Abstract
During development, trithorax group (trxG) chromatin remodeling complexes counteract repression by Polycomb group (PcG) complexes to sustain active expression of key regulatory genes. Although PcG complexes are well characterized in plants, little is known about trxG activities. Here we demonstrate that the Arabidopsis SAND (Sp100, AIRE-1, NucP41/75, DEAF-1) domain protein ULTRAPETALA1 (ULT1) functions as a trxG factor that counteracts the PcG-repressive activity of CURLY LEAF. In floral stem cells, ULT1 protein associates directly with the master homeotic locus AGAMOUS, inducing its expression by regulating its histone methylation status. Our analysis introduces a novel mechanism that mediates epigenetic switches controlling post-embryonic stem cell fates in plants.
Keywords: Stem cell, epigenetics, chromatin, trithorax group, Polycomb group, gene activation
Plants generate their reproductive organs late in development through the formation of flowers. This establishes a requirement for the repression of key floral homeotic genes during vegetative development and their subsequent tissue-specific activation during the reproductive stage. In Arabidopsis thaliana, ULTRAPETALA1 (ULT1) is essential to terminate stem cell activity in the center of the flower meristem through the timely activation of the floral homeotic gene AGAMOUS (AG) (Fletcher 2001; Carles et al. 2004). AG encodes a MADS (MCM1, AGAMOUS, DEFICIENS, SRF) domain transcription factor that specifies reproductive organ identity (Bowman et al. 1989) and acts in a negative feedback loop to limit floral stem cell proliferation (Lenhard et al. 2001; Lohmann et al. 2001; Sun et al. 2009). ULT1 is expressed early during floral meristem initiation (Carles et al. 2005) and is required to activate AG expression at the correct stage of floral development, thus acting as a critical temporal component of the floral meristem termination pathway.
Maintenance of the appropriate AG transcriptional status in nonreproductive versus reproductive tissues involves the opposite activities of Polycomb group (PcG) and trithorax group (trxG) chromatin remodeling factors. The PcG gene CURLY LEAF (CLF), a homolog of the Drosophila Enhancer of zeste E(z) histone methyltransferase, acts as a component of Polycomb-Repressive Complex PRC2 (Chanvivattana et al. 2004). CLF is a direct transcriptional repressor of AG expression in leaves, inflorescences, and the outer whorls of flowers (Goodrich et al. 1997) that mediates trimethylation of histone 3 Lys 27 (H3K27me3) (Schubert et al. 2006). Conversely, ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1), a homolog of Drosophila trithorax, encodes a histone methyltransferase for trimethyl groups on histone 3 Lys 4 (H3K4me3) (Alvarez-Venegas et al. 2003; Alvarez-Venegas and Avramova 2005; Saleh et al. 2007) that sustains high-level AG transcription in flowers (Alvarez-Venegas et al. 2003). These proteins maintain the AG locus in either a repressed or an active state in a tissue-specific fashion, yet nothing is known about the epigenetic factors that mediate the AG switch from a repressed to an active state within the floral stem cell reservoir. Nor is it understood through what mechanism the ULT1 protein, which contains a putative DNA-binding SAND (Sp100, AIRE-1, NucP41/75, DEAF-1) domain (Bottomley et al. 2001) but no evident transcription activation domain, functions in the floral stem cell termination pathway.
Results and Discussion
ULT1 gain-of-function phenotypes resemble those of 35S∷AG and clf plants
To gain insight into the mechanism of ULT1 transcriptional control, we expressed ULT1 from the constitutively active cauliflower mosaic virus (CaMV) 35S promoter in transgenic Arabidopsis plants. The 35S∷ULT1 transgenic plants displayed small rosettes and upward-curled rosette leaves, as well as early flowering, short stature, increased branch outgrowths, and prematurely terminating inflorescence meristems (Fig. 1A–D). The plants produced small flowers with mosaic organs; notably, carpeloid sepals and stamenoid petals (Fig. 1D; Supplemental Fig. S1A–E). RT–PCR showed that the severity of the phenotypes strongly correlated with the level of ULT1 overexpression (Fig. 1B); severely affected class 1 35S∷ULT1 plants were used for further analysis.
Figure 1.
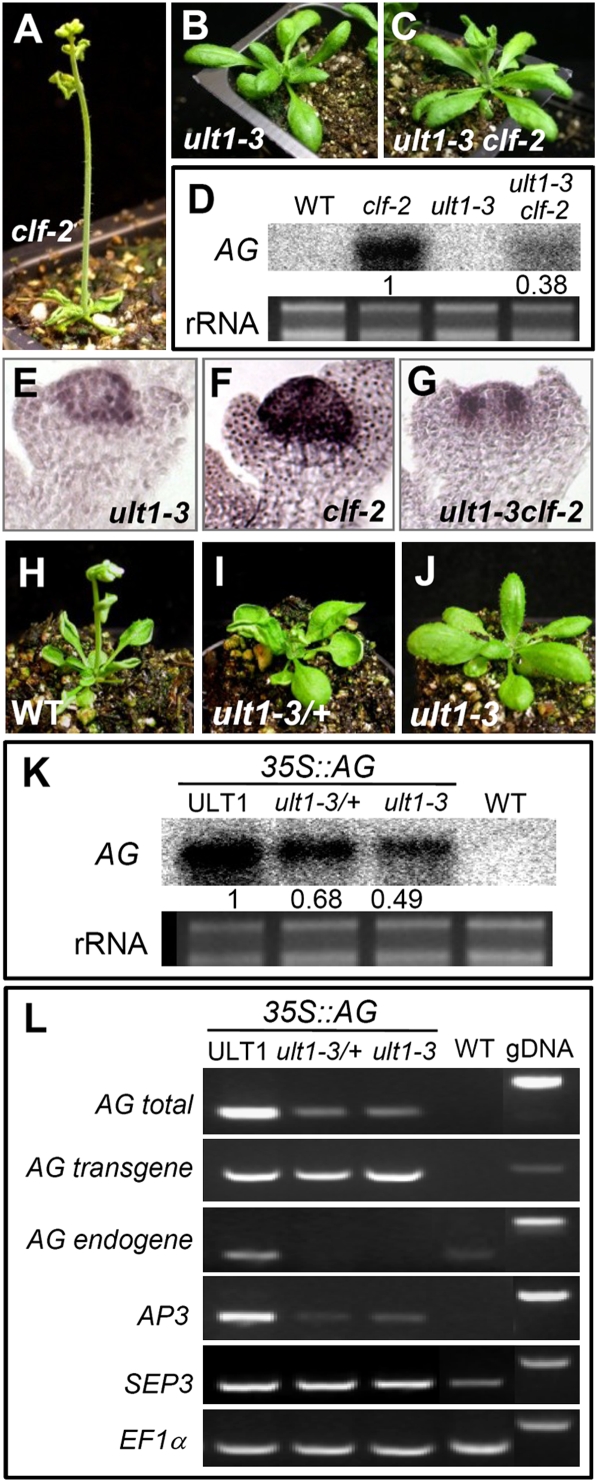
35S∷ULT1 transgenic plants resemble clf plants and ectopically activate AG and AP3. (A,B) Correlation between 35S∷ULT1 phenotype severity and ULT1 expression levels. (A) Population classes range from most (class 1) to least (class 3) severe. (WT) Wild type. (B) RT–PCR with seedling RNA from wild-type and class 1 to class 3, using EF1α as the reference gene. (gDNA) Genomic DNA control. (C,D) 35S∷ULT1 vegetative and inflorescence phenotypes resemble those observed in clf plants. (C) Fifteen-day-old plantlets on soil. (D) Scanning electron micrographs of inflorescences. (E,F) RNA in situ hybridization showing ectopic expression of AG and AP3 in seedlings (E) and inflorescences (F) of wild-type, 35S∷ULT1, and clf-2 plants. (SAM) Shoot apical meristem; (Lp) leaf primordium; (Se) sepal primordium; (Pe) petal primordium; (Ca) carpel primordium. (G–L) Segregating T3 descendents of a 35S∷ULT1; ag-3/+ plant. (G) 35S∷ULT1 AG/AG plant. (H) 35S∷ULT1 ag-3/ag-3 plant. (I) ag-3/ag-3 plant. (J) Wild-type plant. (K) 35S∷ULT1; ap3-3 plant. (L) ap3-3 plant. Bars: C, 5 mm; D, 1 mm; G–L, 1 cm.
The 35S∷ULT1 phenotypes resembled those of 35S∷AG plants (Mizukami and Ma 1992) and of loss-of-function clf plants, which ectopically express AG and APETALA3 (AP3) in leaves and flowers (Figs. 1, 2A,H; Supplemental Fig. S2A; Goodrich et al. 1997). AG and AP3 transcripts accumulated ectopically in 35S∷ULT1 rosette leaves and inflorescence meristems, as well as in sepals and carpels, respectively (Fig. 1E,F). Thus, ULT1 can induce AG expression, consistent with previous results showing that AG induction is delayed in the center of ult1 floral meristems (Fletcher 2001). Introduction of the 35S∷ULT1 construct into either the ag-3 or the ap3-3 background demonstrated that the 35S∷ULT1 phenotypes, like the clf phenotypes (Goodrich et al. 1997), were dependent primarily on ectopic AG activity (Fig. 1G–J; Supplemental Fig. S1F–I). However, the 35S∷ULT1 phenotypes also depended to a lesser extent on AP3 activity (Fig. 1K,L).
Figure 2.
ULT1-dependent expression of phenotypes caused by AG ectopic activation. (A–C) clf-2 phenotypes are rescued in an ult1-3-null mutant background. (D) RNA blot of rosette leaf tissue showing reduction in clf-induced ectopic AG expression in 10-d-old clf-2 ult1-3 plants. The relative amount of AG transcript accumulation is indicated, normalized to rRNA. (E–G) RNA in situ hybridization showing expression of AG in stage 3 floral buds of ult1-3, clf-2, and ult1-3 clf-2 plants. (H–J) Loss of ULT1 rescues 35S∷AG phenotypes in a dosage-dependent fashion. Plants shown are 35S∷AG transgenic in a wild-type (WT), ult1-3/ULT1 heterozygous (ult1-3/+), or ult1-3 homozygous (ult1-3) background. (K) RNA blot of rosette leaf tissue showing ult1-induced reduction of ectopic AG expression in 28-d-old 35S∷AG plants. The relative amount of AG transcript accumulation is indicated, normalized to rRNA. (L) RT–PCR with leaf RNA of 28-d-old plants. Primers were used to detect total AG mRNA, AG mRNA from the 35S∷AG transgene, AG mRNA from the endogenous locus, AP3 mRNA, and SEP3 mRNA. EF1α is shown as the reference gene. Identical results were obtained in experiments using the independent ult1-1 and ult1-2 alleles.
Additional ULT1 regulatory targets were identified by RT–PCR analysis. The AG-related MADS-box genes SEP1, SEP2, and SEP4 were strongly up-regulated in 35S∷ULT1 plants, while SEP3 expression was weakly elevated (Supplemental Fig. S2A). These four SEP genes were also up-regulated in clf-2 plants (Supplemental Fig. S2A). SEP gene activation may reflect a secondary effect of AG ectopic expression because these genes, as well as AP3, are targets of AG transcriptional regulation (Gomez-Mena et al. 2005). Expression of the floral activator AGL19 was unaltered in 35S∷ULT1 leaves (Supplemental Fig. S2A), indicating that ULT1 shows specificity in regulating MADS-box gene expression. We also observed ectopic activation of the class I KNOX homeobox gene STM and up-regulation of the related BP, KNAT2, and KNAT6 genes in 35S∷ULT1 and clf-2 leaves (Supplemental Fig. S2B). These data indicate a substantial degree of overlap between genes positively regulated by ULT1 and genes negatively regulated by CLF. CLF and ULT1 do not significantly regulate one another's transcription (Supplemental Fig. S2C), nor is the CLF expression pattern altered in 35S∷ULT1 inflorescence tissue (Supplemental Fig. S3). These data are inconsistent with ULT1 being a downstream target of CLF or vice versa, or ULT1 inhibiting the activity of the CLF locus, but instead suggest that ULT1 and CLF oppositely regulate a common set of target genes.
ULT1 displays a genetic trxG function
To determine the genetic relationship between ULT1 and CLF, we generated ult1 clf-2 double mutants. Three different ult1 mutant alleles independently rescued all clf-2 vegetative and reproductive defects (Fig. 2A–C; Supplemental Fig. S4). These results show that clf phenotypes are totally dependent on ULT1 activity, and indicate that ULT1 and CLF have opposite, possibly antagonistic, effects on plant development. Drosophila genes belonging to the trxG were originally identified as suppressors of PcG mutant phenotypes (Ingham 1988; Kennison 1995). Because ult1 mutations were able to completely suppress all PcG clf-2 mutant phenotypes, ULT1 meets the genetic definition of a trxG gene.
We investigated the ult1-mediated suppression of the clf-2 phenotypes by examining expression levels of the key CLF target gene, AG. Compared with clf rosette leaves, ult1 clf rosette leaves displayed much lower levels of ectopic AG expression (Fig. 2D), and these levels were insufficient to induce the clf phenotype. Similarly, ult1 clf stage 3 floral buds accumulated AG mRNA at much lower levels than clf stage 3 floral buds, and in a pattern similar to ult1 stage 3 floral buds (Fig. 2E–G; Fletcher 2001). We observed that the expression levels of the AG downstream target genes AP3, SEP1, SEP2, SEP3, and SEP4 were also reduced in ult1 clf plants compared with clf plants (Supplemental Fig. S5), likely contributing to the rescue of the clf phenotypes by mutations in ult1. Conversely, the timing of AG activation in the center of ult1 floral meristems was not affected by the clf mutation. In wild-type and clf-2 floral buds, AG was expressed throughout the central dome by stage 3 (Fig. 2F; Goodrich et al. 1997), whereas the onset of AG expression was delayed in the center of both ult1 (Fig. 2E) and ult1 clf (Fig. 2G) floral meristems.
We next determined whether ectopic induction of AG target genes in 35S∷AG plants was impaired by mutations in ULT1. We observed that 35S∷AG ult1 rosette leaves displayed wild-type morphology (Fig. 2H–J) and had reduced total AG mRNA levels relative to 35S∷AG lines containing functional ULT1 (Fig. 2K). Moreover, loss of ULT1 activity rescued the 35S∷AG phenotypes in a dosage-dependent fashion, because 35S∷AG ult1-3/+ plants displayed developmental and molecular phenotypes intermediate between those of 35S∷AG and 35S∷AG ult1-3 plants (Fig. 2H–K). Whereas the ult1 mutation did not impair AG transgene expression in 35S∷AG plants, ULT1 was required to sustain autoactivation (Gomez-Mena et al. 2005) of the endogenous AG locus (Fig. 2L). This result indicates that ULT1 regulation of AG requires the native regulatory sequence and/or chromatin configuration. In addition, ectopic AG induction of AP3 but not SEP3 was dependent on ULT1 (Fig. 2L). ULT1 activity is therefore required for maximal expression of AG as well as of some AG target genes.
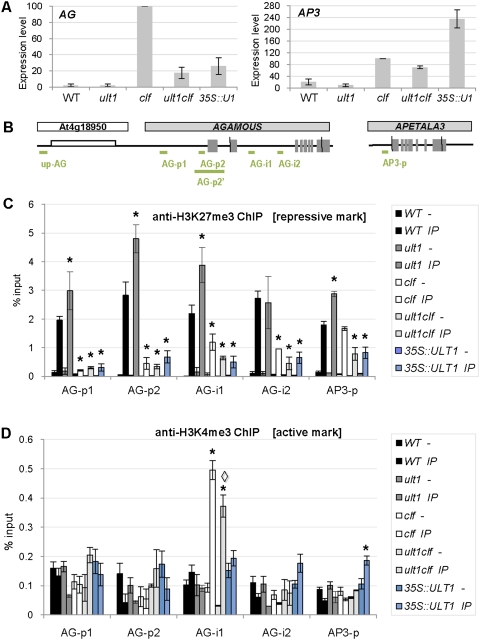
The effect of ULT1 on AG and AP3 transcription levels was confirmed by quantitative real-time PCR (Fig. 3A). In wild-type and ult1 seedlings, AG and AP3 expression was barely detectable, whereas AG expression was dramatically elevated in clf seedlings. In ult1 clf seedlings, AG transcript levels were reduced by 80% relative to clf seedlings, confirming that ULT1 activity is required to obtain high levels of AG expression in clf seedlings. Surprisingly, AG transcription levels in 35S∷ULT1 plants were very similar to those in ult1 clf plants, despite the fact that 35S∷ULT1 seedlings had strong curly leaf phenotypes (Fig. 1A,C), whereas ult1 clf seedlings had normal leaf morphology (Fig. 2C). This discrepancy can be explained by the observation that AP3 expression levels are far higher in 35S∷ULT1 seedlings than in ult1 clf seedlings (Fig. 3A), and AP3 contributes to the 35S∷ULT1 curly leaf phenotype (Fig. 1K,L). We therefore propose that extremely high-level ectopic expression of AP3, as well as other downstream effectors of AG activity such as SEP1 and SEP3 (Supplemental Fig. S2), is sufficient to cause a curly leaf phenotype in 35S∷ULT1 plants, whereas their ectopic expression at much lower levels in ult1 clf plants fails to condition the phenotype.
Figure 3.
Influence of ULT1 and CLF on AG and AP3 expression levels and histone methylation patterns. (A) RT–qPCR analysis of AG and AP3 expression levels in 4-d-old wild-type, ult1-3, clf-2, ult1-3 clf-2, and 35S∷ULT1 seedlings. Graphs represent average values from two biological replicates, each standardized to EF1α, with the value in the clf background set at 100%. Error bars represent standard error. (B) Schematic structures of the AG locus, AG 5′-flanking region, and AP3 locus. For AG and AP3, the exon/intron structure is depicted, with the exons as gray boxes. The transcribed region of the upstream At4g18950 gene is indicated (white box). Green lines denote the regions amplified by qPCR for ChIP analysis. (C,D) H3K27me3 and H3K4me3 patterns at the AG and AP3 loci. ChIP was analyzed by qPCR and results are presented as percentage of input chromatin. Graphs represent average values from two biological replicates. For each locus of interest, the amplification was standardized to the amplification obtained from the region upstream of AG (up-AG). Minus sign (−) indicates ChIP minus antibody and IP indicates ChIP plus antibody. Each set of bars in one color represents minus (−) and IP for the same genotype. Error bars represent standard error. Asterisks (*) indicate values that are significantly different from wild type (P < 0.05 using Student's t-test), and the diamond (◊) indicates that the value for ult1 clf is significantly different from clf (P < 0.05 using Student's t-test).
ULT1 activates the AG locus by controlling its histone methylation status
A genetic trxG function for ULT1 suggested that it may regulate AG expression through chromatin modifications. We tested this hypothesis by analyzing the distribution of histone H3 lysine methylation marks within the promoter and second intron of AG (Fig. 3), regions necessary for the correct spatial and temporal regulation of the gene (Sieburth and Meyerowitz 1997). In seedlings, these regions of the silent AG locus (Fig. 3A,B) carry the H3K27me3-repressive mark deposited by CLF (Schubert et al. 2005).
Comparisons of chromatin immunoprecipitation (ChIP) profiles of wild-type, ult1-3, and 35S∷ULT1 seedlings showed alterations in the deposition of the H3K27me3-repressive mark (Fig. 3C). In ult1-3 seedlings, a 50% to 77% gain in the H3K27me3 mark occurred at the AG-p1, AG-p2, and AG-i1 locations (Fig. 3C). Conversely, in 35S∷ULT1 seedlings, we observed a significant loss of the H3K27me3 mark at all four locations tested. These data indicate that ULT1 limits the deposition of H3K27me3-repressive marks at the AG locus. The AG locus in 35S∷ULT1 seedlings carried a similar level of the active H3K4me3 mark as wild-type seedlings (Fig. 3D) but a reduced amount of the repressive H3K27me3 mark, indicating that the locus was shifted toward a more active state, in accordance with the observation that 35S∷ULT1 seedlings have higher AG transcript levels than wild-type seedlings (Fig. 3A). In 35S∷ULT1 seedlings, the repressive mark was lost even though wild-type CLF activity was still present, demonstrating that excess or ectopic ULT1 is sufficient to prevent deposition of repressive marks by CLF. These results are consistent with ULT1 functioning as an trxG anti-repressor that counteracts PcG-mediated transcriptional silencing, as has been proposed for the Drosophila trxG proteins Trx and Ash1 (Klymenko and Muller 2004; Papp and Muller 2006).
In agreement with previous reports (Goodrich et al. 1997; Chanvivattana et al. 2004), we found that, in clf-2 seedlings, AG expression was highly elevated (Fig. 3A), and that the AG locus was depleted of the H3K27me3-repressive mark in the promoter and second intron (Fig. 3C). Comparison of the H3K27me3-repressive mark in ult1-3, clf-2, and ult1-3 clf-2 seedlings indicated that clf-2 was epistatic to ult1-3 (Fig. 3C), because in clf-2 plants, AG was depleted of H3K27me3 regardless of the presence or absence of ULT1. In clf-2 seedlings, the AG second intron was also enriched for the H3K4me3 activation mark relative to wild-type seedlings (Fig. 3D). However, deposition of H3K4me3 within the second intron decreased by 26% in ult1-3 clf-2 seedlings compared with clf-2 seedlings (Fig. 3D). Thus, although the AG locus was depleted of H3K27me3 in clf-2 seedlings, the absence of ULT1 resulted in modest additional locus-specific depletion of H3K4me3. This reduction in the H3K4me3 activation mark may contribute to the decrease in AG transcript levels detected in ult1-3 clf-2 seedlings (Fig. 3A), as the ratio of the two histone marks is shifted toward a less active state. In contrast to clf-2 seedlings, 35S∷ULT1 seedlings did not show increased deposition of H3K4me3-activating marks at the AG locus (Fig. 3D). A possible reason for this is that ULT1 does not itself have H3K4me3 methyltransferase activity, but rather interacts with such an enzyme as part of a trxG complex, and that the relative abundance of this associated H3K4me3 methyltransferase is rate-limiting in seedlings.
Several additional observations are in accordance with ULT1 not only functioning as an anti-repressor, but also playing a limited role in switching the AG locus to an activated state. First, ult1 flowers, like weak ag flowers, produce a fifth whorl of stamen and/or carpel organs (Fletcher 2001), and this fifth whorl activity is still observed in ult1 clf flowers (Supplemental Table S1). Second, AG is induced normally in the central domain of stage 3 clf floral meristems, but is absent from the center of ult1 and ult1 clf floral meristems (Fig. 2E–G; Fletcher 2001). These data demonstrate that even in the absence of the repressive marks, ULT1 is still required for AG to be transcribed at the correct time during floral meristem development. Taken together, our results suggest that, in addition to restricting the deposition of H3K27 methylation marks at the AG locus by CLF, ULT1 may also play a role in recruiting proteins responsible for local H3K4 methylation and subsequent transcription elongation.
Finally, because AP3 expression levels were exceedingly high in 35S∷ULT1 seedlings compared with wild-type and clf seedlings (Fig. 3A), we analyzed the H3 lysine methylation marks at the AP3 promoter. We found that H3K27me3-repressive marks were significantly reduced and H3K4me3-activating marks were significantly increased in 35S∷ULT1 seedlings compared with wild-type or clf seedlings (Fig. 3C,D). A likely explanation for the observation that AP3 expression levels are higher in 35S∷ULT1 than in clf tissues is that both AG and ULT1 play a role in inducing AP3 transcription. This is consistent with the reduction of AP3 expression in ult1 compared with wild-type plants and in ult1 clf compared with clf plants (Fig. 3A). Thus, we propose that the dramatically increased AP3 transcription levels in 35S∷ULT1 plants are a combined effect of an altered ratio of repressive to activating chromatin marks at the AP3 locus, mediated via elevated ULT1 expression, and elevated expression of AG, a transcriptional activator of AP3 (Gomez-Mena et al. 2005).
ULT1 associates with the AG locus and physically interacts with ATX1
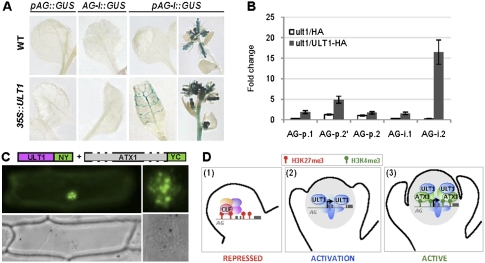
To determine which regions of the AG locus were required for responsiveness to ULT1-mediated transcriptional activation, we analyzed the expression patterns of combinations of AG promoter and second intron sequences driving a β-glucuronidase (GUS) reporter. Both the promoter and the second intron were necessary for ectopic AG activation in 35S∷ULT1 seedlings and inflorescences (Fig. 4A). To assess whether ULT1 could interact directly with these AG regulatory sequences, we generated 35S∷ULT1-HA; ult1 transgenic plants and performed ChIP assays on chromatin extracted from rescued ult1 plants with wild-type phenotypes. We found that ULT1 associated in vivo with specific AG regulatory sites located in the promoter-proximal region and in the second intron (Fig. 4B). Thus, AG is a direct target of ULT1 regulation. Interestingly, we found that ULT1 has the strongest affinity for a region of the AG locus that is slightly downstream from the area carrying enhanced local H3K4me3 in clf plants (Fig. 3D). Physical separation of trxG-binding sites from regions of maximal H3K4me3 has also been reported for the animal trxG factor Ash1 (Papp and Muller 2006).
Figure 4.
ULT1 associates with the AG locus and physically interacts with ATX1. (A) GUS reporter activity driven by various AG regulatory sequences in the presence or absence of 35S∷ULT1. In wild-type plants, pAG-I∷GUS activity was restricted to floral organs. In 35S∷ULT1 plants, ectopic pAG-I∷GUS activity was detected in rosette leaves and inflorescences. (B) Quantification of ULT1 association with AG promoter and second intron sequences by ChIP analysis. qPCR values are presented as fold change relative to the AG upstream region (up-AG). Error bars represent standard error. ult1/HA represents the ult1-2 mutant expressing a HA tag; ult1/ULT1-HA represents the ult1-2 mutant expressing an ULT1-HA fusion. (C) Bimolecular fluorescence complementation assay in which the green signal indicates physical interaction between the ULT1-NY and ATX1-YC fusion proteins inside the nucleus. The right image illustrates the subnuclear pattern of the interaction, and the bottom images show the bright-field exposure of the transformed cell. (NY) N-terminal half of YFP; (YC) C-terminal half of YFP. (D) Mechanistic model for ULT1 regulation of the AG locus. (Panel 1) Floral buds at stage 2, when AG repression is maintained by PcG-mediated CLF deposition of H3K27me3 marks. (Panel 2) At stage 3, the AG locus undergoes activation, with ULT1 protein functioning as an anti-repressor to antagonize PcG-induced histone methylation. (Panel 3) At later stages, AG transcription is maintained by trxG activity, as ULT1 associates with ATX1, which deposits H3K4me3 marks.
The ability of ULT1 to affect histone methylation patterns at the AG locus suggested the potential for ULT1 to physically interact with the ATX1 trxG factor. Indeed, the two proteins showed a strong interaction in the plant nucleus (Fig. 4C; Supplemental Fig. S6), with a subnuclear localization pattern similar to those of chromatin remodeling proteins (Hernandez-Munoz et al. 2005; Calonje et al. 2008). A comparable pattern was reported for the mammalian SAND domain protein AIRE1, a transcriptional activator of tissue-specific genes in the thymus (Pitkänen et al. 2000; Ferguson et al. 2008). This result, together with the methylation mark analyses, indicates that, in addition to restricting the extent to which H3K27 methylation marks are deposited at the AG locus by PcG proteins, ULT1 also associates with proteins such as ATX1 that are responsible for local H3K4 methylation (Alvarez-Venegas et al. 2003). ATX1 is unlikely to be the only factor responsible for H3K4me3 deposition at the AG locus, for several reasons. First, atx1-null mutants display only weak floral homeotic defects and do not show floral meristem indeterminacy phenotypes characteristic of ag flowers (Alvarez-Venegas et al. 2003). Second, 85% of genome-wide H3K4me3 is still present in atx1 plants (Alvarez-Venegas and Avramova 2005), indicating that additional methyltransferases are likely to play key roles in this process, potentially including other members of the ATX1 family (Alvarez-Venegas and Avramova 2002).
Mechanistic model for activation of the AG locus
Together, our data identify the SAND domain protein ULT1 as a trxG factor that binds to AG regulatory sequences during flower development and that regulates the deposition of the epigenetic marks, preventing inappropriate PcG silencing of the AG locus in the center of the flower, where it must become transcriptionally active (Fig. 4D). During the switch of the AG locus from a repressed to an active state, ULT1 may function as a coactivator to recruit additional trxG proteins such as ATX1 that, by analogy with animal systems (Schuettengruber et al. 2007, 2009), are involved in subsequent local H3K4 methylation and/or reading of the chromatin marks for transcription initiation and elongation (Petruk et al. 2006; Li et al. 2007).
Conclusion
Flexible regulation of gene expression through chromatin remodeling is critical for the correct development of eukaryotic organisms, and may be the source of plant developmental plasticity (Pfluger and Wagner 2007). Whereas PcG-mediated repression is considered to be very stable in animal systems, it is often reversible in plants, allowing rapid and dynamic responses to environmental stimuli or developmental cues (Pien et al. 2008). The plasticity of plant cell fate highlights the importance of plant trxG factors that counteract PcG complex repression and promote target gene transcription in a spatially and temporally restricted manner. Identification of the trxG activity of the Arabidopsis SAND domain protein ULT1 expands the repertoire of epigenetic regulators of development and uncovers a chromatin-mediated pathway controlling the dynamic switch of transcriptional states that occurs in floral stem cells. It also suggests a molecular mechanism for the activity of animal SAND domain proteins such as AIRE and Sp100, two transcriptional activators associated with human autoimmune regulation (Gibson et al. 1998; Bloch et al. 2000).
Materials and methods
All Arabidopsis thaliana genotypes were in the Landsberg erecta (Ler) ecotype and were grown on either Murashige and Skoog medium or soil under continuous light. RNA extraction and RT–PCR were performed as described previously (Carles et al. 2005). Quantification of AG and AP3 cDNAs by real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems) and an ABI 7000 Thermocycler (Applied Biosystems). RNA blot hybridization was conducted using a 32P-labeled DNA probe corresponding to the full-length AG ORF. For RNA in situ hybridization, AG and AP3 antisense probes were generated using a digoxigenin-labeling mix (Roche). ChIP was performed following a procedure detailed in the Supplemental Material. Immunoprecipitated DNA was analyzed by real-time PCR, and relative enrichments were calculated as the percentage of the obtained values for the immunoprecipitated and input fractions. Bimolecular fluorescence complementation assays were conducted in onion epidermal cells after transformation by particle bombardment using a Biolistic PDS-1000/He unit (Bio-Rad). Epidermal peels were visualized 24 and 36 h after bombardment using a Zeiss Axiophot microscope. Further details and other methods can be found in the Supplemental Material.
Acknowledgments
We thank Chris Day, Justin Goodrich, Bassem Al Sady, Zoya Avramova, Ivan Ndamukong, and Ian Small for materials and constructs. We are grateful to Daniel Schubert and Justin Goodrich for communicating primer sequences, Elisa Fiume and Erin Osborne for assistance with phenotypic analysis, and Sasha Preuss for insights about the ChIP procedure. We thank Renee Sung, Sheila McCormick, Sarah Hake, Vincent Colot, François Roudier, Robert Blanvillain, and members of the Fletcher laboratory for critical reading of the manuscript. We acknowledge Syngenta, the Salk Institute for Biological Studies, and the Arabidopsis Biological Resource Center for providing clones and insertion lines. This work was supported by the National Science Foundation (IBN-0110667).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1812609.
Supplemental material is available at http://www.genesdev.org.
References
- Alvarez-Venegas R, Avramova Z. The SET-domain proteins of the Su(var)3-9, E(z), and Trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–5207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- Bloch DB, Nakajima A, Gulick T, Chiche JD, Orth D, de La Monte SM, Bloch KD. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol Cell Biol. 2000;20:6138–6146. doi: 10.1128/mcb.20.16.6138-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8:626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen L, Sung ZR. EMBRYONIC FLOWER1 participates in Polycomb Group-mediated AG gene silencing in Arabidopsis. Plant Cell. 2008;20:277–291. doi: 10.1105/tpc.106.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Lertpiriyapong K, Reville K, Fletcher JC. The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity, and acts through WUSCHEL to regulate floral meristem determinacy. Genetics. 2004;167:1893–1903. doi: 10.1534/genetics.104.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC. ULTRAPETALA1 encodes a putative SAND domain transcription factor that controls shoot and floral meristem activity in Arabidopsis. Development. 2005;132:897–911. doi: 10.1242/dev.01642. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon Y-H, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Alexander C, Rossi SW, Liiv I, Rebane A, Worth CL, Wong J, Laan M, Peterson P, Jenkinson EJ, et al. AIRE's CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J Biol Chem. 2008;283:1723–1731. doi: 10.1074/jbc.M707211200. [DOI] [PubMed] [Google Scholar]
- Fletcher JC. The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development. 2001;128:1323–1333. doi: 10.1242/dev.128.8.1323. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Ramu C, Gemund C, Aasland R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci. 1998;23:242–244. doi: 10.1016/s0968-0004(98)01231-6. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires the histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes & Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by Trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol. 2007;10:645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, Dean C, Grossniklaus U. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkänen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, Will H, Vähämurto P, Ollila J, Vihinen M, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275:16802–16809. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007;35:6290–6296. doi: 10.1093/nar/gkm464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e1000013. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng K-H, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes & Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]