Abstract
The Saccharomyces cerevisiae CLASP (CLIP-associated protein) Stu1 is essential for the establishment and maintenance of the mitotic spindle. Furthermore, Stu1 localizes to kinetochores. Here we show that, in prometaphase, Stu1 assembles in an Ndc80-dependent manner exclusively at kinetochores that are not attached to microtubules. Stu1 relocates to microtubules when a captured kinetochore reaches a spindle pole. This relocation does not depend on kinetochore biorientation, but requires a functional DASH complex. Stu1 at detached kinetochores facilitates kinetochore capturing. Furthermore, since most of the nuclear Stu1 is sequestered by one or a few detached kinetochores, the presence of detached kinetochores prevents Stu1 from localizing the spindle, and therefore from stabilizing the spindle. Thus, the sequestering of Stu1 by detached kinetochores serves as a checkpoint that keeps spindle poles in close proximity until all kinetochores are captured. This is likely to facilitate kinetochore biorientation. In agreement with the findings described above, a kinetochore mutant (okp1-52) that fails to release Stu1 from the kinetochore displays a severe spindle defect upon spindle pole body separation, and this defect can be rescued by destroying the okp1-52 kinetochore.
Keywords: Kinetochore, spindle, Stu1, CLASP, Okp1
The coordination of kinetochore (KT)–microtubule (MT) attachment with sister chromatid separation and spindle elongation is crucial for chromosome segregation. In Saccharomyces cerevisiae, KT–MT interaction is established during S phase. After centromere DNA replication, reassembled KTs are captured via lateral MT attachment and move to one of the spindle poles by the minus end-directed kinesin Kar3 and via MT plus end tracking (Tanaka et al. 2005, 2007; Kitamura et al. 2007). Subsequently, catalyzed by the protein kinase Ipl1 (Biggins and Murray 2001; Tanaka et al. 2002), initial syntelic attachments are replaced by bipolar attachments. The duplicated spindle poles remain in close proximity at this point and form very short (0.6-μm) nuclear MTs (Winey and O'Toole 2001). Controlling the amount of Cin8 and Kip1, two MT plus end-directed kinesins, is important for maintaining spindle poles in close proximity; a failure to do so delays bipolar sister KT attachment (Liu et al. 2008). Upon bipolar attachment of all KTs, the formation of a 2-μm metaphase spindle with anti-parallel interpolar MTs (ipMTs) moves the spindle pole bodies to distant sites within the nuclear membrane (Winey and O'Toole 2001). Cin8 and Kip1 (Saunders and Hoyt 1992), as well as a MT-binding protein Stu1 (Yin et al. 2002), are important for establishing and maintaining this metaphase spindle; however, it is unclear how these proteins get activated upon prometaphase-to-metaphase transition. Astral MTs, emanating from the cytosolic side of the spindle pole bodies, interact with the cell cortex and position the metaphase spindle in the mother–bud axis (McCarthy and Goldstein 2006). Bipolar attachment of all sister KTs also satisfies the spindle assembly checkpoint (SAC). The SAC relies on six proteins (Mad1, Mad2, Mad3, Bub1, Bub3, and Mps1) that recognize unattached KTs and generate an activity that inhibits the anaphase-promoting complex (APC) via its activator, Cdc20 (Musacchio and Salmon 2007). Without SAC signaling, APC targets Pds1, an inhibitor of the protease Esp1, for degradation. Consequently, Esp1 is free to resolve sister chromatid cohesion. Deprived of the restriction presented by the cohesion of bipolar-attached sister chromatids, spindle poles move apart (anaphase B). The forces that power the anaphase B movement are probably generated partly via astral MTs (Amaro et al. 2008) and partly by elongating the ipMT array to form an (ultimately 9-μm-long) anaphase spindle. The latter requires that the spindle midzone (the overlap region of the interdigitizing ipMTs) is reinforced by a group of proteins that either facilitate MT bundling or stabilize MT plus ends (Khmelinskii and Schiebel 2008). Crucial to this is the dephosphorylation of several MT-binding proteins by Cdc14, which relocalizes these proteins to the midzone (Pereira and Schiebel 2003; Higuchi and Uhlmann 2005; Khmelinskii et al. 2007). Among those, Ase1 apparently serves as the core component that determines the midzone localization of all other midzone proteins (Khmelinskii et al. 2007). Sequestering and inhibiting Cdc14 in the nucleolus is the primary way to regulate its activity (D'Amours and Amon 2004). At the beginning of anaphase, a signaling network (FEAR [Cdc14 early anaphase release]) releases Cdc14 from the nucleolus and thus alters the dynamic stability of the ipMTs through midzone reinforcement. Esp1 is one of the activators of FEAR, and consequently of Cdc14 (Sullivan and Uhlmann 2003). As Esp1 activity, controlled by the SAC (see above), starts anaphase by cleaving the cohesion complex, the appearance of free Esp1 apparently coordinates sister chromatid separation with spindle elongation. After anaphase is complete, spindle disassembly is induced by a second wave of Cdc14 release from the nucleolus that is orchestrated by the mitotic exit network (Stegmeier and Amon 2004).
The S. cerevisiae KT is a complex structural entity with >60 different proteins (Westermann et al. 2007) that are frequently organized in protein complexes. Models that organize KT subcomplexes in a hierarchical way have been presented (De Wulf et al. 2003). Currently, S. cerevisiae KT components are usually categorized as follows: (1) the inner components that directly interact with the centromere DNA, such as the CBF3 complex, Mif2, Cbf1, and the Cse4 nucleosome; (2) linker components such as the COMA complex, the MIND complex, and the Spc105 complex that connect the KT–MT interface to the inner KT; and (3) the KT–MT interface. This includes the Ndc80 complex, the DASH complex, Slk19, MT plus end-tracking proteins (Bim1, Bik1, Stu2, and Stu1), kinesins (Cin8, Kip1, Kip3, and Kar3), and the Ipl1 complex (Ma et al. 2007; Westermann et al. 2007). With the exception of the Ndc80 complex, the DASH complex, Kip1, and Kar3, all of these proteins also localize to the spindle midzone during anaphase (Khmelinskii et al. 2007) and are required to form stable anaphase spindles. Furthermore, as mentioned above, Kip1, Cin8, and Stu1 are also required to establish and maintain metaphase spindles.
Stu1 belongs to the CLASP (CLIP-associated protein) family (Akhmanova and Hoogenraad 2005). CLASPs are considered to behave as plus end-tracking proteins and to stabilize MT plus ends by facilitating the incorporation of tubulin subunits (Maiato et al. 2005). In addition, by localizing at specialized MT regions distinct from plus ends, at least one CLASP member contributes to the stability of MT bundles in interphase (Bratman and Chang 2007). Furthermore, CLASPs have important mitotic and meiotic functions. Human CLASP1 and CLASP2 (Maiato et al. 2003; Mimori-Kiyosue et al. 2006), Drosophila MAST/Orbit (Maiato et al. 2002), Xenopus Xorbit (Hannak and Heald 2006), Caenorhabditis elegans CLASPCLS-2 (Cheeseman et al. 2005), and Schizosaccharomyces pombe cls1p (Grallert et al. 2006; Bratman and Chang 2007) are, like Stu1, essential for the formation and maintenance of a bipolar mitotic or meiotic spindle. Furthermore, metazoan CLASPs bind to KTs, where they regulate the dynamic behavior of KT MTs (kMTs) and, consequently, chromosome congression (Maiato et al. 2002, 2003; Cheeseman et al. 2005; Mimori-Kiyosue et al. 2006). In contrast to metazoan CLASPs, the role of KT-localized Stu1 is unclear.
Here we show that, in S. cerevisiae, the primary function of KT-attached Stu1 is to facilitate capturing of detached KTs. Initial findings that a severe spindle defect observed in mutants affecting the COMA complex is due to a failure of Stu1 relocalization from the mutant KTs to MTs prompted further investigation on Stu1 localization and function. This revealed that Stu1 binds exclusively and with high affinity to detached KTs, where it is required for KT capturing. Relocalization of Stu1 occurs when a captured KT reaches the pole. However, association of Stu1 with MTs only prevails in the absence of detached KTs. Thus, Stu1 localization at detached KTs also indirectly regulates the formation of a stable metaphase spindle.
Results
Mutations in Okp1 and Ame1 cause a severe spindle defect
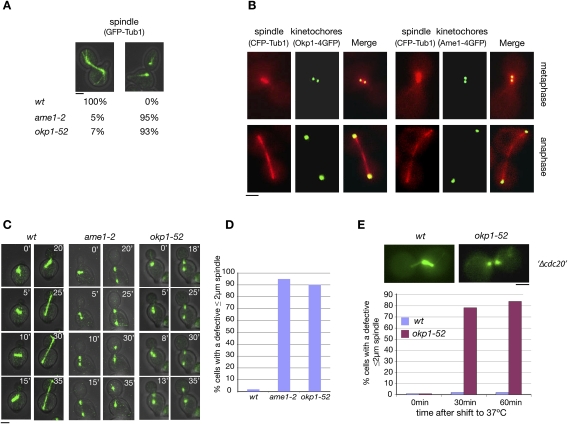
Two temperature-sensitive COMA complex mutants, okp1-52 and ame1-2, exhibited a severe defect in the formation of the mitotic spindle. One-hundred-eighty minutes after the release from a G1 arrest at 37°C, in 70% of the mutants, the spindle poles had separated 3–6 μm. However, in most cases, no detectable anaphase spindle was observed (Fig. 1A). This is in contrast to wild-type cells that showed stable spindles at 3- to 6-μm spindle length. In anaphase, kMTs are short and close to the poles. Thus, the spindle defect in the mutants reflects a failure to establish or maintain ipMTs. Spindle defects have also been observed in mutants with mutations in other KT components. These include Slk19 (Zeng et al. 1999), the CBF3 complex (Bouck and Bloom 2005), and the DASH complex (Cheeseman et al. 2001; Janke et al. 2002; Li et al. 2002), which represent KT components that also localize to the spindle. Consequently, in these cases, the spindle defect might be due to a compromised spindle-related function of the mutated protein. Okp1 and Ame1, however, do not localize to the spindle (Fig. 1B). Therefore, ame1-2 and okp1-52 spindle defects are due to KT defects.
Figure 1.
okp1-52 and ame1-2 mutants display a severe spindle defect, although Okp1 and Ame1 do not localize to spindles. (A) ame1-2 and okp1-52 display a severe spindle defect. Spindles were quantified 180 min after the release from G1 at 37°C. n > 100. Bar, 2 μm. (B) Okp1 and Ame1 do not localize to the spindle. (C–E) Most okp1-52 and ame1-2 cells display a spindle defect at a spindle length ≤2 μm. (C) Starting 60 min after the release from G1 at 37°C (0′), spindles (GFP-Tub1) were visualized by time-lapse microscopy. Individual frames for the indicated times are shown. Bar, 2μm. (D) Quantification of cells visualized in C with a defective 2-μm (or shorter) spindle. n > 50. (E) GAL1-CDC20 cells were arrested in metaphase by Cdc20 depletion (Δcdc20) at 25°C and then shifted to 37°C. At the indicated time points after the temperature shift, spindles (Tub1-GFP) were quantified as intact (as shown for wild type) or defective (as shown for okp1-52). Only cells that had separated spindle poles 2 μm or less were included in the count. n > 100. Bar, 2 μm.
Lack of Cdc14 may contribute to the okp1-52 and ame1-2 spindle defect
One possible explanation for the observed spindle defect is that okp1-52 and ame1-2 mutants fail to activate the protein phosphatase Cdc14 in early anaphase that is required for midzone formation and spindle stability (Higuchi and Uhlmann 2005; Khmelinskii et al. 2007). As described above, in S. cerevisiae, Cdc14 is activated in early anaphase via a partial and temporal release from the nucleolus, and this requires, among other factors of the FEAR pathway, Esp1. Thus, cells that separate their spindle poles in the absence of Esp1 display spindle defects (Higuchi and Uhlmann 2005). Furthermore, since an active SAC inhibits Esp1, cells that separate the spindle poles while SAC is active can also be expected to display spindle defects. As explained and shown in the Supplemental Material, there is evidence that ame1-2 and okp1-52 cells separate spindle poles while the SAC is partially active (Supplemental Material; Supplemental Fig. S1). Furthermore, ame1-2 cells fail to release Cdc14 from the nucleolus during spindle pole separation (Supplemental Material; Supplemental Fig. S2; Supplemental Movies S1, S2). Thus the lack of Cdc14 activity during spindle pole separation is likely to contribute to the okp1-52 and ame1-2 spindle defect.
Lacking Cdc14 activity is not the sole cause for the okp1-52 and ame1-2 spindle defect
Cdc14 is required for the stability of anaphase but not metaphase spindles (Higuchi and Uhlmann 2005). As observed by time-lapse microscopy (Fig. 1C,D; Supplemental Movies S3, S4), the spindle defects in the okp1-52 and ame1-2 mutants, however, were occurring already at a spindle length (≤2 μm) that resembles metaphase spindles. Furthermore, when the okp1-52 mutation was induced during a metaphase arrest, the spindle defect was observed at the metaphase spindle length (Fig. 1E). Thus, apparently the absence of Cdc14 activity during spindle pole separation is not the sole cause for the okp1-52 and ame1-2 spindle defect. This was further supported by the observation that neither SAC inactivation (permitting Esp1 and thus Cdc14 activation) nor Cdc14 overexpression rescued the okp1-52 and ame1-2 spindle defect (see the Supplemental Material; Supplemental Fig. S3).
Stu1 fails to localize to MTs in okp1-52 cells
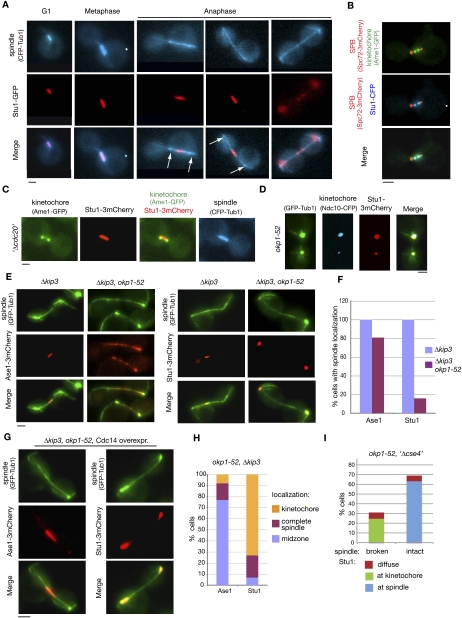
To explain the okp1-52 and ame1-2 spindle defects, we focused on proteins that localize to the KT as well as to the spindle and that are essential for prometaphase/metaphase spindle stability. Stu1 fulfilled these criteria (Yin et al. 2002; Ma et al. 2007). We confirmed and extended the current knowledge on Stu1 localization in wild-type cells. Early in mitosis, Stu1 localized predominantly either as two foci that represent the KT (Fig. 2B) or with short spindles (Fig. 2A). When cells were arrested in metaphase, Stu1 always localized to the ipMTs in between the two KT clusters (Fig. 2C). Thus, although Stu1 localizes to MTs in metaphase, there appears to be a short period before metaphase when Stu1 binds exclusively to KTs. During anaphase, the majority of Stu1 dissociated from the KTs and relocalized to the spindle midzone (Fig. 2A; Yin et al. 2002). In telophase, Stu1 dissociated from the spindle (Fig. 2A). In contrast to the wild type, a large quantity of Stu1 remained at KTs at least up to 300 min after the release from G1 in okp1-52 mutants (Fig. 2D). Thus, the okp1-52 mutation might interfere with Stu1 relocalization, and this could cause the spindle defect. Alternatively, the okp1-52 cells might have a spindle defect for other reasons, and the absence of ipMTs would block the dissociation of Stu1 from the KTs. To distinguish between these two possibilities, we restabilized ipMTs in okp1-52 cells by deleting KIP3 that codes for a MT-depolymerizing kinesin (Gupta et al. 2006). Two-hundred minutes after the release from G1, 60% of okp1-52 Δkip3 double mutants displayed ipMTs between spindle poles that were 3–9 μm apart (Fig. 2E). These anaphase spindles appeared somewhat weaker than those in wild-type cells, but they clearly interacted with various known spindle-associated proteins such as Ase1 (Fig. 2E,F), Spc34, Bim1, and Stu2 (Supplemental Fig. S4). In contrast to this, they did not display Stu1 localization. Instead, Stu1 remained at the KTs (Fig. 2E,F). Notably, Ase1 localization is not focused to the midzone in okp1-52 Δkip3 cells (Fig. 2E). Since Cdc14 facilitates midzone formation, this indicates again a lack of Cdc14 activity during spindle formation in these cells. We therefore asked whether this lack of Cdc14 is the reason for the Stu1 localization defect. Upon overexpression of Cdc14 in the okp1-52 Δkip3 cells, Ase1 localization is restricted to the midzone (Fig. 2G). This supports the published data (Khmelinskii et al. 2007) that Cdc14 is required for midzone localization of Ase1, and shows that, upon overexpression, there is sufficient Cdc14 to overcome the Cdc14 deficiency in the okp1-52 Δkip3 cells. Despite this, Stu1 did not localize to the midzone or to the spindle in general, even upon overexpression of Cdc14. It remained at the KTs (Fig. 2G,H). Taken together, we thus conclude that okp1-52 KTs fail to release Stu1 and thus interfere with Stu1 localization to the ipMTs. If this is the reason for the spindle defect, then eliminating the okp1-52 KTs should cure it. This is indeed the case. When okp1-52 KTs were further compromised by Cse4 depletion, Stu1 localized to ipMTs, and the occurrence of intact spindles increased considerably (Fig. 2I).
Figure 2.
okp1-52 KTs fail to release Stu1. (A–C) Stu1 localization in wild-type cells. Bar, 2 μm. (A) Stu1 and spindle were visualized at the indicated cell cycle phases. Arrows indicate the position of KTs. (B) Stu1 localizes at the KT early in mitosis. Cells were analyzed 80 min after the release from G1. The phenotype shown was observed in 24% of the cells. n > 100. (C) Stu1 localizes to the spindle in metaphase. GAL-CDC20 cells were arrested in metaphase by Cdc20 depletion. (D) Stu1 remains at okp1-52 KTs during spindle pole separation. Cells were analyzed 120 min after the release from G1 at 37°C. Bar, 2 μm. (E) In okp1-52 cells, Stu1, in contrast to Ase1, fails to locate to ipMTs stabilized by KIP3 deletion. Δkip3 cells were analyzed 150 min after the release from G1 at 37°C. Bar, 2 μm. (F) Quantification of Ase1 and Stu1 localization as observed in E. n > 100. (G) In okp1-52 cells, Stu1, in contrast to Ase1, does not localize to the midzone of Δkip3-stabilized ipMTs upon overexpression of Cdc14. Cells were released from G1 at 25°C in raffinose, the temperature was shifted to 37°C, Cdc14 overexpression was induced (2% galactose) 90 min after the release, and spindles were analyzed 210 min after the release. Bar, 2 μm. (H) Quantification of Ase1 and Stu1 localization as observed in G. n > 100. (I) Compromising okp1-52 KTs by Cse4 depletion rescues spindle formation. GAL-CSE4 cells were depleted of Cse4 during a G1 arrest. One-hundred-twenty minutes after the release at 37°C, spindle phenotypes (GFP -Tub1) and Stu1-3mCherry localization were quantified.
Stu1 associates with detached KTs, and relocalizes to spindle MTs when captured KTs reach a spindle pole
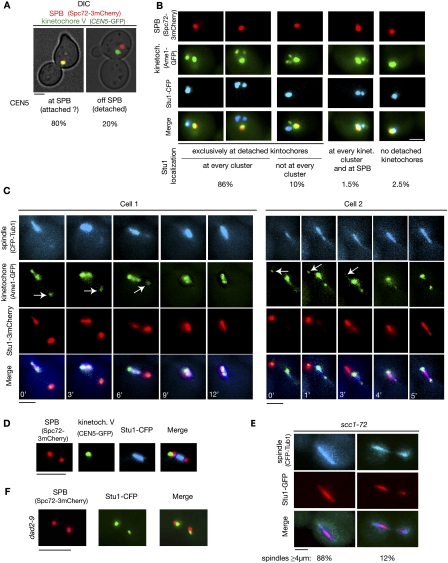
The okp1-52 phenotype included the possibility of detached KTs. We therefore wondered whether Stu1 selectively interacts with unattached KTs. During the S. cerevisiae cell cycle, unattached KTs occur as a consequence of centromere DNA replication, and are subsequently captured via lateral MT interaction (Kitamura et al. 2007). To enrich for unattached wild-type KTs, we therefore released cells from G1 into nocodazole. Spindle poles failed to separate under these conditions, revealing that the nocodazole treatment prevented the formation of ipMTs. Short MTs emanating from the poles, however, are thought to resist nocodazole treatment. Thus, a high percentage of KTs remained close to the spindle poles (Fig. 3A,B; Gillett et al. 2004). Twenty percent of the KTs, however, did not colocalize with the spindle poles, and thus could be considered clearly detached KTs (Fig. 3A). In 96% of the cells, Stu1 colocalized exclusively with detached KTs, but did not localize to the majority of KTs that remained at the poles (Fig. 3B). In 10% of the cells, Stu1 only localized to a fraction of detached KTs (see also below). In a small amount of cells, Stu1 localized (probably to MTs) at the spindle poles. The majority of these cells did not display detached KTs.
Figure 3.
Stu1 selectively interacts with detached KTs and relocalizes to the spindle when captured KTs reach a spindle pole. Relocalization depends on a functional DASH complex but not on biorientation. (A,B) Wild-type cells were analyzed 150 min after the release from a G1 arrest into nocodazole. (A) Occurrence of clearly detached KTs. Cells with a detached KT V, as shown, were quantified. n > 100. Bar, 2 μm. (B) Stu1 interacts selectively with detached KTs. Stu1 localization was quantified as indicated. n > 100. Bar, 2 μm. (C,D) Wild-type cells were released from G1 into nocodazole. One-hundred-twenty minutes after the release, nocodazole was removed. (C) Stu1 moves with the captured KTs to a spindle pole and then relocalizes to spindle MTs. Starting 50 min after the nocodazole washout, the cells were visualized by time-lapse microscopy (3 or 1 min per frame). Individual frames with the indicated times are shown for two different cells. Arrows indicate detached KTs. Bar, 2 μm. (D,E) Biorientation is not required for Stu1 relocalization to the spindle. (D) Stu1 localization in respect to spindle poles and KTs of chromosome V in 8% of the cells (n > 50) 60 min after the nocodazole washout. Bar, 2 μm. Note that Stu1 localizes to the spindle and not to the KTs, although the latter have not achieved biorientation. (E) Stu1 localizes to spindles in scc1-72 cells. Cells were analyzed 120 min after the release from G1 at 37°C. Stu1 localized to the spindle in 100% of the cells. Phenotypes for spindles with a length of ≥4 μm were quantified as shown. n > 100. Bar, 2 μm. (F) Stu1 relocalization to the spindle fails in dad2-9 cells. Cells were processed as in C, but shifted to 37°C when released from G1 and analyzed 60 min after the nocodazole washout. The phenotype shown was observed in 92% of the cells. n > 100. Bar, 2 μm.
When we removed nocodazole, the detached KTs were captured and relocated to one of the spindle poles (Fig. 3C). For the majority of cells, the KTs were too close to the poles to clarify the fate of Stu1 during capturing. However, in the seven events in which we observed the capturing of detached KTs that were >2 μm away from the poles, we found that Stu1 relocated together with the captured KTs to one of the poles (Fig. 3C). In any case, when the captured KTs reached the pole, Stu1 was swiftly relocated from the KTs to the re-established ipMTs. To further support Stu1 localization as observed by microscopy, we performed chromatin immunoprecipitation (ChIP). This revealed (Supplemental Fig. S5) that Stu1 clearly localized to KTs in nocodazole-treated cells when ∼20% of kintochores are detached (see above). However, Stu1 was barely detectable at KTs in cells that were arrested in anaphase by the induction of the cdc15-1 mutation. Thus, the ChIP results support the microscopy data.
Biorientation is not a prerequisite for the spindle localization of Stu1, but the intactness of the DASH complex is
Following the nocodazole washout and KT capturing, 25% of the cells displayed a phenotype as seen in Figure 3C (cell 2, 5 min). Stu1 localized to the spindle, while KTs devoid of visible Stu1 localize either with the spindle poles or to the side of the poles, but not within the array of ipMTs. This phenotype could be particularly well evaluated when KT capturing was observed with cells that had CEN5 and the spindle pole body labeled (Fig. 3D). KTs that have achieved a bipolar attachment are observed as two lobes in between the spindle poles on the spindle axes. Therefore, the KTs observed in the cells described above are clearly not bipolar-attached. Consequently, establishing bipolar attachment and tension appears not to be required for Stu1 to leave the KTs. To support this, we analyzed Stu1 localization in scc1-72 cells in which defective sister chromatid cohesion compromises the establishment of bipolarity. scc1-72 cells released from G1 at 37°C established spindles that (probably due to SAC activity and a lack of Cdc14) failed to limit Stu1 localization to the midzone and exhibited fissures once they extended (Fig. 3E). But the fact that Stu1 localized to the spindles confirms that the establishment of bipolarity and tension is not the signal for Stu1 relocalization.
When transported to the poles by Kar3, captured KTs sooner or later encounter a MT-bound DASH complex, which then facilitates transport via plus end tracking (Tanaka et al. 2007). We therefore wondered whether the DASH complex is required for Stu1 relocalization. Indeed, Stu1 remained at KTs in the vicinity of spindle poles in a dad2-9 mutant (harboring a defect in a DASH complex component) when released from G1 at 37°C (Fig. 3F). This is in agreement with the assumption that Stu1 relocalization requires the interaction of the DASH complex with the core KT.
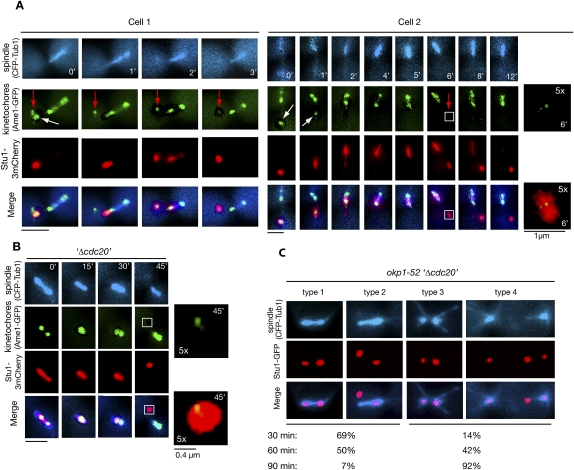
The localization of Stu1 to spindles is reversible
In four events of KT capturing, we observed a detached KT (cluster) that failed to relocate to a pole while other KTs did (Fig. 4A). This KT (cluster) was completely devoid of Stu1. All of the detectable Stu1 localized to the captured KTs and, as described above, moved with them to a spindle pole. Only after Stu1 had been relocated to the spindle did it become available to associate with the remaining uncaptured KTs. This shows that, in prometaphase, the spindle-associated Stu1 can be relocated back to detached KTs. Next we asked whether Stu1 could also be relocated from MTs to KTs after metaphase has been achieved. We therefore treated metaphase-arrested cells with nocodazole to produce unattached KTs. Upon this treatment, cells first lost bipolar KT attachment, and in some cases revealed (according to the very weak signal, probably only one) detached KT distant to the spindle poles (Fig. 4B). Before the ipMTs had depolymerized completely, all of Stu1 relocalized to that detached KT (Fig. 4B). To exclude that this result was triggered by the nocodazole treatment, we also produced detached KTs by inducing the okp1-52 mutation in metaphase. As seen when okp1-52 cells were released from G1 (Fig. 2D), the okp1-52 KTs localizing close to the poles acquired Stu1 (see the Discussion; Fig. 4C). In addition, we frequently observed Stu1 signals (i.e., completely detached KTs) distant from the poles and the spindle axes. Thirty minutes after the temperature shift, Stu1 had dissociated from the ipMTs and had relocated to detached KTs in most cells, although ipMTs were clearly still present (Fig. 4C, types 1 and 2). Subsequently, ipMTs disappeared, while the poles moved apart and Stu1 remained at the detached KTs (Fig. 4C, types 3 and 4). Thus, MT-associated Stu1 could relocate back to detached KTs also in metaphase, and this correlated with the occurrence of a spindle defect.
Figure 4.
Spindle-associated Stu1 relocates to detached KTs. (A) Stu1 relocates from captured KTs to the spindle and then returns to the remaining detached KTs. Cells were processed as in C. Magnification of the boxed area is as indicated. (White arrows) Captured KTs; (red arrows) detached KTs that acquired Stu1 only in the second round. Bar, 2 μm or as shown. (B) Stu1 relocates to detached KTs in metaphase. GAL-CDC20 cells were arrested in metaphase by Cdc20 depletion, treated with nocodazole, and visualized by time-lapse microscopy. (0′) Nocodazole addition. Magnification of the boxed area is as indicated. Bar, 2 μm or as shown. (C) Stu1 relocates to KTs when the okp1-52 mutation is induced in metaphase. GAL-CDC20 cells were arrested in metaphase by Cdc20 depletion, then shifted to 37°C and analyzed at the indicated time after the temperature shift. Cells with KT-localized Stu1 were quantified (n > 100) and classified into type 1 + 2 (intact spindle) and type 3 + 4 (defective spindle). Bar, 2 μm.
Unattached KTs stably sequester Stu1
Our data suggest that, at least up to metaphase, a detached KT sequesters nuclear Stu1 so efficiently that Stu1 localization at MTs is prevented or reversed. Thus, most of the nuclear Stu1 should interact firmly with one or a few detached KTs. This assumption is in agreement with the observation that the intensity of the Stu1 signal dramatically exceeds that of the KT signal when Stu1 localizes to detached KTs (Figs. 3B,C, 4A,B). To test whether Stu1 sequestering by detached KTs is limited by a defined number of direct binding sites at the KT or is facilitated by Stu1 oligomerization, we overexpressed Stu1 in nocodazole-treated cells. We found that, upon overexpression, Stu1 formed crescent-shaped structures at the detached KTs (Fig. 5A). Thus, the additional Stu1 produced by the Stu1 overexpression also accumulated at detached KTs. This and the fact that Stu1 interacts with itself (Yin et al. 2002) support the idea that Stu1 oligomerizes at KTs.
Figure 5.
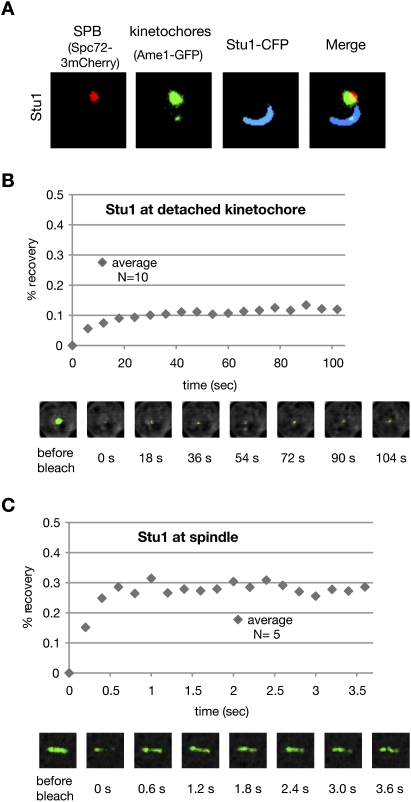
Most of the nuclear Stu1 associates firmly with detached KTs. (A) Stu1 polymerizes at detached KTs. Stu1 was overexpressed from a GAL promoter in a wild-type STU1 background during a G1 arrest. Cells were analyzed 240 min after the release from G1 into nocodazole. (B) Stu1 localization at detached KTs is stable. FRAP analysis of Stu1-GFP. (C) Stu1 localization at metaphase spindles is dynamic. FRAP analysis of Stu1-GFP in cells arrested in metaphase by Cdc20 depletion.
To test the dynamic behavior of KT-localized Stu1, we performed FRAP (fluorescence recovery after photobleaching) analysis. The low and slow recovery (∼12% in 30 sec) after photobleaching (Fig. 5B) indicates that the Stu1–KT interaction is stable, and that a detached KT sequesters most of the nuclear Stu1. To understand how spindle-localized Stu1 relocates to KTs, we bleached Stu1 along the complete metaphase spindle and performed FRAP. The recovery of metaphase spindle-associated Stu1 (Fig. 5C) was higher and much faster (∼29% in 0.6 sec) than that of Stu1 localized at the detached KTs. This indicates that Stu1 at metaphase spindles is more dynamic than Stu1 at detached KTs and that there is a larger pool of free Stu1 in the absence of detached KTs. Thus, it appears possible that the relocation of spindle-associated Stu1 to detached KTs is at least partly driven by an equilibrium-based mechanism that exploits different dissociation constants for Stu1–MT and Stu1–KT interactions. Irrespective of the exact mechanism, the fact that detached KTs prevent spindle localization of Stu1 and that Stu1 is essential for spindle formation (Yin et al. 2002) provides a novel checkpoint that prevents stable spindle formation in the presence of detached KTs.
Stu1 is essential for KT capturing
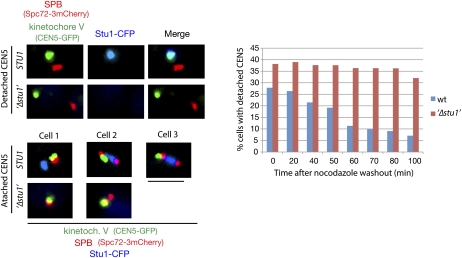
Our data suggest that Stu1 is involved in KT capturing. We therefore analyzed KT capturing in Stu1-depleted cells. One-hundred-eighty minutes after the release from G1 into nocodazole, Stu1-depleted cells did not reveal any Stu1 at detached KTs, proving the effective depletion (Fig. 6), and displayed ∼40% more detached KTs than the control cells (Fig. 6, time 0). Following a nocodazole washout, the control cells decreased the number of detached KTs dramatically within 100 min, whereas the Stu1-depleted cells did not (Fig. 6). Thus, Stu1 is essential for KT capturing. Considering that a detached KT accumulates an abundance of Stu1 molecules (see above) that can interact with MTs, a Stu1-marked KT should be a very efficient target for MTs. Stu1 thus appears to be a landmark for capturing. This is a completely novel function for a member of the CLASPs.
Figure 6.
Stu1 is required for KT capturing. Cells were depleted of Stu1 during a G1 arrest and otherwise processed as in Figure 3C. KT capturing was assayed by quantifying cells with a detached KT V (as shown) at the indicated time after the nocodazole washout. n > 100. Bar, 2 μm.
Ndc80, in contrast to Mad2 or Bub1, is essential for Stu1 recruitment at detached KTs
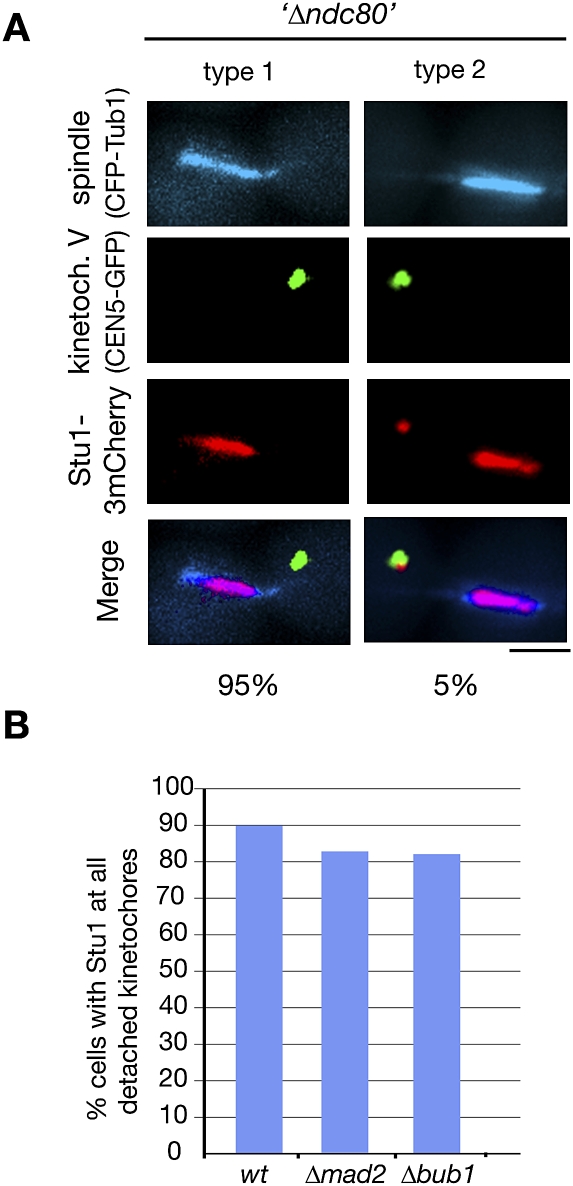
okp1-52 KTs apparently can still interact with Stu1 (see above). However, as shown by ChIP, the okp1-52 defect severely compromises the KT structure (Supplemental Fig. S6A). With the exception of components of the inner KT complexes (Ndc10 and Cse4), the KT localization of all other KT proteins tested was severely compromised in okp1-52 cells. We therefore wondered whether Stu1 binding was facilitated directly by the inner KT components or whether the remaining fraction of central and outer KT components was sufficient to support Stu1 interaction (like they support SAC function) (see above). When okp1-52 cells were released from G1 into nocodazole at 37°C, quite in contrast to wild-type cells (Fig. 3B), Stu1 localized exclusively at the few KTs residing close to the spindle poles but not to the abundance of KTs that were completely detached from the poles (Supplemental Fig. S6B). This confirms the observation described above that okp1-52 KTs in the vicinity of the spindle pole body recruit Stu1 (see the Discussion). Importantly, it also shows that most unattached okp1-52 KTs are not competent to recruit Stu1. This strongly indicates that the KTs at the poles are the ones that have sufficient central and outer KT components to execute partial KT functions, whereas the completely detached KTs have lost this ability. Consequently, Stu1 requires more than just the inner KT components for KT binding. Next, we investigated Stu1 localization in the absence of Ndc80. As described for ndc80-1 (Wigge et al. 1998), KTs of cells released from G1 after Ndc80 depletion were predominantly detached. Importantly, Stu1 did not localize to these detached KTs (Fig. 7A), but associated with ipMTs and consequently promoted the formation of a stable spindle. Thus, Ndc80 is essential for Stu1 recruitment to detached KTs. In contrast to this, and although it is reminiscent of the behavior of SAC components, Stu1 localization at detached KTs does not depend on Mad2 or Bub1 (Fig. 7B).
Figure 7.
KT localization depends on Ndc80 but not on Mad2 or Bub1. (A) Localization of Stu1 to detached KTs depends on Ndc80. GAL-NDC80 cells were depleted of Ndc80 during a G1 arrest, and Stu1 localization was quantified 105 min after the release. n > 100. Bar, 2 μm. (B) Localization of Stu1 to detached KTs does not depend on Mad2 or Bub1. Ninety minutes after the release from G1 into nocodazole, spindle poles (Spc72-3mCherry), KTs (Ame1-GFP), and Stu1-CFP were visualized, and Stu1 localization was quantified. n > 100.
The Stu1 C terminus is required for KT localization
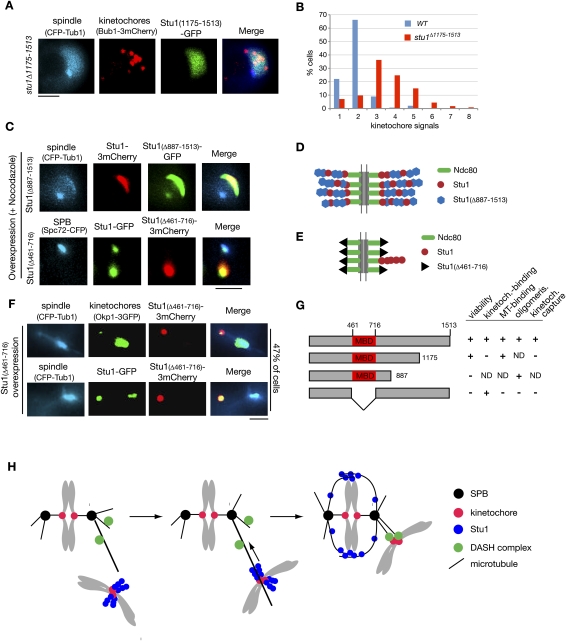
Deletion of the 339 C-terminal amino acids of Stu1 (Stu1Δ1175–1513) severely compromises the localization of Stu1 to detached KTs (Fig. 8A), but allows interaction with MTs (data not shown). Thus, although no apparent homology exists, Stu1, similar to its human orthologs (Maiato et al. 2003, 2005), uses C-terminal sequences for KT localization. However, they are probably not sufficient, since we were unable to observe KT localization of a C-terminal Stu1 fragment (amino acids 887–1513) (data not shown). stu1Δ1175–1513 cells exhibit slow growth but are viable. This could indicate that KT localization of Stu1 is not essential, or that some residual KT binding of Stu1Δ1175–1513 suffices for its KT function. The fact that the deletion of amino acids 887–1513 is lethal (data not shown) supports the latter interpretation, but more detailed analysis is required to clarify this point. When released from a G1 arrest into nocodazole, the stu1Δ1175–1513 mutant displayed an increase in detached KT signals in comparison with wild-type cells (Fig. 8B). This is consistent with the assumption that KT localization of Stu1 is required for KT capturing.
Figure 8.
Stu1 domain analysis. (A) C-terminal sequences are required for KT localization of Stu1. stu1Δ1175–1513 cells were analyzed 150 min after the release from G1 into nocodazole. Bar, 2 μm. (B) stu1Δ1175–1513 cells display an increased number of detached KTs per cell. Quantification of detached KTs or KT clusters in wild-type and stu1Δ1175–1513 cells processed as in A. n > 100. (C) Stu1 polymerization at detached KTs depends on Stu1's MBD (amino acids 461–716). Stu1Δ887–1513 and Stu1Δ461–716 were overexpressed from a GAL promoter in a wild-type STU1 background during a G1 arrest. Cells were analyzed 240 min after the release from G1 into nocodazole. Note that Stu1Δ887–1513 can still copolymerize with wild-type Stu1, whereas Stu1Δ461–716 cannot. Bar, 2 μm. (D,E) Putative models illustrating the phenotypes observed in C. (F) Overexpression of Stu1Δ461–716 results in detached KTs. Stu1Δ461–716 overexpression was as in C. Cells were analyzed 120 min after the release from G1. Bar, 2 μm. (G) Summary of Stu1 domain functions. (ND) Not done. (H) Model describing the role of Stu1 during KT capturing. For details, see the text.
The MT-binding domain (MBD) of Stu1 is required for Stu1 oligomerization and KT capturing
Amino acids 461–716 are important for Stu1–MT interaction in vitro (Yin et al. 2002). We constructed a mutant Stu1 (Stu1Δ461–716) that lacked this MBD, and found that Stu1Δ461–716 does not support viability (data not shown) and cannot bind to MTs in vivo (Fig. 8F). Next, we asked whether the MBD is required for the oligomerization of Stu1 at detached KTs. We first overexpressed Stu1Δ887–1513 that still has the MBD but lacks C-terminal sequences required for KT interaction (see above) in G1-arrested cells with a wild-type STU1 background and released them into nocodazole. Apparently initiated by wild-type Stu1 associated with detached KTs, mutant and wild-type Stu1 copolymerized (Fig. 8C,D) and formed structures similar to cells overexpressing wild-type Stu1 (see above; Fig. 5A). Thus, the Stu1 oligomerization domain does not reside within the 626 C-terminal amino acids. In contrast, upon overexpression in a wild-type background, Stu1Δ461–716 failed to produce the polymeric structures, but colocalized with wild-type Stu1 at detached KTs (Fig. 8C). This indicates that the MBD of Stu1 is essential for Stu1 oligomerization. Furthermore, overexpression of Stu1Δ461–716 allowed wild-type Stu1 to localize to (probably) MTs at the spindle poles. Therefore, Stu1Δ461–716 apparently competes with wild-type Stu1 for KT binding, and this indicates that Stu1Δ461–716 interacts with KTs independent of wild-type Stu1. Notably, we were not able to remove all wild-type Stu1 from the KTs by overexpression of Stu1Δ461–716. However, being dependent on Ndc80, Stu1 should have multiple binding sites at the KT (Joglekar et al. 2006). Therefore, even if only one of these sites is occupied by the oligomer-forming wild-type Stu1 and the other sites are occupied by the overexpressed, but oligomerization-incompetent, Stu1Δ461–716, a substantial amount of wild-type Stu1 may reside at the detached KTs (Fig. 8E).
Finally, we asked whether the MBD is important for KT capturing. Overexpression of Stu1Δ461–716 before a G1 release resulted in a high number of cells (47% of the population) with detached KTs (Fig. 8F). Furthermore, the cells arrested at metaphase, probably due to SAC activity. Overexpression of wild-type Stu1 during the G1 release or overexpression of Stu1Δ461–716 after metaphase did not result in detached KTs (data not shown). As observed above, wild-type Stu1 colocalized with Stu1Δ461–716 at detached KTs, but (due to competition at the KT) also associated with the spindle (Fig. 8F). Taken together, these data support that Stu1Δ461–716 (lacking MDB) is able to bind to KTs and to compete with wild-type Stu1 for KT localization. Furthermore, it reveals that the MBD of Stu1 is important for KT capturing (Fig. 8G).
Discussion
The role of Stu1 at unattached KTs
Summarizing our data, we suggest a model that describes the role of Stu1 at S. cerevisiae KTs (Fig. 8H).
Detached KTs present high-affinity, Ndc80-dependent, binding sites for Stu1. Several lines of evidence suggest that KT-bound Stu1, possibly via a conformation change, triggers Stu1 oligomerization. (1) Even one or few KTs compete for the pool of nuclear Stu1 so efficiently that Stu1 binding to MTs is not detectable. (2) As observed by fluorescent microscopy, the Stu1 signal dramatically exceeds the KT signal. (3) Upon overexpression in nocodazole-treated cells, Stu1 or Stu1Δ887–1513 (in a wild-type STU1 background) forms polymeric structures at detached KTs. This is quite in agreement with the idea that the additional Stu1 produced by overexpression extends oligomeric Stu1 structures that are present already at normal cellular Stu1 levels, although an aggregation artifact cannot be completely excluded.
Stu1 association with KTs is essential for KT capturing by MTs, since Stu1-depleted cells fail to capture detached KTs. Also, compromising the KT localization of Stu1 (as in stu1Δ1175–1513 cells) results in an increased number of detached KTs. The frequent occurrence of detached KTs in cells overexpressing Stu1Δ461–716 indicates that KT capturing is facilitated by the MBD of Stu1, either directly and/or, since this domain is also required for oligomerization, via Stu1 oligomerization. In any case, KT-associated Stu1 oligomers could provide a lattice for MT interaction and turn a detached KT into an efficient target for lateral capturing.
After capturing, Stu1 and KTs apparently comigrate to a spindle pole. This does not prove conclusively that Stu1 remains at the captured KT(s). In principle, it could be displaced upon KT–MT interaction, assembled onto an unattached sister KT, and transported to the pole.
Stu1 relocates from the captured KTs to MTs when the KTs reach the poles. This transition does not require a bipolar attachment of sister KTs, since captured KTs close to a pole but lacking bipolar attachment frequently revealed no Stu1 localization. Furthermore, scc1-72 cells that fail to achieve biorientation of KTs did not withhold Stu1 at their KTs. So what triggers the dissociation of Stu1 from the KT? (1) KT–MT interaction may be the sole factor that causes Stu1 dissociation. We consider this unlikely because we observed in several cases that, within a cluster of detached KTs, a few failed to recruit Stu1, and consequently were not captured. If Stu1 is released during the capturing process, one might expect that these remaining detached KTs receive some of the free Stu1. (2) Association of both sister KTs (syntelic or bipolar) with MTs may be the trigger. This would imply that even a syntelic KT–MT interaction causes alterations in the KT structure that promote Stu1 dissociation. (3) Interaction of the DASH complex with the core KT could displace Stu1 from the KT. For the most part, this should occur for a pair of sister KTs collectively only in the vicinity of the spindle poles (when a possibly unattached sister is captured). Thus, Stu1 dissociation from the KT would take place close to the spindle. This model is in agreement with our finding that Stu1 does not relocate from the captured KTs to the spindle when the DASH complex is defective (dad2-9 cells). Also, the persistent KT localization in okp1-52 cells might result from a compromised interaction of the DASH complex with a defective KT. Furthermore, it might explain why captured KTs remain at the plus ends of MTs, and prevent these MTs from depolymerizing when Kar3 and the DASH complex are compromised (Tanaka et al. 2007). Stu1 at these KTs would maintain KT–MT interaction and stabilize MT plus ends.
Localization of Stu1 to spindle MTs is a reversible step. Irrespective of whether detached KTs appeared in prometaphase, metaphase, or late anaphase (our unpublished data), spindle-localized Stu1 relocated to the detached KTs. At least for prometaphase and metaphase, this did not depend on the depolymerization of the spindle. Thus, spindle-localized Stu1 is competent to interact with detached KTs. Furthermore, since detached KTs effectively keep Stu1 from interacting with MTs, they regulate the stability of ipMTs. This may be important when newly formed KTs are captured and bipolar-attached in prometaphase. In the absence of stable ipMTs, spindle pole bodies will stay in close proximity, and thus increase the chance for biorientation of sister KTs. In agreement with this is the fact that overexpression of Cin8 in prometaphase, which results in an abnormally large distance between the spindle poles, delays bipolar attachment of KTs (Liu et al. 2008).
Stu1 at okp1-52 KTs and the spindle defect
Even when ipMTs were restabilized by KIP3 deletion, okp1-52 cells released from G1 at 37°C withheld Stu1 at KTs. Irrespective of whether okp1-52 cells were released in the presence or absence of nocodazole, Stu1 localized only to KTs close to the spindle poles, although detached KTs distant from the poles clearly existed, at least in the nocodazole-treated cells. This is in contrast to wild-type cells, where Stu1 only localized to detached KTs distant from the poles. Apparently, the okp1-52 KTs distant from the poles predominantly lost the structural requirements for Stu1 interaction, except when the mutation was induced during metaphase. The latter causes, according to ChIP (our unpublished data), considerably less KT damage than if the mutation is induced during a G1 release. The okp1-52 KTs close to the poles had at least one sister KT that was sufficiently intact to interact with a MT. We envision two possibilities as to why Stu1 interacts with these KTs. (1) One sister KT fails to attach to a MT and recruits Stu1. This would imply that the second KT is compromised to a point that prohibits MT attachment, but still allows Stu1 interaction. (2) Sister KTs may be in syntelic/lateral attachment with the poles, but the mechanism that releases Stu1 from attached KTs is defective. This mechanism may involve the DASH complex.
The okp1-52 KT causes a severe spindle defect largely because it allows spindle pole separation while sequestering Stu1. In strong support of this is the fact that interfering with Stu1 localization to okp1-52 KTs by Cse4 depletion or via the ndc80-1 mutation (our unpublished data) allows Stu1 to localize to ipMTs and thus to form stable metaphase or anaphase spindles. In contrast to the okp1-52 cells, the stu1-5 mutant (Yin et al. 2002) fails to separate the spindle poles. A possible explanation is that sufficient KTs in stu1-5 cells achieve biorientation, whereas in okp1-52 cells, they don't. Thus the spindle poles are subject to an inward force in stu1-5 cells and require Stu1-stabilized ipMTs for separation. This force is absent in okp1-52 cells, therefore allowing spindle poles to separate in the absence of stable ipMTs.
ame1-2 cells fail to execute the FEAR-dependent Cdc14 activation. Although not directly shown, this is most likely also the case for the okp1-52 mutant, since overexpression of Cdc14 is required to locate Ase1 to the spindle midzone in okp1-52, Δkip3 cells. Consequently, a lack of Cdc14 activity during spindle pole separation probably contributes to the ame1-2 and okp1-52 spindle defect. However, it is clearly not the sole cause, since the spindle defect occurs already at metaphase spindle length and cannot be rescued by Cdc14 overexpression. One reason for the lack of FEAR signaling may be the prolonged SAC activation in ame1-2 and okp1-52 cells. However, the mutants maintain an active SAC only ∼40 min longer than control cells, whereas Cdc14 activation in ame1-2 cells was not observed for at least 2 h after Cdc14 activation in wild-type cells. Thus, the ame1-2 and okp1-52 defects may interfere with FEAR signaling not only via SAC activation.
The mitotic function of CLASP proteins
From yeast to humans, CLASPs are required for the stability of mitotic spindles. Interfering with CLASP function results in various aberrant spindle phenotypes, including monopolar spindles originating from collapsed spindle poles (Maiato et al. 2002, 2003). As shown for humans and yeast (Yin et al. 2002; Maiato et al. 2003), CLASPs localize to the spindle midzone during anaphase, where they contribute to spindle stability by MT rescue. KT localization/function of CLASPs has been demonstrated for metazoans and yeast (see above). In metazoans, KT-bound CLASP regulates the dynamic stability of kMTs by promoting MT growth (Maiato et al. 2002, 2003, 2005). This facilitates chromosome biorientation and congression. In agreement with these functions, CLASPs remain at KTs in metazoans throughout metaphase and at least early anaphase. The S. cerevisiae CLASP Stu1, however, dissociates from KTs before it achieves biorientation in prometaphase. Also, the S. pombe CLASP cls1p only localizes to KTs up to prometaphase (Bratman and Chang 2007). Thus, in yeasts, the regulation of kMTs dynamics via a KT-associated CLASP may not be required for bipolar attachment and congression. However, it cannot be completely excluded that a small amount of Stu1 resides at the S. cerevisiae KTs (or is localized there via MT plus ends) during metaphase. For S. cerevisiae, we established a novel role of KT-bound CLASP. By accumulating at a detached KT, it marks this KT and facilitates capturing. At the same time, this accumulation at a detached KT prevents stabilization of the spindle and probably facilitates timely biorientation. Whether these functions are conserved in metazoans is not clear. When CLASP1 function was compromised in human cells, MTs were still able to attach to one of the sister KTs (Maiato et al. 2003). However, also in S. cerevisiae, a majority of KTs remained in the vicinity of spindle poles after Stu1 depletion. If these KTs are attached to MTs, then Stu1 may only be required to capture KTs that got further displaced from the spindle poles and, due to the low MT density, need additional help to be captured. This “emergency mechanism” could be a role for CLASPs in all eukaryotes. Intriguingly, KT localization of human CLASP1 is most pronounced in prometaphase (Maiato et al. 2003). On the other hand, the way in which Stu1 and metazoan CLASPs interact with KTs might be different, although C-terminal sequences are important in both cases. Consistent with the fact that Stu1 physically interacts with Nuf2 (a component of the Ndc80 complex) (Tarassov et al. 2008), it requires an intact Ndc80 complex for KT localization. In contrast to this, interaction of C. elegans CLASPCLS-2 with the KT is independent of Ndc80 (Cheeseman et al. 2005). This could indicate that only S. cerevisiae uses Stu1 for capturing.
Metazoan CLASPs interact with CLIPs (Akhmanova and Hoogenraad 2005), and it is thus intriguing that the human CLIP-170 and the Drosophila CLIP-190 localize specifically to unattached KTs (Dujardin et al. 1998; Dzhindzhev et al. 2005). The KT localization of human CLASPs does not depend on CLIP170 (Maiato et al. 2003; Mimori-Kiyosue et al. 2006). Nevertheless, it will be of interest to test whether CLIPs serve as a capturing device in metazoans. The S. cerevisiae CLIP Bik1 localizes to KTs throughout mitosis and is important for KT–MT interaction in polyploids (Lin et al. 2001), but it is currently unclear whether Bik1 is important for capturing. Also, there is currently no evidence that Stu1 interacts with Bik1.
A +TIP that has been described in the context of KT capture in S. cerevisiae is Stu2 (Tanaka et al. 2005). Stu2 localizes to attached and detached KTs. Upon capturing, a fraction of it is transported from the captured KT to the plus end of the capturing MT and facilitates MT rescue. Whether Stu2 is required for capturing beyond the stabilization of capturing MTs is unclear. Thus, Stu1 is currently the only +TIP that facilitates capturing while bound to the KT.
How Stu1 sequestering is initiated by unattached KTs; how it facilitates capturing, an initial step in chromosome segregation; and whether these functions are conserved in other eukaryotes are intriguing future questions.
Materials and methods
Yeast strains
Yeast strains and usage links are shown in Supplemental Table S1. Construction and growth conditions are provided in the Supplemental Material.
Microscopy
Live-cell imaging, including the time-lapse experiments in Figures 3C and 4, A and B, were performed on a Life Science Imaging Station Olympus Cell R as described (Kemmler et al. 2009). For the experiment in Figure 4B, the medium on the microscope dish was adjusted to 15 μg/mL nocodazole during imaging.
For the time-lapse imaging shown in Figure 1C and Supplemental Figure S2A as well as Supplemental Movies S1–S4, cells were released from G1 and spotted on a concavalin A-coated culture dish. After 60 min at 37°C, the rich medium was exchanged for nonfluorescent medium, and images were collected at 37°C on an inverted Zeiss LSM510 Meta microscope, using a ×63/1.40 NA objective lens. Time-lapse sequences were taken every minute by scanning through five Z stacks and processed as maximal intensity projections.
FRAP experiments were done on an inverted Zeiss LSM510 Meta microscope with a Plan-Apochromat 100×/1.40 NA objective lens, a 158-nm pinhole, and a 30-mW, 488-nm Argon laser adjusted to bleach an area of 1-μm radius. Three prebleach images were acquired followed by five to 10 laser iterations (80% intensity) for 50 scans with 0.2- to 6-sec intervals. Images were composed of two to five Z stacks of 0.45-μm optical section spacing. Fluorescence intensities were measured and corrected for autobleaching by using ImageJ in combination with the Zeiss LSM import panel and FRAP profiler plug-ins from MBF (http://www.macbiophotonics.ca).
Acknowledgments
We thank Maria Knapp, Eileen Dietzel, and Nicole Grau for excellent technical assistance; Dimitris Liakopoulos for discussions and help with the microscope; Maren Scharfenberger-Schmeer for time-lapse microscopy; and Elmar Schiebel and Frank Uhlman for strains and/or plasmids. C.F. is a member of the MD/PhD program of the Universität Heidelberg. This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.541309.
Supplemental material is available at http://www.genesdev.org.
References
- Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: Mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Amaro IA, Costanzo M, Boone C, Huffaker TC. The Saccharomyces cerevisiae homolog of p24 is essential for maintaining the association of p150Glued with the dynactin complex. Genetics. 2008;178:703–709. doi: 10.1534/genetics.107.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, MacLeod I, Yates JR, III, Oegema K, Desai A. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Amon A. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes & Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes & Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin D, Wacker UI, Moreau A, Schroer TA, Rickard JE, De Mey JR. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J Cell Biol. 1998;141:849–862. doi: 10.1083/jcb.141.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Rogers SL, Vale RD, Ohkura H. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J Cell Sci. 2005;118:3781–3790. doi: 10.1242/jcs.02504. [DOI] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes & Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1–Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Schiebel E. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 2008;7:283–286. doi: 10.4161/cc.7.3.5349. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Lawrence C, Roostalu J, Schiebel E. Cdc14-regulated midzone assembly controls anaphase B. J Cell Biol. 2007;177:981–993. doi: 10.1083/jcb.200702145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes & Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes & Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore–microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liang F, Jin F, Wang Y. The coordination of centromere replication, spindle formation, and kinetochore–microtubule interaction in budding yeast. PLoS Genet. 2008;4:e1000262. doi: 10.1371/journal.pgen.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, McQueen J, Cuschieri L, Vogel J, Measday V. Spc24 and Stu2 promote spindle integrity when DNA replication is stalled. Mol Biol Cell. 2007;18:2805–2816. doi: 10.1091/mbc.E06-09-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Sampaio P, Lemos CL, Findlay J, Carmena M, Earnshaw WC, Sunkel CE. MAST/Orbit has a role in microtubule–kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J Cell Biol. 2002;157:749–760. doi: 10.1083/jcb.200201101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EK, Goldstein B. Asymmetric spindle positioning. Curr Opin Cell Biol. 2006;18:79–85. doi: 10.1016/j.ceb.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Sasaki H, Matsui C, Akhmanova A, Tsukita S, Vorobjev I. Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells. 2006;11:845–857. doi: 10.1111/j.1365-2443.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Separase regulates INCENP–Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–254. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O'Toole ET. The spindle cycle in budding yeast. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Yin H, You L, Pasqualone D, Kopski KM, Huffaker TC. Stu1p is physically associated with β-tubulin and is required for structural integrity of the mitotic spindle. Mol Biol Cell. 2002;13:1881–1892. doi: 10.1091/mbc.01-09-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J, Saunders WS. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]