritance of kinetochores during the first mitotic divisions. This technique reveals that the mother cells preferentially inherit old kinetochores, whereas the budding daughter cells will inherit the newly synthesized copy. Although this would provide an elegant manner to distinguish sister chromatids, the phenomenon is observed only in the “mother cell lineage,” but not in any cell that results from a newly formed bud. Although it is unlikely to be a general phenomenon, it could provide a beautiful explanation for immortal strand segregation.
Thus, we are still far away from understanding mitotic asymmetry of DNA segregation. Although the immortal strand hypothesis is attractive, experimental evidence is limited, and, if it is correct, it will certainly apply to only a subset of stem cell lineages.
Asymmetric inheritance of vesicular compartments
Cell fate determinants that are asymmetrically inherited during mitosis can act as regulators of vesicular trafficking. Numb binds to α-adaptin (Berdnik et al. 2002) and regulates the endocytosis of Sanpodo (O'Connor-Giles and Skeath 2003; Hutterer and Knoblich 2005). Neuralized is an E3 ubiquitin ligase that controls the endocytosis of the notch ligand Delta (Lai et al. 2001). While these proteins regulate the trafficking of specific transmembrane proteins, asymmetric cell divisions can also result in daughter cells with different composition of membrane compartments, or even involve the asymmetric inheritance of vesicular structures into one of the two daughter cells.
The possibility to perform live imaging (Bellaiche et al. 2001a) and the development of whole-mount endocytosis assays (Le Borgne and Schweisguth 2003) have made Drosophila SOP cells the best model system to study vesicular trafficking during asymmetric cell division. Differences in protein trafficking are largely responsible for the different levels of Notch activity that establish pIIa and pIIb fates in the two daughter cells. In particular, the Notch ligand Delta is found mostly in intracellular vesicles, and the numbers of these vesicles are higher in the signal-sending pIIb cell (Le Borgne and Schweisguth 2003). This is because Neuralized is segregated into the pIIb cell, where it stimulates endocytosis by acting as an E3 ubiquitin ligase for Delta. After division, Delta traffics to an apical membrane compartment whose formation requires the exocyst component Sec15 (Jafar-Nejad et al. 2005). Mutations in sec15 cause a transformation of pIIa into pIIb, consistent with a loss of Delta signaling activity. Both pIIa and pIIb contain an apical actin-rich structure (ARS) that consists of numerous microvilli, and this is where most of the Delta-rich vesicles reside. Formation of this structure requires actin but also the Arp2/3 (actin-rich protein 2/3) complex and the actin regulator WASp (Rajan et al. 2009). Like sec15 mutants, arp3 mutants cause cell fate transformations consistent with a loss of Delta signaling activity. Taken together, these data suggest that the ARS is required for formation of the apical membrane compartment through which Delta has to pass in order to activate Notch on the neighboring cell. Although the ARS is found in both pIIa and pIIb, Delta trafficking is distinct. The Rab11-positive recycling endosomal compartment through which endocytosed proteins are transported back to the plasma membrane is much more prominent in pIIb (Emery et al. 2005). This is due to a higher activity of the Rab11-binding protein Nuclear fallout (Nuf) in this cell. Surprisingly, this asymmetry is not established by the Par-3/6/aPKC complex, but follows an independent unidentified mechanism of asymmetric cell division. Thus, differential regulation of whole endocytic compartments can regulate signaling in the daughter cells of an asymmetric cell division.
Cell fate can also be regulated by the differential inheritance of vesicular structures. The SARA endosome is an early endocytic compartment that is enriched for molecules required for Dpp (the Drosophila homolog of TGFb) signaling. It accumulates the membrane-associated protein SARA that is involved in targeting the transcription factor Mad (Mothers against Dpp) to the membrane where it is phosphorylated by the Dpp receptor Tkv (Thick veins) (Bokel et al. 2006). In epithelial cells of the developing Drosophila wing, the SARA endosome transiently associates with the central spindle during cytokinesis and is equally distributed between the two daughter cells, and signaling levels are equal in both daughter cells after division (Bokel et al. 2006). This mechanism allows tissues to maintain constant signaling levels even during active proliferation (Furthauer and Gonzalez-Gaitan 2009). In asymmetrically dividing SOP cells, however, the SARA endosome is excluded from the anterior spindle region during mitosis so that it preferentially segregates into the posterior pIIa daughter cell (Coumailleau et al. 2009). In this case, it does not regulate Dpp signaling, but carries the Notch receptor together with its ligand, Delta. In the pIIa cell, SARA endosomes contain just the extracellular domain of Notch, whereas the intracellular domain is cleaved off and translocates into the nucleus. Thus, the asymmetric segregation of activated receptor ligand complexes ensures that signaling commences immediately after asymmetric cell division in one of the two daughter cells.
Asymmetric segregation of vesicular compartments is not restricted to Drosophila. In C. elegans, early endosomes are asymmetrically localized in an actin–myosin-dependent process during the first division of the zygote, and this leads to their asymmetric segregation into the anterior AB cell (Andrews and Ahringer 2007). Like segregation of the SARA endosome, this process requires the Par-3/6/aPKC complex, and the vesicles are enriched on the side where those Par proteins are. Despite these exciting parallels, the precise vesicular localization pattern during mitosis suggests that different underlying mechanisms act to achieve vesicle asymmetry in Drosophila and C. elegans.
Whether endocytic compartments are also segregated asymmetrically in vertebrates is unclear. SARA-positive signaling endosomes exist in vertebrates as well, but neither precisely symmetric nor asymmetric segregation has been described in mitosis. On the other hand, a number of transmembrane receptors segregate asymmetrically in cultured mitotic hematopoietic stem cells (Beckmann et al. 2007) or in T lymphocytes (Chang et al. 2007). Among these proteins are several markers for early endosomal compartments (Giebel and Beckmann 2007), and it will be interesting whether their asymmetric segregation is functionally important or even uses a mechanism related to what is seen in Drosophila or C. elegans.
Asymmetric cell division and its implications for stem cell biology
Stem cells can generate both self-renewing and differentiating daughter cells. Although different fates could arise in a purely stochastic manner, several studies have shown that intrinsically asymmetric cell divisions in mammalian stem cell lineages do occur (Knoblich 2008). Homologs of the genes regulating asymmetric cell division in flies and worms are involved, but they often act in very different ways. Numb, for example, has a variety of seemingly unrelated functions in different tissues. In muscle stem cells (Shinin et al. 2006) and T lymphocytes (Chang et al. 2007), Numb segregates asymmetrically during mitosis. During brain development, however, Numb regulates E-cadherin recycling and is therefore required for maintaining the integrity of the neuroepithelium (Rasin et al. 2007). In mammary glands, Numb acts as a tumor suppressor (Pece et al. 2004) by binding to the E3 ubiquitin ligase MDM2 and preventing degradation of p53 (Colaluca et al. 2008). Understanding how these various functions of Numb are coordinated within a cell is a major challenge for the future.
The best-studied example for an intrinsically asymmetric division in vertebrates is provided by the developing mouse forebrain (Gotz and Huttner 2005). The developing brain is organized in a layered structure in which proliferating stem cells are apical, facing the lumen of the lateral ventricle, while differentiating neurons are located on the more basal side. Early during development, stem cells divide symmetrically to enlarge the progenitor pool. Starting from day 10 of embryonic development, progenitor cells start expressing glia markers and are therefore called radial glia cells. Their divisions become asymmetric, with one daughter cell retaining radial glia fate and the other migrating toward the basal side of the neuroepithelium to differentiate into a neuron. Alternatively, the more differentiated daughter cell can become a so-called basal progenitor, which typically divides once more to form two differentiating neurons (Haubensak et al. 2004; Noctor et al. 2004).
It was originally thought that the orientation of the mitotic spindle determines the symmetric or asymmetric outcome of the division (Chenn and McConnell 1995), but this hypothesis is no longer accepted in the field. In fact, most asymmetric radial glia divisions occur in a planar orientation, resulting in two daughter cells that are initially located at the apical neuroepithelial surface (Kosodo et al. 2004; Konno et al. 2008). Despite their planar orientation, however, these divisions are highly asymmetric. A basal process extends from the dividing progenitor all the way to the pial surface. During early symmetric divisions, this process is split during cytokinesis (Kosodo et al. 2008). During asymmetric divisions, the process is inherited by just one of the two daughter cells, but the data on its fate-determining role are controversial (Miyata et al. 2001; Noctor et al. 2001). The apical domain of radial glia cells is very narrow, and even slight tilting of the cleavage plane can lead to its asymmetric inheritance by only one of the two daughter cells (Fig. 10; Kosodo et al. 2004). Apical adherens junctions display a layered structure with aPKC and Par-3 on the apical-most side, followed by the zonula adherens protein ZO-1/Afadin, and finally the adherens junction proteins N-cadherin and β-catenin (Marthiens and ffrench-Constant 2009). As a result, spindles can be oriented so that the apical-most proteins Par-3 (Bultje et al. 2009) and aPKC (Marthiens and ffrench-Constant 2009) are asymmetrically inherited, while junctional proteins can still attach both daughter cells to the apical web of the neuroepithelium. In mice lacking the Pins homolog LGN, spindles are oriented randomly, and daughter cells become abnormally localized at a distance from the apical surface because they lose their junctional complexes (Konno et al. 2008). Several studies have demonstrated that the asymmetric inheritance of aPKC and/or Par-3 can contribute to the asymmetric outcome of neural glia divisions (Costa et al. 2008; Bultje et al. 2009).
Figure 10.
Model for cell fate specification in the vertebrate neural stem cells. Neural progenitor cells in the developing brain undergo either neurogenic divisions (in which another progenitor cell and a differentiating neuron are produced) or proliferative divisions (which result in the formation of two progenitor cells). Par-3 is partitioned unequally during neurogenic divisions through cleavage plane orientation (dashed line), and induces differential Notch signaling in the daughter cells by inhibiting Numb, resulting in high levels of Notch signaling in the future progenitor cell and low levels in the future neuron (DNA, blue).
How could the Par proteins act? Par-3 can activate Notch signaling in a Numb-dependent way (Fig. 10; Bultje et al. 2009). In Drosophila, Par-3 acts as an adaptor protein, allowing Numb to be phosphorylated by aPKC (Wirtz-Peitz et al. 2008). Similar observations have been made for mammalian Numb as well (Nishimura and Kaibuchi 2007). It is plausible that Par-3 allows aPKC to phosphorylate Numb in one daughter cell. This could prevent Numb from inhibiting Notch, and the different levels of Notch activity in the two daughter cells could establish different fates. Alternatively, Par-3 could act in localizing a segregating determinant, as it does in Drosophila. The mouse Brat homolog TRIM32 is asymmetrically inherited in dividing radial glia cells (Schwamborn et al. 2009), and it will be interesting to determine whether Par-3 is involved in generating this asymmetry.
The analysis of brain progenitor division demonstrates that biological mechanisms identified in invertebrate models are conserved in vertebrates, but are often used in a more complex manner. Indications for intrinsically asymmetric cell divisions exist in muscle stem cells (Shinin et al. 2006), in skin stem cells (Lechler and Fuchs 2005), and in the hematopoietic system (Chang et al. 2007; Wu et al. 2007), but certainly, some time will pass before we understand the underlying mechanisms at the same level of detail.
Asymmetric cell division and tumor formation
Stem cells seem to play an important role in the formation and maintenance of human tumors (Reya et al. 2001; Clarke and Fuller 2006). When transplanted into immunocompromised mice, only a small subset of the cells in a human tumor can reinitiate tumor formation, and these cells often have stem cell character (Lapidot et al. 1994; Bonnet and Dick 1997; Al-Hajj et al. 2003; Singh et al. 2003). This observation has led to the so-called cancer stem cell hypothesis, which predicts that tumors are maintained by a small population of stem cells that can give rise to all other cells and are the only cells capable of tumor reinitiation (Reya et al. 2001). In an extended version, this hypothesis predicts that stem cells might even be at the origin of a tumor, and, therefore, mutations converting normal to cancerous stem cells might be a root cause of tumor formation (Clarke and Fuller 2006). Although this is currently very speculative for human tumors, evidence for a stem cell origin of colon cancer (Barker et al. 2009) and brain tumors (Kwon et al. 2008) has been obtained recently in mouse model organisms. Thus, it will be important to understand how proliferation is controlled in stem cell lineages and how defects in these controls could potentially lead to tumor formation.
In Drosophila, defects in asymmetric cell division can turn neural stem cells into tumor-initiating cells that recapitulate several of the steps typical for mammalian tumors as well. In fact, many of the key regulators of asymmetric cell division act as tumor suppressors (Caussinus and Gonzalez 2005; Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006a,c; Wang et al. 2006, 2007; Gonzalez 2007; Castellanos et al. 2008), and, in some cases, those genes have actually been identified due to their tumor phenotype (Gateff 1978). Mutations in these tumor suppressor genes typically lead to an overproliferation of neuroblasts during larval stages. As a consequence, brains become enormous in size and the animals eventually die. However, tumor progression can be followed over long times by transplanting tumor tissue into the abdomen of female host flies, where it will continue to grow and kill the host as well (Caussinus and Gonzalez 2005). This transplantation process can be repeated for years, and, in fact, becomes more efficient with each round of transplantation. Although the original tumors have normal karyotypes (Caussinus and Gonzalez 2005; Castellanos et al. 2008), cells eventually become aneuploid and, at the same time, start to metastasize and acquire other tissue identities (Caussinus and Gonzalez 2005). Thus, Drosophila can serve as a model organism to study tumor formation from stem cell lineages.
Typically, tumors form because excess neuroblasts are generated at the expense of differentiating neurons. Therefore, all genes that encode for proteins segregating into the basal GMC (like brat [Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c], numb [Lee et al. 2006a; Wang et al. 2006], pros [Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006c], mira [Betschinger et al. 2006; Lee et al. 2006c], and pon [Wang et al. 2007]) and those that specify the basal domain (like lgl [Lee et al. 2006b]) act as tumor suppressors. Genes that specify the apical domain (like aPKC [Lee et al. 2006b] or par-3 [Wirtz-Peitz et al. 2008]) have the opposite phenotype, leading to neuroblast loss and lineage termination.
One simple interpretation would be that symmetric division of neuroblasts leads to exponential rather than linear proliferation, and this is why neuroblast numbers explode. However, this view is too simple. First, all larval neuroblasts become quiescent during pupal stages of development (Ito and Hotta 1992) or when transplanted into the abdomen of another fly (Caussinus and Gonzalez 2005). Tumor neuroblasts, however, continue to proliferate indefinitely, even during adult stages (Bello et al. 2006) or upon transplantation (Caussinus and Gonzalez 2005). Second, the asymmetry of neuroblast division is not affected per se, since other determinants still segregate normally when one of them is missing and since one daughter cell is still larger than the other, at least in most tumors. Third, tumor formation actually starts with a cell cycle delay in the misspecified GMC, rather than an immediate rapid exponential proliferation (Lee et al. 2006c; Bowman et al. 2008). And finally, a detailed survey of the immediate events leading to tumor formation in brat mutants (Bowman et al. 2008) reveals a stereotyped series of events that lead to the generation of abnormally proliferating tumor-initiating neuroblasts in brat mutants.
In brat mutants, tumors arise from type II CB neuroblasts (Bowman et al. 2008). Unlike all other CB neuroblasts, these do not express the transcription factor ase. Upon division, ase is transcribed in the INP before and during transit-amplifying divisions. In brat mutants, type II neuroblasts still divide asymmetrically, but ase transcription is never initiated. Instead, the newly born INP does not enter mitosis, but remains in interphase for ∼24 h. Between 24 and 48 h however, the INP enters mitosis without transcribing ase, and it seems that this mitosis of the immature precursor is the tumor-initiating event. Afterward, cell proliferation proceeds at a rapid pace, and, eventually, cells no longer respond to growth-inhibitory signals. Thus, the mechanisms leading to tumor formation seem to involve much more than a simple cell fate transformation. It seems as if defects in asymmetric cell division lead to the formation of a cell type that does not occur in wild-type flies and no longer obeys the signals for proliferation control.
What could those signals be? Neuroblasts express a temporal series of transcription factors during embryonic and larval stages (Isshiki et al. 2001; Maurange et al. 2008). During pupal stages, they become smaller in size and, ultimately, they undergo a symmetric division in which Pros enters both daughter cells, leading to terminal differentiation. When certain larval transcription factors of the neuroblast clock are missing (for example, seven up), this does not happen, and neuroblasts proliferate into adulthood. It is quite possible that defects in asymmetric cell division perturb the neuroblast clock, and this is what immortalizes the tumor cells. Alternatively, a symmetric neuroblast division could generate a “rejuvenated” neuroblast that has an immature identity and fails to terminate in pupal stages. Finally, the tumor cells could have the correct temporal identity but simply lack molecules required to receive the hormonal signals that determine the pupal stage. It is interesting that neuroblasts are different in the timing of their cell cycle exit. Mushroom body neuroblasts, a subset of CB neuroblasts that generate the learning and memory centers of the fly brain, stop proliferating only at the end of the pupal period (Ito and Hotta 1992). Interestingly, mushroom body neuroblasts specifically express the transcription factor tailless (tll), and this is required for their prolonged proliferation (Kurusu et al. 2009). tll is an inhibitor of pros expression and is tumorigenic when expressed in other CB neuroblast lineages. Thus, changes in these transcriptional programs could well alter cell cycle control in tumor neuroblasts.
To what extent are those results from flies relevant for mammalian tumor biology? In vertebrates, genetic aberrations are responsible for tumor formation. As early as 1914, Boveri claimed that aneuploidy caused by missegregation of the genetic material is responsible for tumor formation (Harris 2008), and described that changes in centrosome number could be responsible for this effect. Only recently, this hypothesis has been experimentally verified. In mammalian tissue culture cells, the presence of extra centrosomes leads to misattachment of chromosomes to the mitotic spindle and to chromosome missegregation and chromosomal instability (Ganem et al. 2009). In Drosophila, an increase in centrosome number can lead to tumor formation as well (Basto et al. 2008; Castellanos et al. 2008), but, surprisingly, this effect is seen only in neuroblasts and not in symmetrically dividing cells (Castellanos et al. 2008). It is not enhanced by mutations causing genomic instability (Castellanos et al. 2008), and therefore is attributed to a defect in asymmetric cell division rather than chromosome missegregation. Excess centrosomes cause defects in aligning the spindle with polarity axis (Basto et al. 2008), and, consequently, to missegregation of determinants or of aPKC (Cabernard and Doe 2009) during mitosis. Precisely how the resulting cell fate changes lead to tumor formation is not understood, but it will certainly be interesting to test whether a comparable effect is seen in mammalian stem cell lineages as well. Vertebrate homologs of genes regulating asymmetric cell division in Drosophila like Numb (Pece et al. 2004) and Lgl (Schimanski et al. 2005) have been demonstrated to act as tumor suppressors. Whether or not they act on stem cell lineages and their asymmetry is currently unclear. It is known that tumor stem cells generate more stem cell-like daughters than their “normal” counterparts (Singh et al. 2004). Finding out whether this is due to defects in asymmetric cell division and whether those defects are crucial for tumor formation in vertebrates as well will certainly be an exciting challenge for the future.
Future perspectives
Over the past years, many of the key questions in asymmetric cell division have been solved. In C. elegans and in Drosophila, we know how polarity is set up during mitosis, and we have a good understanding of the mechanisms that align the mitotic spindle and result in spindle displacement. Recently, it also became clear that asymmetric phosphorylation is the key mechanism behind the asymmetric segregation of cell fate determinants in mitosis. Does this mean the puzzle is solved?
Certainly not! Despite these major advances, we still lack a molecular understanding of many of the processes involved. How is polarity inherited over several divisions? How do G proteins connect to molecular motors and increase their force-generating capacity? How are cytoskeletal asymmetries of the actin cytoskeleton coupled to asymmetric phosphorylation events that seem to act on segregating determinants? And finally, how is one of the daughter nuclei reprogrammed to become different from the sister cell? Determinants like Numb disappear some time after mitosis, but cell fate does not revert. What are the mechanisms that send one cell down a distinct pathway, and how do those become irreversible? And what goes wrong when defects in asymmetric cell division lead to the formation of a tumorous cell type that proliferates indefinitely and kills the whole organism?
Finally, however, we still have no real clue of how asymmetric cell division is regulated in mammalian adult stem cell lineages. One major problem is that, even in model organisms, all experiments have been done during development so far. Adult stem cells in flies have only recently been discovered in the midgut (Micchelli and Perrimon 2006; Ohlstein and Spradling 2006), and it will be interesting to test whether they use mechanisms similar to neuroblast division as well. In fact, gut stem cells can react dynamically to damage and infection (Jiang et al. 2009), and purely intrinsic mechanisms seem too inflexible, so that interesting variations of the classical theme might be expected.
Understanding the contribution of asymmetric cell division to mammalian development and organ homeostasis will be the primary goal, and, with better understanding and improved lineage analysis and live-imaging tools, the goal of uncovering the key mechanisms comes within reach.
Acknowledgments
We thank all of the members of the Knoblich laboratory for helpful discussions and comments on the topics reviewed in this paper. Work in J.A.K.’s laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund (FWF), the EU network ONCASYM, and the EU Framework 7-funded EUROSYSTEM.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1850809.
References
- Abrash EB, Bergmann DC. Asymmetric cell divisions: A view from plant development. Dev Cell. 2009;16:783–796. doi: 10.1016/j.devcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gonczy P. RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Afshar K, Willard FS, Colombo K, Siderovski DP, Gonczy P. Cortical localization of the Gα protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132:4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Protein oxidation in G0 cells of Saccharomyces cerevisiae depends on the state rather than rate of respiration and is enhanced in pos9 but not yap1 mutants. J Biol Chem. 2001;276:35396–35404. doi: 10.1074/jbc.M101796200. [DOI] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews R, Ahringer J. Asymmetry of early endosome distribution in C. elegans embryos. PLoS One. 2007;2:e493. doi: 10.1371/journal.pone.0000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Prehoda KE. aPKC phosphorylates miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: What's the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Le Borgne R, Schweisguth F. Asymmetric localization and function of cell-fate determinants: A fly's view. Curr Opin Neurobiol. 2004;14:6–14. doi: 10.1016/j.conb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle Myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–840. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Scheitza S, Wernet P, Fischer JC, Giebel B. Asymmetric cell division within the human hematopoietic stem and progenitor cell compartment: Identification of asymmetrically segregating proteins. Blood. 2007;109:5494–5501. doi: 10.1182/blood-2006-11-055921. [DOI] [PubMed] [Google Scholar]
- Beers M, Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development. 2006;133:3745–3754. doi: 10.1242/dev.02544. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Gho M, Kaltschmidt JA, Brand AH, Schweisguth F. Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol. 2001a;3:50–57. doi: 10.1038/35050558. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, Radovic A, Woods DF, Hough CD, Parmentier M, O'Kane CJ, Bryant PJ, Schweisguth F. The partner of inscuteable/discs-large complex is required to establish planar polarity during asymmetric cell division in Drosophila. Cell. 2001b;106:355–366. doi: 10.1016/s0092-8674(01)00444-5. [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1156/1749-8104-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetka W, Mehlmer N, Maurer-Stroh S, Sammer M, Koranda M, Neumuller R, Betschinger J, Knoblich JA, Teige M, Eisenhaber F. Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle. 2008;7:3709–3719. doi: 10.4161/cc.7.23.7176. [DOI] [PubMed] [Google Scholar]
- Benton R, Johnston DS. Drosophila PAR-1 and 14–3–3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein α-adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Eisenhaber F, Knoblich JA. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol. 2005;15:276–282. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bokel C, Schwabedissen A, Entchev E, Renaud O, Gonzalez-Gaitan M. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 2006;314:1135–1139. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Piel M. Centrosome inheritance: Birthright or the privilege of maturity? Curr Biol. 2002;12:R71–R73. doi: 10.1016/S0960-9822(01)00678-9. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA homolog mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Bray D, White JG. Cortical flow in animal cells. Science. 1988;239:883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- Budirahardja Y, Gonczy P. PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development. 2008;135:1303–1313. doi: 10.1242/dev.019075. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein A, Shi S-H. Mammalian Par3 regulates progenitor cell asymmetric division via Notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Carmena A, Murugasu-Oei B, Menon D, Jimenez F, Chia W. Inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes & Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–1214. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- Chabu C, Doe CQ. Twins/PP2A regulates aPKC to control neuroblast cell polarity and self-renewal. Dev Biol. 2009;330:399–405. doi: 10.1016/j.ydbio.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Stearns T. δ-Tubulin and ɛ-tubulin: Two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: Cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cismowski MJ, Takesono A, Bernard ML, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-proteins. Life Sci. 2001;68:2301–2308. doi: 10.1016/s0024-3205(01)01019-0. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: Two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin EG. The organization and cell-lineage of the ascidian egg. J Acad Nat Sci Phila. 1905;13:1–119. [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004a;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004b;431:92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- Daniels BR, Perkins EM, Dobrowsky TM, Sun SX, Wirtz D. Asymmetric enrichment of PIE-1 in the Caenorhabditis elegans zygote mediated by binary counterdiffusion. J Cell Biol. 2009;184:473–479. doi: 10.1083/jcb.200809077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NB, Martin CA, Segalen M, Rosenfeld F, Schweisguth F, Bellaiche Y. Drosophila Ric-8 regulates Gαi cortical localization to promote Gαi-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat Cell Biol. 2005;7:1083–1090. doi: 10.1038/ncb1319. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: Balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Develop. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric rab11 endosomes regulate Delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Erben V, Waldhuber M, Langer D, Fetka I, Jansen RP, Petritsch C. Asymmetric localization of the adaptor protein Miranda in neuroblasts is achieved by diffusion and sequential interaction of Myosin II and VI. J Cell Sci. 2008;121:1403–1414. doi: 10.1242/jcs.020024. [DOI] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant JA, Bellaiche Y, Huynh JR. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat Cell Biol. 2009;11:685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Roth MB. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J Cell Biol. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: Two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Gonzalez-Gaitan M. Endocytosis, asymmetric cell division, stem cells and cancer: Unus pro omnibus, omnes pro uno. Mol Oncol. 2009;4:339–353. doi: 10.1016/j.molonc.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: Aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- Giebel B, Beckmann J. Asymmetric cell divisions of human hematopoietic stem and progenitor cells meet endosomes. Cell Cycle. 2007;6:2201–2204. doi: 10.4161/cc.6.18.4658. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cleavage furrow positioning. J Cell Biol. 2004;164:347–351. doi: 10.1083/jcb.200310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- Gotta M, Ahringer J. Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B, Knoblich JA. Heterotrimeric G proteins: New tricks for an old dog. Cell. 2004;119:453–456. doi: 10.1016/j.cell.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Hampoelz B, Hoeller O, Bowman SK, Dunican D, Knoblich JA. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nat Cell Biol. 2005;7:1099–1105. doi: 10.1038/ncb1318. [DOI] [PubMed] [Google Scholar]
- Hao Y, Boyd L, Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. Standing on Boveri's shoulders. J Cell Sci. 2008;121:3. doi: 10.1242/10.1242/jcs.017483. [DOI] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps NR, Brewis ND, Lineruth K, Davis T, Kaiser K, Cohen PT. Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J Cell Sci. 1998;111:1331–1340. doi: 10.1242/jcs.111.10.1331. [DOI] [PubMed] [Google Scholar]
- Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess HA, Roper JC, Grill SW, Koelle MR. RGS-7 completes a receptor-independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell. 2004;119:209–218. doi: 10.1016/j.cell.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Horiuchi S, Araki N, Morino Y. Immunochemical approach to characterize advanced glycation end products of the Maillard reaction. Evidence for the presence of a common structure. J Biol Chem. 1991;266:7329–7332. [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Hutterer A, Knoblich JA. Numb and α-adaptin regulate Sanpodo endocytosis to specify cell fate in Drosophila external sensory organs. EMBO Rep. 2005;6:836–842. doi: 10.1038/sj.embor.7400500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: A cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Itoh-Furuya A, Fuse N, Matsuzaki F. Differential functions of G protein and Baz–aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol. 2004;164:729–738. doi: 10.1083/jcb.200309162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of drosophila sensory organ precursors. Dev Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Johansson A, Driessens M, Aspenstrom P. The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J Cell Sci. 2000;113:3267–3275. doi: 10.1242/jcs.113.18.3267. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Cheng V, van der Kooy D. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: A tale of destruction and degradation. Trends Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- Kirilly D, Xie T. The Drosophila ovary: An active stem cell community. Cell Res. 2007;17:15–25. doi: 10.1038/sj.cr.7310123. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. The N terminus of the Drosophila Numb protein directs membrane association and actin-dependent asymmetric localization. Proc Natl Acad Sci. 1997;94:13005–13010. doi: 10.1073/pnas.94.24.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Toida K, Dubreuil V, Alexandre P, Schenk J, Kiyokage E, Attardo A, Mora-Bermudez F, Arii T, Clarke JD, et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008;27:3151–3163. doi: 10.1038/emboj.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical–basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16:901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Maruyama Y, Adachi Y, Okabe M, Suzuki E, Furukubo-Tokunaga K. A conserved nuclear receptor, Tailless, is required for efficient proliferation and prolonged maintenance of mushroom body progenitors in the Drosophila brain. Dev Biol. 2009;326:224–236. doi: 10.1016/j.ydbio.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe JC, Maddox PS, Salmon ED, Goldstein B. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr Biol. 2003;13:707–714. doi: 10.1016/s0960-9822(03)00251-3. [DOI] [PubMed] [Google Scholar]
- Lai EC. Protein degradation: Four E3s for the notch pathway. Curr Biol. 2002;12:R74–R78. doi: 10.1016/S0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Unequal segregation of neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes & Dev. 2006a;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006b;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006c;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Li SC, Zwahlen C, Vincent SJ, McGlade CJ, Kay LE, Pawson T, Forman-Kay JD. Structure of a Numb PTB domain–peptide complex suggests a basis for diverse binding specificity. Nat Struct Biol. 1998;5:1075–1083. doi: 10.1038/4185. [DOI] [PubMed] [Google Scholar]
- Lin H. The stem-cell niche theory: Lessons from flies. Nat Rev Genet. 2002;3:931–940. doi: 10.1038/nrg952. [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- Lu B, Rothenberg M, Jan LY, Jan YN. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- Lu B, Ackerman L, Jan LY, Jan YN. Modes of protein movement that lead to the asymmetric localization of partner of Numb during Drosophila neuroblast division. Mol Cell. 1999;4:883–891. doi: 10.1016/s1097-2765(00)80218-x. [DOI] [PubMed] [Google Scholar]
- Marthiens V, ffrench-Constant C. Adherens junction domains are split by asymmetric division of embryonic neural stem cells. EMBO Rep. 2009;10:515–520. doi: 10.1038/embor.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Mayer B, Emery G, Berdnik D, Wirtz-Peitz F, Knoblich JA. Quantitative analysis of protein dynamics during asymmetric cell division. Curr Biol. 2005;15:1847–1854. doi: 10.1016/j.cub.2005.08.067. [DOI] [PubMed] [Google Scholar]
- Mechler BM, McGinnis W, Gehring WJ. Molecular cloning of lethal(2)giant larvae, a recessive oncogene of Drosophila melanogaster. EMBO J. 1985;4:1551–1557. doi: 10.1002/j.1460-2075.1985.tb03816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FL, Scheres B. Plant asymmetric cell division, vive la difference! Cell. 2009;137:1189–1192. doi: 10.1016/j.cell.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 gene encodes a 14–3–3 protein required for cellular asymmetry in the early embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- Motegi F, Sugimoto A. Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat Cell Biol. 2006;8:978–985. doi: 10.1038/ncb1459. [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport par proteins to establish and maintain anterior–posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and Gα proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- O'Connor-Giles KM, Skeath JB. Numb inhibits membrane localization of sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohno S. Intercellular junctions and cellular polarity: The PAR–aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155:1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p–Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN. The Drosophila myosin VI jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Dev Cell. 2003;4:273–281. doi: 10.1016/s1534-5807(03)00020-0. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- Qiu RG, Abo A, Steven Martin G. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCζ signaling and cell transformation. Curr Biol. 2000;10:697–707. doi: 10.1016/s0960-9822(00)00535-2. [DOI] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA. The immortal strand hypothesis: Segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Roldan M, Gonzalez C. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 2009;136:3393–3397. doi: 10.1242/dev.041822. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Rodrigues V. Sibling cell fate in the Drosophila adult external sense organ lineage is specified by prospero function, which is regulated by Numb and Notch. Development. 1999;126:2083–2092. doi: 10.1242/dev.126.10.2083. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Rivers DM, Moreno S, Abraham M, Ahringer J. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol. 2008;180:877–885. doi: 10.1083/jcb.200710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Gα-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schimanski CC, Schmitz G, Kashyap A, Bosserhoff AK, Bataille F, Schafer SC, Lehr HA, Berger MR, Galle PR, Strand S, et al. Reduced expression of Hugl-1, the human homologue of Drosophila tumour suppressor gene lgl, contributes to progression of colorectal cancer. Oncogene. 2005;24:3100–3109. doi: 10.1038/sj.onc.1208520. [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Schonegg S, Hyman AA. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY, Jan YN. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes & Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–682. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced pins/Gαi cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Skwarek LC, Garroni MK, Commisso C, Boulianne GL. Neuralized contains a phosphoinositide-binding motif required downstream of ubiquitination for Delta endocytosis and Notch signaling. Dev Cell. 2007;13:783–795. doi: 10.1016/j.devcel.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Drosophila brain tumor is a translational repressor. Genes & Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Chia W, Somers WG. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes & Dev. 2009;23:359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Spana EP, Kopczynski C, Goodman CS, Doe CQ. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- Spike CA, Strome S. Germ plasm: Protein degradation in the soma. Curr Biol. 2003;13:R837–R839. doi: 10.1016/j.cub.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Zheng Y. Developmental biology. The mother of all stem cells? Science. 2007;315:469–470. doi: 10.1126/science.1138237. [DOI] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, Van Den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes & Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. The PAR–aPKC system: Lessons in polarity. J Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes & Dev. 2001;15:1031–1040. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- Thorpe PH, Bruno J, Rothstein R. Modeling stem cell asymmetry in yeast. Cold Spring Harb Symp Quant Biol. 2008;73:81–88. doi: 10.1101/sqb.2008.73.010. [DOI] [PubMed] [Google Scholar]
- Thorpe PH, Bruno J, Rothstein R. Kinetochore asymmetry defines a single yeast lineage. Proc Natl Acad Sci. 2009;106:6673–6678. doi: 10.1073/pnas.0811248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Ahringer J. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J Cell Biol. 2007;179:397–402. doi: 10.1083/jcb.200708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Ng KH, Qian H, Siderovski DP, Chia W, Yu F. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat Cell Biol. 2005;7:1091–1098. doi: 10.1038/ncb1317. [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes & Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H. Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells. Development. 2009;136:2287–2296. doi: 10.1242/dev.035758. [DOI] [PubMed] [Google Scholar]
- Watts JL, Morton DG, Bestman J, Kemphues KJ. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev Biol. 1978;65:296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MD, Jin Z, Xie T. Molecular mechanisms of germline stem cell regulation. Annu Rev Genet. 2005;39:173–195. doi: 10.1146/annurev.genet.39.073003.105855. [DOI] [PubMed] [Google Scholar]
- Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, Jackson TL, Gaiano N, Oliver T, Reya T. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Izumi N, Suzuki A, Mizuno K, Ohno S. Lgl mediates apical domain disassembly by suppressing the PAR-3–aPKC–PAR-6 complex to orient apical membrane polarity. J Cell Sci. 2006;119:2107–2118. doi: 10.1242/jcs.02938. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Yu F, Cai Y, Kaushik R, Yang X, Chia W. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Wang H, Qian H, Kaushik R, Bownes M, Yang X, Chia W. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes & Dev. 2005;19:1341–1353. doi: 10.1101/gad.1295505. [DOI] [PMC free article] [PubMed] [Google Scholar]