Abstract
Cholesterol homeostasis is required to maintain normal cellular function and avoid the deleterious effects of hypercholesterolemia. Here we show that the Drosophila DHR96 nuclear receptor binds cholesterol and is required for the coordinate transcriptional response of genes that are regulated by cholesterol and involved in cholesterol uptake, trafficking, and storage. DHR96 mutants die when grown on low levels of cholesterol and accumulate excess cholesterol when maintained on a high-cholesterol diet. The cholesterol accumulation phenotype can be attributed to misregulation of npc1b, an ortholog of the mammalian Niemann-Pick C1-like 1 gene NPC1L1, which is essential for dietary cholesterol uptake. These studies define DHR96 as a central regulator of cholesterol homeostasis.
Keywords: Gene regulation, nuclear receptor signaling, cholesterol metabolism, Niemann-Pick disease gene
Cholesterol is an essential component of cell membranes that influences the permeability and fluidity of the lipid bilayer. Cholesterol also acts as a precursor for steroid hormone biosynthesis and contributes to cell–cell signaling pathways. These critical cellular functions are supported by regulatory mechanisms that maintain normal cholesterol levels and prevent hypercholesterolemia, which is a major risk factor for cardiovascular disease in humans. Cholesterol homeostasis in vertebrates is achieved primarily through de novo synthesis and dietary uptake (Ikonen 2008). Although extensive studies have defined a central role for the sterol regulatory element-binding protein (SREBP) family of transcription factors in controlling cholesterol synthesis (Brown and Goldstein 1997), the mechanisms that regulate dietary cholesterol absorption remain more poorly understood. One central component of this pathway is the Niemann-Pick C1-like 1 gene NPC1L1, which encodes a plasma membrane protein that mediates the uptake of dietary cholesterol by the intestine (Wang 2007; Ge et al. 2008). Mouse mutants for NPC1L1 display significantly reduced levels of cholesterol absorption and are insensitive to treatment with the anti-hypercholesterolemia drug ezetimibe, which acts as a specific NPC1L1 inhibitor (Davis et al. 2008). Another major regulator of cholesterol homeostasis is the liver X receptor α (LXRα) nuclear receptor, which binds cholesterol metabolites and regulates the transcription of genes that control cholesterol transport and metabolism, including NPC1L1 (Duval et al. 2006; Kalaany and Mangelsdorf 2006; Valasek et al. 2007).
We used the fruit fly, Drosophila, as a model system to study the regulation of cholesterol homeostasis. Unlike vertebrates, insects are cholesterol auxotrophs that are unable to synthesize this essential compound (Va'nt Hoog 1936). Little is known, however, about the mechanisms that regulate the uptake of dietary cholesterol in Drosophila. A recent study showed that the fly ortholog of NPC1L1, npc1b, is expressed specifically in the midgut and is essential for dietary cholesterol absorption (Voght et al. 2007). Other NPC disease gene homologs in Drosophila also contribute to cholesterol homeostasis. The Drosophila ortholog of vertebrate NPC1, npc1a, and two of the eight fly NPC2 homologs, npc2a and npc2b, play important roles in intracellular cholesterol trafficking and synthesis of the steroid hormone 20-hydroxyecdysone (20E) (Huang et al. 2005, 2007; Fluegel et al. 2006). Other predicted regulators of cholesterol metabolism in Drosophila, however, remain unstudied, and upstream factors that might sense cholesterol levels and control cholesterol homeostasis are undefined.
In this study, we show that the Drosophila DHR96 nuclear receptor binds cholesterol, is essential for survival on a low-cholesterol diet, and is required to maintain cholesterol homeostasis when animals are grown on a high-cholesterol diet. We further show that dietary cholesterol regulates the transcription of many genes that are expressed in the midgut and that act in lipid metabolism, and that this transcriptional response fails to occur in DHR96 mutants. Misregulation of one of these genes, npc1b, is sufficient to explain the cholesterol accumulation defect seen in DHR96 mutants, defining npc1b as a critical functional target of the receptor. This study provides a new framework for understanding the molecular mechanisms that regulate cholesterol homeostasis.
Results and Discussion
DHR96 binds cholesterol
Mass spectrometry (MS) was used to identify potential ligands for DHR96. The DHR96 ligand-binding domain (LBD) was overexpressed, purified from insect cells, and subjected to electrospray ionization (ESI) MS, under both denaturing and nondenaturing conditions. The full mass range spectrum of the sample from the denaturing condition had a series of peaks corresponding to the DHR96 LBD, with a measured molecular weight (MW) of 31,831.92 Da, close to the predicted theoretical mass (31,830.84 Da for His6-DHR96S471-H723) (data not shown). Under nondenaturing conditions, the full mass range scan detected an additional series of peaks corresponding to a MW of 32,218.44 Da. This mass shift indicates that the DHR96 LBD copurifies with a molecule of 386.52 Da in a stoichiometry of 1:1, indicative of specific ligand binding.
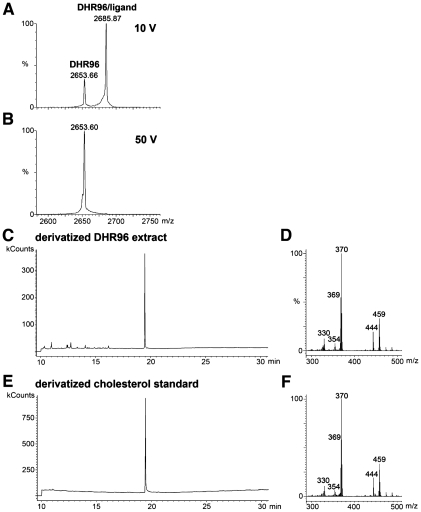
One of the presumptive DHR96/ligand complex peaks (2685.87 m/z) was then selected for collision-induced dissociation (CID) with a collision voltage of 10 V, resulting in the generation of two ions in the 12+ charge state (Fig. 1A). One ion represents the mass of the intact DHR96 LBD/ligand complex (2685.87 m/z; 32218.44 Da), while the other represents the unbound DHR96 LBD (2653.66 m/z; 31831.92 Da). Using a higher collision voltage of 50 V completely disrupted the DHR96 LBD/ligand complex to generate a CID spectrum with only an unbound receptor (2653.60 m/z; 31831.2 Da) (Fig. 1B). The charge state of the receptor ion did not change upon loss of the ligand and no peak was observed in the lower mass range, indicating that the ligand is a neutral molecule. These observations, along with the mass shift of 386.52 Da, suggest that the bound molecule may be cholesterol (MW, 386.6 Da).
Figure 1.
Mass spectrometry identifies cholesterol bound to the DHR96 LBD. (A) CID mass spectrum of the DHR96/cholesterol complex under nondenaturing conditions with a collision voltage of 10 V. At this voltage, a portion of the ions representing the 12+ charge state of the DHR96/ligand complex (2685.87 m/z) fragment to generate ions of the 12+ charge state of DHR96 (2653.66 m/z), losing the ligand as a free molecule. The charge states of the ions at 2685.87 and 2653.66 m/z were determined from full-range MS scan spectra (data not shown). (B) CID mass spectrum of the DHR96/ligand complex with a collision voltage of 50 V caused complete dissociation of the receptor/ligand complex into unbound receptor. (C–F) The elution time of the major peak on a gas chromatogram of a derivatized chloroform–methanol extraction of the DHR96 LBD (C) matches that of a derivatived cholesterol standard (E). The corresponding electron ionization spectrum from the major peak of the DHR96 LBD at 19 min generates major fragmentation ions (D) that correspond closely to the major fragmentation ions generated from the major peak of the derivatived cholesterol standard (F).
To further verify the identity of the molecule bound to the DHR96 LBD, purified receptor was extracted with chloroform–methanol and a portion of the extract was derivatized and analyzed by gas chromatography/MS (GC/MS) with electron ionization (EI). A single major peak at 19 min was observed in the GC/MS chromatogram (Fig. 1C). The retention time of this peak was identical to that of a derivatized cholesterol standard (Fig. 1E). Moreover, the major peak of the DHR96 sample on GC/MS produced an EI spectrum (Fig. 1D) that matched a previously described spectrum for derivatized cholesterol (Fig. 1F; Bitsch et al. 2003). Based on these two independent modes of MS—ESI/MS and GC/MS—we conclude that the DHR96 LBD is capable of binding a single molecule of cholesterol. GC/MS analysis of several other Drosophila nuclear receptors indicated that this interaction is specific to DHR96 (Supplemental Fig. S1). In addition, although this interaction is stable through three rounds of protein chromatography, the partial dissociation of ligands at 10 V and complete dissociation at 50 V suggests that the bound cholesterol is exchangeable.
DHR96 regulates the transcriptional response to cholesterol
The observation that DHR96 binds cholesterol raises the possibility that it may mediate transcriptional responses to this compound. To test this possibility, control and DHR96 mutants were grown on low-cholesterol medium in the absence or presence of 0.03% cholesterol, and were subjected to microarray analysis. From this analysis, 117 genes were identified that are up-regulated at least 1.4-fold in response to cholesterol in wild-type larvae, along with 270 genes that are down-regulated (Fig. 2A,B; Supplemental Table S1). This response appears to be rapid, occurring within 1–2 h of cholesterol treatment, and displays similar cholesterol dose response profiles (Supplemental Fig. S2). We also see a response to sitosterol treatment, indicating that it is not specific to cholesterol (Supplemental Fig. S3). Interestingly, a number of the cholesterol-regulated genes are predicted to play central roles in cholesterol metabolism and transport. These include CG32186, which encodes a predicted ABCA3-like transporter; CG6472, which encodes a lipoprotein lipase (LPL) homolog; and CG8112, which encodes acyl-CoA:cholesterol acetyltransferase (ACAT) (Supplemental Table S5). This enzyme plays a critical role in esterifying cholesterol, which is the primary stored form of intracellular cholesterol (Ikonen 2008). Four Drosophila homologs of NPC genes are also regulated by cholesterol, including npc1b and three NPC2 homologs: npc2b, npc2c, and npc2d (Huang et al. 2007).
Figure 2.
Most cholesterol-regulated genes depend on DHR96 for their proper expression. (A,B) Heat maps are depicted representing the top 50 genes that are either up-regulated by cholesterol (A) or down-regulated by cholesterol (B) in CanS wild-type larvae, along with the responses of these same genes in DHR961 mutants, as determined by microarray analysis. The heat maps are arranged from top to bottom by their fold response to cholesterol in wild-type larvae. The expression levels in columns 2–4 of each heat map are normalized to the expression level in column 1 (CanS −, cholesterol). Red represents increased transcript levels relative to the transcript level in column 1, while green represents lower transcript levels. (C) RNA isolated from CanS control larvae and DHR961 mutant larvae, grown in either the absence (−) or presence (+) of cholesterol, was analyzed by Northern blot hybridization for expression of npc2c, CG14745, npc1b, CG5932, CG31148, and npc2e. Hybridization to detect rp49 was used as a control for loading and transfer.
Importantly, this transcriptional response is almost entirely dependent on DHR96 function. Only 13% of the genes that are up-regulated by cholesterol in wild-type larvae (15 genes), and 10% of the genes that are down-regulated (27 genes), display a similar profile of expression in DHR96 mutants treated with cholesterol (Fig. 2A,B; Supplemental Fig. S4). Moreover, many additional genes become responsive to cholesterol in DHR96 mutants, with 355 genes up-regulated and 446 genes down-regulated at least 1.4-fold, indicating that DHR96 normally plays a key role in suppressing this transcriptional program (Supplemental Fig. S4; Supplemental Table S2). These DHR96-regulated genes include four of the remaining five NPC2 family members: npc2e, npc2f, npc2g, and npc2h (Fig. 2C; Supplemental Table S5). DHR96 also regulates Lip3 (CG8823), which encodes a predicted cholesterol ester hydrolase; CG9663, which encodes an ABCG1 homolog; CG11162, which encodes a sterol-C4-methyl oxidase; and several genes that encode predicted stearoyl-CoA-desaturases. Many genes involved in other aspects of lipid metabolism are also misregulated in DHR96 mutants (Supplemental Table S5). In addition, DHR96 itself is down-regulated approximately twofold by cholesterol in wild-type larvae, and this response fails to occur in DHR96 mutants, suggesting that there is autoregulation. Further examination of the cholesterol-regulated genes in both wild-type and DHR96 mutant larvae revealed that many of these genes are expressed in the midgut, consistent with the critical role of dietary cholesterol uptake in a cholesterol auxotroph such as Drosophila (Supplemental Tables S5, S6).
Northern blot hybridizations confirmed that genes such as npc2c and CG14745 (which encodes a predicted peptidoglycan recognition protein) are induced by cholesterol in wild-type larvae, while genes such as npc1b, CG5932 (which encodes a gastric lipase), and CG31148 (which encodes a predicted enzyme in sphingolipid metabolism) are repressed by cholesterol (Fig. 2C). Other genes, such as npc2e, are not responsive to cholesterol in wild-type larvae. All of these genes, however, are misregulated in DHR96 mutants.
DHR96 mutants are unable to survive on a low-cholesterol diet
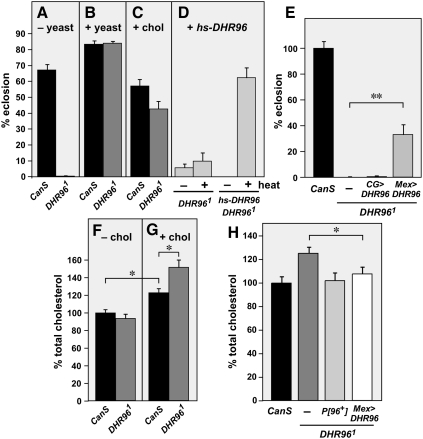
The central role of DHR96 in mediating transcriptional responses to cholesterol suggests that it may contribute to the regulation of cholesterol homeostasis. As an initial test of this possibility, we examined how control and DHR961 mutants respond to growth on a low-cholesterol diet. Whereas most wild-type larvae grown on this medium develop through to adulthood, DHR96 mutants arrest their development primarily as second instar larvae and die within several days (Fig. 3A). Supplementation with a complete nutrient source, yeast, was sufficient to rescue this lethality (Fig. 3B). Efficient rescue was also achieved by supplementing with cholesterol, demonstrating that the lack of this essential nutrient is a cause of the lethality (Fig. 3C). Widespread heat-induced expression of a wild-type DHR96 transgene, or specific expression of DHR96 in the midgut, is also sufficient to rescue the lethality of DHR96 mutants grown on the low-cholesterol diet (Fig. 3D,E). Specific expression of DHR96 in the fat body of DHR96 mutants had no effect. Taken together, these observations indicate that DHR96 mutants are unable to survive under limiting cholesterol conditions, and that this phenotype is due to a specific loss of DHR96 function in the midgut. Similar results were obtained when control and DHR96 mutants were raised on a chloroform-extracted medium that is deprived of all sterols (data not shown). A dose response study on the low-cholesterol diet revealed that 0.03% cholesterol is the ideal concentration for rescue (Supplemental Fig. S5). This amount is identical to the optimal amount required for Drosophila larval survival on a minimal defined medium (Sang 1956). A number of other sterols are able to substitute for cholesterol in these experiments, including 7-dehydrocholesterol, ergosterol, dehydroergosterol, sitosterol, and stigmasterol (data not shown), consistent with their ability to support normal growth of wild-type Drosophila (Cooke and Sang 1970). Supplementing the low-cholesterol medium with other lipids, however, including triacylglycerol (TAG) and oleic acid, had no effect, indicating that the rescue is specific to sterols (data not shown).
Figure 3.
DHR96 regulates cholesterol homeostasis. (A–C) DHR96 mutants arrest development on a low-cholesterol medium. CanS control and DHR961 mutant larvae were maintained on a low-cholesterol medium without supplementation (−yeast) (A), supplemented with yeast (+yeast) (B), or supplemented with 0.03% cholesterol (+chol) (C), and scored for the percent of adults that eclosed. (D) DHR961 mutants carrying a heat-inducible wild-type DHR96 transgene (+hs-DHR96) were grown on the low-cholesterol medium without supplementation in either the absence (−heat) or presence (+heat) of heat treatment, and scored for the percent of adults that eclosed. (E) The lethality of DHR96 mutants maintained on the low-cholesterol medium is rescued by expressing DHR96 in the midgut. CanS control and DHR961 mutant larvae, maintained either without any transgenes (−), with CG-GAL4;UAS-DHR96 (CG > DHR96; fat body-specific), or with Mex-GAL4;UAS-DHR96 (Mex > DHR96; midgut-specific) were grown on the low-cholesterol medium without supplementation and were scored for the percent of adults that eclosed. (F,G) DHR96 mutants accumulate cholesterol when grown on a high-cholesterol diet. CanS control and DHR961 mutant larvae were grown on the low-cholesterol medium either without supplementation (−chol) (F) or in the presence of 0.03% cholesterol (+chol) (G). Total cholesterol levels were measured in larvae collected 2 d after hatching and normalized for total protein. Data were pooled from two experiments and are presented as normalized to a wild-type (minus added cholesterol) level of 100%. (H) CanS control and DHR961 mutant larvae, maintained either without any transgenes (−), with a wild-type DHR96 genomic construct (P[96]+), or with Mex-GAL4;UAS-DHR96 (Mex > DHR96) were grown in the presence of 0.03% cholesterol. Total cholesterol levels were measured in larvae collected 2 d after hatching and were normalized for total protein. Data are presented as normalized to a wild-type level of 100%. Error bars are ±SE. (*) P < 0.05; (**) P < 5 × 10−4.
Several lines of evidence indicate that proper cholesterol metabolism is essential for larval development. Drosophila larvae grown in the absence of sterols arrest development at the first or second instar, similar to the lethal phase of DHR96 mutants grown under low-cholesterol conditions (Cooke and Sang 1970). Similarly, mutants for npc1a die as first instar larvae due to defects in intracellular cholesterol trafficking and reduced production of the molting hormone 20E (Huang et al. 2005; Fluegel et al. 2006). Double mutants for npc2a and npc2b also fail to progress through development and can be rescued by feeding 20E (Huang et al. 2007). DHR96 mutants grown on a high-cholesterol diet fail to show any clear defects in intracellular cholesterol localization, as detected by filipin staining (Supplemental Fig. S6). In addition, feeding 20E to DHR96 mutants maintained on the low-cholesterol diet does not rescue their lethality (data not shown). These observations do not support the hypothesis that the lethality in DHR96 mutants is due to a defect in cholesterol trafficking that affects 20E production. Rather, this phenotype is likely to arise from other defects, such as inefficient cholesterol utilization, changes in cholesterol storage, or disruption of other lipid metabolic pathways.
DHR96 mutants accumulate cholesterol
To determine whether DHR96 mutants display defects in cholesterol homeostasis, we measured total cholesterol levels in control and DHR961 mutant larvae grown on the low-cholesterol medium, in either the absence or presence of 0.03% cholesterol. DHR96 mutants have the same level of cholesterol as wild-type larvae when propagated without cholesterol supplementation (Fig. 3F). In the presence of added cholesterol, however, DHR96 mutants display cholesterol levels that are significantly higher than the 10%–20% increase seen in control larvae (Fig. 3G). This cholesterol accumulation defect can be rescued by either a wild-type genomic DHR96 transgene or expression of wild-type DHR96 in the midgut of mutant larvae, indicating that it arises from a specific loss of DHR96 function in this tissue (Fig. 3H).
An npc1b mutation rescues the cholesterol accumulation defect in DHR96 mutants
Expression of npc1b, which is required for dietary cholesterol absorption, is down-regulated when wild-type larvae are treated with cholesterol (Fig. 2C). The observation that this switch fails to occur in DHR96 mutants could explain why these animals accumulate excess cholesterol when grown on a high-cholesterol diet. Consistent with this, we found that DHR96 is expressed in the region of the larval midgut, where npc1b exerts its functions (Fig. 4A; King-Jones et al. 2006; Voght et al. 2007). This overlap in DHR96 and npc1b expression also appears to be functionally significant because specific expression of wild-type DHR96 in the midgut of DHR96 mutants is sufficient to restore appropriate npc1b transcriptional repression in response to cholesterol (Fig. 4B). Importantly, the accumulation of cholesterol in DHR96 mutants is dependent on npc1b function because npc1b;DHR96 double mutants have wild-type levels of cholesterol (Fig. 4C). This is consistent with the normal cholesterol levels seen in npc1b mutants at this stage of development, which likely derive from maternal loading of cholesterol during oogenesis (Voght et al. 2007).
Figure 4.
npc1b contributes to the cholesterol accumulation phenotype in DHR96 mutants. (A) DHR96 and npc1b are both expressed in the midgut. Midguts were dissected from NPC1b-GAL4;UAS-nGFP third instar larvae (NCP1b > nGFP) (Voght et al. 2007) and stained with affinity-purified antibodies to detect DHR96 protein. DHR96 protein is shown in red, and NPC1b expression is shown in green. All images were taken from the same field and focal plane. (B) DHR96 regulates npc1b transcription. RNA was extracted from CanS control and DHR961 mutant larvae, maintained in the presence of 0.03% cholesterol either without any transgenes (−) or with Mex-GAL4;UAS-DHR96 (Mex > DHR96), and analyzed by quantitative RT–PCR for levels of npc1b transcript. npc1b expression levels are presented as normalized to the level in CanS. (C) An npc1b mutation rescues the cholesterol accumulation defect in DHR96 mutants. CanS control, DHR961 mutant larvae, and npc1b1;DHR961 double mutants were grown in the presence of 0.03% cholesterol. Total cholesterol levels were measured in larvae collected 2 d after hatching and were normalized for total protein. Data are presented as normalized to a wild-type level of 100%. The cholesterol levels in CanS and npc1b1;DHR961 double mutants are not significantly different (P = 0.19). (D) Sterol absorption, but not fatty acid absorption, is blocked by an npc1b mutation in a DHR96 mutant background. CanS control, DHR961 mutants, npc1b1 mutants, and npc1b1;DHR961 double mutants were grown on a low-cholesterol medium supplemented with either 3H-cholesterol, 3H-sitosterol, or 3H-oleic acid along with 14C-glucose. Levels of radioactive lipid were normalized to the 14C-glucose and are presented as normalized to a wild-type level of 100%. Error bars are SE. (*) P < 0.05; (**) P < 0.01; (***) P < 1 × 10−4.
We tested whether DHR96 mutants require npc1b function for their ability to absorb dietary cholesterol. Control larvae, DHR961 mutants, npc1b1 mutants, and npc1b1;DHR961 double mutants were grown on the low-cholesterol medium in the presence of 3H-cholesterol. Radioactive glucose was also added to the food in order to normalize the levels of cholesterol absorption and to control for different feeding rates (Voght et al. 2007). Under these conditions, DHR96 mutants display a similar level of cholesterol uptake as wild-type larvae (Fig. 4D). This result is consistent with the wild-type levels of total cholesterol seen in DHR96 mutants grown on a low-cholesterol diet (Fig. 3F). In contrast, both npc1b mutants and npc1b;DHR96 double mutants display a dramatic reduction in cholesterol absorption (Fig. 4D). This result agrees with the original study of npc1b mutants, and indicates that DHR96 mutants require NPC1b to take up dietary cholesterol (Voght et al. 2007). A similar result was seen with a key plant sterol, 3H-sitosterol, suggesting that this pathway mediates general sterol absorption (Fig. 4D). Moreover, the npc1b mutation has no effect on the ability of DHR96 mutants to absorb oleic acid, consistent with its specific role in sterol uptake (Fig. 4D). Taken together, these observations indicate that the misregulation of npc1b expression in DHR96 mutants is sufficient to explain their inability to maintain proper cholesterol homeostasis when grown on a high-cholesterol diet.
It is important to note, however, that the cholesterol accumulation defect seen in DHR96 mutants does not appear to be related to the lethality that is observed when the mutant is grown on a low-cholesterol diet. DHR96 mutants that arrest their development when maintained on a low-cholesterol diet display normal levels of cholesterol (Fig. 3F). In addition, the npc1b;DHR96 double mutants are small and die earlier than npc1b mutants alone when raised in either the presence or absence of cholesterol, in spite of their wild-type cholesterol levels (M Horner and CS Thummel, unpubl.). Thus, it is likely that the DHR96 mutants suffer from additional metabolic defects beyond their inability to properly regulate npc1b transcription. This conclusion is supported by the widespread effects of the DHR96 mutation on the expression of genes involved in lipid metabolism and midgut physiology, as well as the expression of DHR96 in tissues outside the midgut (King-Jones et al. 2006). Further studies of DHR96 mutants should help to uncover its other key metabolic activities.
DHR96 coordinates the uptake of dietary lipids
The studies reported here define essential functions for DHR96 in maintaining cholesterol homeostasis during larval development. A parallel set of studies in our laboratory has also identified nonessential functions for DHR96 in lipid metabolism during adult stages due to their reduced levels of stored energy in the form of TAG (Sieber and Thummel 2010). Interestingly, microarray analysis of DHR96 mutant adults raised on normal growth medium revealed widespread effects on the transcription of genes expressed in the midgut, many of which are identical to the DHR96 regulatory targets described in this study. Among this core set of DHR96 target genes is CG5932, which encodes a gastric lipase that is required for the breakdown of dietary fat. Misregulation of CG5932 in the midgut is sufficient to explain the defects that we observed in TAG homeostasis in DHR96 mutants, analogous to the important role of DHR96 in regulating npc1b expression to maintain proper cholesterol homeostasis. Interestingly, CG5932 transcription is also regulated by cholesterol, raising the possibility that this dietary sterol, acting through DHR96, may coordinate dietary TAG breakdown with cholesterol absorption (Fig. 2C). This convergence of our results points to a central role for DHR96 in midgut physiology and provides a strong foundation to refine our understanding of its critical regulatory functions in maintaining lipid homeostasis.
DHR96 is a nuclear protein, in the presence or absence of cholesterol, consistent with its predicted role as a direct regulator of transcription (Fig. 4A; data not shown). It has a unique P-box sequence within its DNA-binding domain (DBD), which determines its DNA-binding specificity. This sequence is shared by only three Caenorhabditis elegans nuclear receptors: DAF-12, NHR-48, and NHR-8. Although a preferred binding site has been identified for DAF-12, it not known whether DAF-12 contacts this sequence as a homodimer or a heterodimer with another nuclear receptor (Shostak et al. 2004). Similarly, it is unclear whether DHR96 binds DNA as a monomer, homodimer, or heterodimer with the Drosophila retinoid X receptor (RXR) ortholog, like its mammalian homologs. In addition, the observation that DHR96 target genes display all possible combinations of regulatory responses to cholesterol and the DHR96 mutation indicates that there is further complexity in this pathway. Studies are under way to define the molecular mechanisms by which DHR96 regulates target gene transcription.
DHR96 represents an ancestral regulator of cholesterol metabolism
Ligands have been identified for only two of the 18 canonical nuclear receptors in Drosophila: ecdysone receptor (EcR) and E75 (Koelle et al. 1991; Reinking et al. 2005). This study adds a third such adopted orphan receptor to the list: DHR96. The identification of cholesterol as a DHR96 ligand fits with its membership in the NR1I subfamily of nuclear receptors (Nuclear Receptors Nomenclature Committee 1999). The closest mammalian and C. elegans homologs of DHR96—pregnane X receptor (PXR), constitutive androstane receptor (CAR), vitamin D3 receptor (VDR), and DAF-12—are all transcriptionally responsive to cholesterol derivatives. Similar responses to sterol derivatives are seen for members of the next most closely related group of receptors, the NR1H group, which includes LXR, farnesoid X receptor (FXR), and EcR. These observations support the proposal that the NR1H and NR1I groups arose from an ancestral progenitor that acted as a sterol receptor in primitive organisms.
It is important, however, to note that although cholesterol copurifies with the DHR96 LBD, it may not be the natural ligand for this receptor. In our efforts to address this issue, we were unable to detect changes in DHR96 activity in response to exogenous cholesterol, dietary factors, or genetic backgrounds that disrupt cholesterol absorption or intracellular trafficking (data not shown). Further studies are required to determine whether cholesterol or a related metabolite acts as a regulatory ligand to modulate the activation status of DHR96.
Although the sequence of DHR96 is most closely related to mammalian PXR and CAR, it shares significant functional characteristics with its more distant cousin, LXRα. LXRα binds oxysterols and, similar to DHR96 mutants, LXRα mutant mice fail to respond properly to dietary cholesterol and accumulate hepatic cholesterol when maintained on a high-cholesterol diet (Peet et al. 1998). LXRs also play a central role in the transcriptional response to dietary cholesterol in mice, and directly or indirectly control a number of genes that are related to DHR96-regulated genes, including ABCG1, LPL, steroyl-CoA desaturase, NPC1, and NPC2 (Maxwell et al. 2003; Rigamonti et al. 2005; Kalaany and Mangelsdorf 2006). Finally, like DHR96, LXRs repress NPC1L1 expression, although it is not clear whether this regulation is direct (Duval et al. 2006; Valasek et al. 2007). Taken together, these results indicate that DHR96 and mammalian LXRs act through similar regulatory pathways to control cholesterol homeostasis. This work establishes a new framework for understanding how cholesterol levels are sensed in Drosophila, and the molecular mechanisms by which cholesterol homeostasis is maintained.
Materials and methods
Drosophila stocks
All studies were performed using Canton-S (CanS) wild-type flies and DHR961-null mutants that had been crossed to CanS for nine generations (King-Jones et al. 2006). The following stocks were used in this study: npc1a1 (from M. Scott), npc1b1 and NPC1b-GAL4 (both from L. Pallanck), CG-Gal4 (Hennig et al. 2006), and Mex-Gal4 (Phillips and Thomas 2006). Flies were maintained on standard Bloomington Stock Center medium with malt at 25°C.
Cholesterol assays
Total cholesterol levels were determined using an Amplex Red Cholesterol Assay Kit (Invitrogen). Embryos staged at 0–3 h were collected and allowed to hatch overnight in the absence of food at 25°C. First instar larvae were transferred to low-cholesterol medium with either vehicle alone or 0.03% cholesterol and grown for 2 d at 25°C. After 2 d, 30 larvae per sample were collected and homogenized in 100 μL of 1× buffer included in the kit. The homogenate was centrifuged at 5000 rpm for 5 min and the supernatant was transferred to new tubes. A 50-μL aliquot (15 larvae equivalents) was assayed according to the kit instructions and measured in a Molecular Devices SpectraMax M2 fluorometer. Cholesterol levels were normalized to protein in each homogenate using a Bradford assay (Bio-Rad). Samples were prepared in triplicate, each experiment was repeated at least three times, and the resulting data was pooled. Data was analyzed using an unpaired two-tailed Student's t-test with unequal variance. All data are reported as the mean with 1× SEM.
See the Supplemental Material for additional materials and methods.
Acknowledgments
We thank M. Scott and L. Pallanck for stocks, A.-F. Ruaud and J. Metherall for helpful discussions, B. Milash for advice on microarray analysis, M. Carvalho and S. Eaton for the sterol-depleted food protocol, A. Kukis for advice on lipid extraction and small molecule MS, and A.-F. Ruaud, K. Schuske, M. Sieber, and J. Tennessen for comments on the manuscript. This research was supported by NIH grant 1R01DK075607.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1833609.
Supplemental material is available at http://www.genesdev.org.
References
- Bitsch F, Aichholz R, Kallen J, Geisse S, Fournier B, Schlaeppi JM. Identification of natural ligands of retinoic acid receptor-related orphan receptor α ligand-binding domain expressed in Sf9 cells–a mass spectrometry approach. Anal Biochem. 2003;323:139–149. doi: 10.1016/j.ab.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Cooke J, Sang J. Utilization of sterols by larvae of Drosophila melanogaster. J Insect Physiol. 1970;16:801–812. doi: 10.1016/0022-1910(70)90214-3. [DOI] [PubMed] [Google Scholar]
- Davis HR, Jr, Basso F, Hoos LM, Tetzloff G, Lally SM, Altmann SW. Cholesterol homeostasis by the intestine: Lessons from Niemann-Pick C1 Like 1 (NPC1L1) Atheroscler Suppl. 2008;9:77–81. doi: 10.1016/j.atherosclerosissup.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Duval C, Touche V, Tailleux A, Fruchart JC, Fievet C, Clavey V, Staels B, Lestavel S. Niemann-Pick C1 like 1 gene expression is down-regulated by LXR activators in the intestine. Biochem Biophys Res Commun. 2006;340:1259–1263. doi: 10.1016/j.bbrc.2005.12.137. [DOI] [PubMed] [Google Scholar]
- Fluegel ML, Parker TJ, Pallanck LJ. Mutations of a Drosophila NPC1 gene confer sterol and ecdysone metabolic defects. Genetics. 2006;172:185–196. doi: 10.1534/genetics.105.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Wang J, Qi W, Miao HH, Cao J, Qu YX, Li BL, Song BL. The cholesterol absorption inhibitor ezetimibe acts by blocking the sterol-induced internalization of NPC1L1. Cell Metab. 2008;7:508–519. doi: 10.1016/j.cmet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Suyama K, Buchanan J, Zhu AJ, Scott MP. A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development. 2005;132:5115–5124. doi: 10.1242/dev.02079. [DOI] [PubMed] [Google Scholar]
- Huang X, Warren JT, Buchanan J, Gilbert LI, Scott MP. Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: A model of human neurodegenerative disease. Development. 2007;134:3733–3742. doi: 10.1242/dev.004572. [DOI] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Kalaany N, Mangelsdorf D. LXRS and FXR: The yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR α. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Phillips MD, Thomas GH. Brush border spectrin is required for early endosome recycling in Drosophila. J Cell Sci. 2006;119:1361–1370. doi: 10.1242/jcs.02839. [DOI] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Samson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. The Drosophila nuclear receptor E75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Rigamonti E, Helen L, Lestavel S, Mutka AL, Lepore M, Fontaine C, Bouhel MA, Bultel S, Fruchart JC, Ikonen E, et al. Liver X receptor activation controls intracellular cholesterol trafficking and esterification in human macrophages. Circ Res. 2005;97:682–689. doi: 10.1161/01.RES.0000184678.43488.9f. [DOI] [PubMed] [Google Scholar]
- Sang J. The quantitative nutritional requirements of Drosophila melanogaster. J Exp Biol. 1956;33:45–72. [Google Scholar]
- Shostak Y, Van Gilst MR, Antebi A, Yamamoto KR. Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes & Dev. 2004;18:2529–2544. doi: 10.1101/gad.1218504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2010 doi: 10.1016/j.cmet.2009.10.010. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Va'nt Hoog EG. Aseptic culture of insects in vitamin research (continued). Zt. Vitaminforsch. 1936;5:118–126. [Google Scholar]
- Valasek MA, Clarke SL, Repa JJ. Fenofibrate reduces intestinal cholesterol absorption via PPARα-dependent modulation of NPC1L1 expression in mouse. J Lipid Res. 2007;48:2725–2735. doi: 10.1194/jlr.M700345-JLR200. [DOI] [PubMed] [Google Scholar]
- Voght SP, Fluegel ML, Andrews LA, Pallanck LJ. Drosophila NPC1b promotes an early step in sterol absorption from the midgut epithelium. Cell Metab. 2007;5:195–205. doi: 10.1016/j.cmet.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Wang DQ. Regulation of intestinal cholesterol absorption. Annu Rev Physiol. 2007;69:221–248. doi: 10.1146/annurev.physiol.69.031905.160725. [DOI] [PubMed] [Google Scholar]