Abstract
Vasa (Vas) is a DEAD-box RNA-binding protein required in Drosophila at several steps of oogenesis and for primordial germ cell (PGC) specification. Vas associates with eukaryotic initiation factor 5B (eIF5B), and this interaction has been implicated in translational activation of gurken mRNA in the oocyte. Vas is expressed in all ovarian germline cells, and aspects of the vas-null phenotype suggest a function in regulating the balance between germline stem cells (GSCs) and their fate-restricted descendants. We used a biochemical approach to recover Vas-associated mRNAs and obtained mei-P26, whose product represses microRNA activity and promotes GSC differentiation. We found that vas and mei-P26 mutants interact, and that mei-P26 translation is substantially reduced in vas mutant cells. In vitro, Vas protein bound specifically to a (U)-rich motif in the mei-P26 3′ untranslated region (UTR), and Vas-dependent regulation of GFP-mei-P26 transgenes in vivo was dependent on the same (U)-rich 3′ UTR domain. The ability of Vas to activate mei-P26 expression in vivo was abrogated by a mutation that greatly reduces its interaction with eIF5B. Taken together, our data support the conclusion that Vas promotes germ cell differentiation by directly activating mei-P26 translation in early-stage committed cells.
Keywords: Oogenesis, RNA-binding protein, DEAD-box, patterning, development
Molecular asymmetries underlying embryonic axis patterning and germ cell specification are established in Drosophila largely by position-dependent translational regulation of maternally expressed mRNAs (Kugler and Lasko 2009). Translation of mRNAs encoding key patterning molecules such as Oskar (Osk), Nanos (Nos), Gurken (Grk), Hunchback, and Caudal is repressed by RNA-binding proteins that bind to specific sequence elements in their untranslated regions (UTRs). These repressors, directly or indirectly, often block recruitment of eukaryotic initiation factor 4E (eIF4E), the cap-binding subunit of eIF4F (Filardo and Ephrussi 2003; Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004), or recruit 4E homology protein (4EHP), which binds unproductively to the 5′ cap structure (Cho et al. 2005, 2006). In some cases, repressors act instead by sequestering target mRNAs into large silencing messenger ribonucleoproteins (mRNPs) (Nakamura et al. 2001; Chekulaeva et al. 2006). Still others negatively regulate their target mRNAs by recruiting the CCR4 deadenylase complex (Semotok et al. 2005; Zaessinger et al. 2006; Chicoine et al. 2007; Kadryova et al. 2007). Targeting also requires mechanisms to alleviate repression and activate translation in the appropriate spatial domain. Relatively less is known about these positive regulatory processes. Osk contributes to derepressing nos by preventing its association with Smaug, and thus with CCR4 deadenylase (Zaessinger et al. 2006). Activation of grk mRNA translation at the antero–dorsal cortex of the developing oocyte is mediated by poly(A)-binding protein 55D (PABP55D) in association with Encore (Clouse et al. 2008).
Vasa (Vas) is a DEAD-box RNA-binding protein that localizes during oogenesis to the posterior pole plasm (Hay et al. 1988; Lasko and Ashburner 1990). It is expressed in all germline cells except mature sperm, and was initially implicated in posterior embryonic patterning and germ cell specification (Schüpbach and Wieschaus 1986). Later work showed that vas-null females produce rare tumorous egg chambers in which nurse cells and oocytes have not differentiated, and also rare egg chambers with supernumerary, misplaced, or no oocytes, suggesting that Vas may also function in restricting cell fate during germline stem cell (GSC) differentiation (Styhler et al. 1998; Tomancak et al. 1998). A DNA damage checkpoint that functions in the oocyte during meiotic prophase, after the 16-cell cyst has formed, regulates Vas activity (Ghabrial and Schüpbach 1999; Abdu et al. 2002; Findley et al. 2003; Theurkauf et al. 2006; Chen et al. 2007; Pane et al. 2007). Vas binds to eIF5B, an essential translation initiation factor that functions in ribosomal subunit joining (Carrera et al. 2000; Pestova et al. 2000). In vas-null oocytes, grk translation is greatly reduced (Styhler et al. 1998; Tomancak et al. 1998), an effect also observed in oocytes that express only a form of Vas (Vas△617) that is specifically compromised for eIF5B binding (Johnstone and Lasko 2004). These results have led to the hypothesis that Vas positively regulates specific mRNAs such as grk by recruiting eIF5B. However, the question as to how Vas distinguishes among potential target RNAs has remained unsolved, as structural analysis of a fragment of Vas in complex with RNA argues against sequence-specific binding (Sengoku et al. 2006). Vas also associates with Piwi, Aubergine, and other components of the repeat-associated siRNA (rasiRNA) pathway (Megosh et al. 2006; Vagin et al. 2006), and has been linked recently to retrotransposon silencing (Shpiz et al. 2009), suggesting possible additional functions for it in RNAi pathways.
In this study, we identify mei-P26 as a novel target mRNA for Vas. We demonstrate that Vas associates with mei-P26 mRNA in vivo, and that full-length Vas binds specifically in vitro to a tract of 30 nucleotides (nt) of the mei-P26 3′ UTR that contains a (U)-rich motif. Vas binding to this segment of mei-P26 mRNA is specifically competed by unlabeled poly(U). Unlike full-length Vas, a Vas fragment identical to that used in a previous structural study (Sengoku et al. 2006) did not show specific binding, indicating that sequence specificity is conferred by a region of Vas outside of the conserved DEAD-box domains. In vivo expression of Mei-P26 is reduced in vas-null or vas△617 ovaries relative to wild type, but expression from GFP:mei-P26 transgenes that lack the Vas-binding site is unaffected by vas activity. Taken together, these results demonstrate that Vas is a sequence-specific RNA-binding protein that positively regulates translation of mei-P26 in early-stage committed germline cells, thus promoting their further differentiation.
Results
Isolation of Vas-associated mRNAs
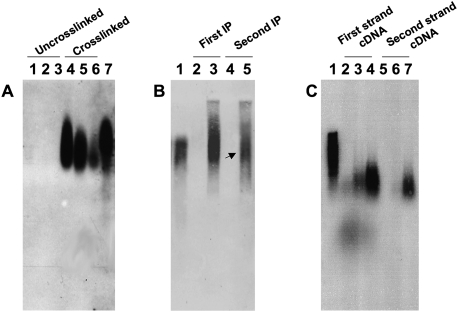
To explore more fully the associations that exist between Vas and mRNA in vivo, we identified mRNAs that copurify with Vas in extracts prepared from early Drosophila embryos. We developed a method based on the in vivo cross-linking and immunoprecipitation approach (see the Supplemental Material for a detailed description; Niranjanakumari et al. 2002; Ule et al. 2003). Embryos (0–2 h) were collected, permeabilized, and in situ cross-linked with formaldehyde. Extracts were prepared, and mRNAs were purified by oligo(dT) chromatography from cross-linked and control uncross-linked embryos. Aliquots of each eluted sample were resolved on agarose gels and used for immunoblot analysis with anti-Vas antibody. Vas, bound to mRNA, was present in the cross-linked sample and not in the control (Fig. 1A). Vas–mRNA complexes purified by oligo(dT) chromatography were further purified by tandem immunoprecipitation with anti-Vas antibody (Liu et al. 2003). Purified extracts were analyzed again for the presence of Vas by immunoblotting (Fig. 1B). The cross-linking was then reversed by heat treatment. mRNA was isolated and used to synthesize double-strand cDNA (Fig. 1C). The synthesized cDNA sequences represent the sequences of the mRNA in the Vas–mRNA complex. The cDNAs were cloned and sequenced.
Figure 1.
Purification of Vas–mRNA complexes. (A) Aliquots (1/100) of eluates from oligo(dT) chromatography were resolved on a 0.75% TAE agarose gel, blotted, and treated with anti-Vas antibody. (Lanes 1–3) First, second, and third eluates from uncross-linked controls. (Lanes 4–6) First, second, and third eluates from cross-linked samples. (Lane 7) Cross-linked lysate before oligo(dT) chromatography, a positive control. (B) Purification of Vas–mRNA complexes by tandem Vas immunoprecipitation, analyzed by immunoblot as in A. (Lane 1) Cross-linked sample before immunoprecipitation, a positive control. (Lanes 2,3) Aliquots of material recovered from the first immunoprecipitation, using rabbit IgG (control, lane 2) and α-Vas (lane 3). (Lanes 4,5) Aliquots of material recovered from the second immunoprecipitation using rabbit IgG (control, lane 4) and α-Vas (lane 5). The arrows indicate the purified Vas–mRNA complexes. (C) Synthesis of double-strand cDNA. Lane 1 is a positive control. Lanes 2–4 are the first-strand cDNA synthesis, and represent buffer control (lane 2), uncross-linked control (lane 3), and cross-linked sample (lane 4). Lanes 5–7 are the second-strand cDNA synthesis, and represent buffer control (lane 5), uncross-linked control (lane 6), and cross-linked sample (lane 7).
From these purifications, we recovered cDNAs representing 221 different mRNAs (Supplemental Table 1). As Vas is enriched in primordial germ cells (PGCs), we investigated whether this population of mRNAs is enriched for those that also accumulate in PGCs, as we might predict that Vas target mRNAs would more frequently show such a distribution than mRNAs from a general sampling. By searching in situ hybridization data that were generated by the Berkeley Drosophila Genome Project (Tomancak et al. 2007) and by Lécuyer et al. (2007), and by carrying out our own in situ hybridizations, we obtained expression pattern data for 180 of the recovered mRNAs, and 24 (13.3%) are clearly enriched in pole cells at the cellular blastoderm stage (Supplemental Table 1). This compares to estimates of 5.0% (223 out of 4496) (Tomancak et al. 2007) or 5.9% (199 out of 3370) (Lécuyer et al. 2007) from large-scale screens.
vas and mei-P26 interact genetically
Among the mRNAs we recovered was mei-P26, which has been implicated in germ cell differentiation in both sexes, and which also functions somatically in the nervous system (Page et al. 2000; Glasscock et al. 2005; Neumüller et al. 2008). Mei-P26, a TRIM-NHL domain protein, interacts with Argonaute-1 (AGO1), inhibiting the microRNA (miRNA) pathway and promoting differentiation of early-stage committed germline cells (Neumüller et al. 2008). To test for a common function, we investigated whether vas and mei-P26 mutations interact. The hypomorphic maternal effect lethal vas1 allele allows oogenesis to proceed to completion even when in trans with the vasPH165-null allele (Styhler et al. 1998). The hypomorphic allelic combination mei-P261/mei-P26fs1 primarily affects chromosome segregation at meiosis (Page et al. 2000). Oogenesis proceeds to completion in these mei-P26 mutants, with only very rare cases of tumorous egg chambers and occasional defects in nurse cell number (Page et al. 2000). We confirmed these earlier data for these two genotypes, and noted that virtually all egg chambers produced by these two genotypes contain oocytes and differentiated polyploid nurse cells (Fig. 2A–D). However, combining vas and mei-P26 mutations gives a strikingly more severe phenotype; vas1/vasPH165; mei-P261/mei-P26fs1 ovaries are comprised mostly of tumorous egg chambers (Fig. 2E,F). We used mAb 1B1, which is directed against an Adducin isoform (Zaccai and Lipshitz 1996), to determine the developmental stage of these tumorous cells by investigating their fusome structure. As cells with punctate spectrosomes as well as cells with branched fusomes were observed, we conclude that the vas1/vasPH165; mei-P261/mei-P26fs1 egg chambers contain mostly early-stage cystocytes, a phenotype also characteristic of strong mei-P26 alleles (Fig. 2G,H; Neumüller et al. 2008). Thus reduction of vas activity enhances the phenotypic consequences of a hypomorphic mei-P26 allelic combination.
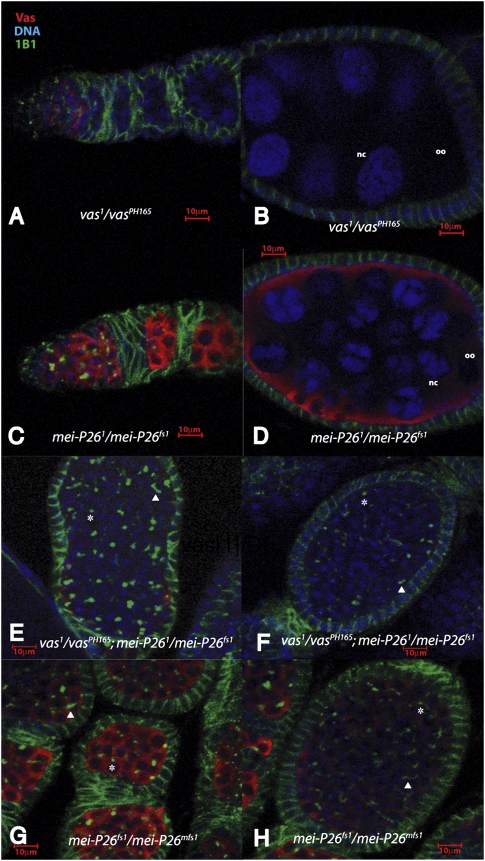
Figure 2.
mei-P26 and vas mutations genetically interact. Nuclei were visualized by DAPI staining (blue), germline cells were labeled with α-Vas, and fusomes were labeled with mAb 1B1. (A,B) vas1/vasPH165 egg chambers are similar to wild-type, composed of 15 nurse cells (nc) and an oocyte (oo), surrounded by a layer of follicle cells. (C,D) mei-P261/mei-P26fs1 ovaries, a relatively mild allelic combination, are mostly wild-type in appearance. Occasional egg chambers (<5%) still differentiate nurse cells and oocytes, but have fewer or more than 15 nurse cells, or are incorrectly patterned (data not shown). (E,F) vas1/vasPH165; mei-P261/mei-P26fs1 ovaries have a much more severe phenotype, with most (>50%) egg chambers appearing tumorous and failing to differentiate nurse cells and oocytes. The germline cells of these ovaries show either punctate spectrosomes (asterisks) or branched fusomes (solid triangles), indicating they are cystocyte-derived. (G,H) ovaries from mei-P26fs1/mei-P26mfs1, a strong allelic combination, resemble vas1/vasPH165; mei-P261/mei-P26fs1 ovaries.
Vas activates mei-P26 translation
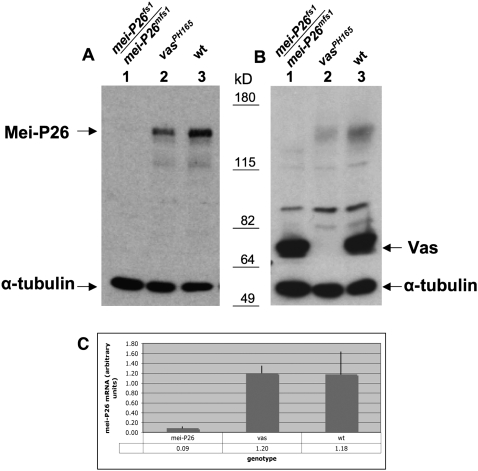
We prepared an anti-Mei-P26 antiserum to investigate whether vas activity affected endogenous Mei-P26 accumulation. Immunoblotting experiments showed that the Mei-P26 protein level is clearly reduced in extracts prepared from homozygous vasPH165 ovaries as compared with wild type (Fig. 3A). Using Quantity ONE software (Bio-Rad), we quantitated the intensity of the Mei-P26 signal from vasPH165 ovaries in comparison with that of wild-type ovaries, and determined it to be 49.4% ± 6.1% (n = 6). Mei-P26 signal was essentially absent in mei-P26fs1/mei-P26mfs1 ovarian extracts, confirming the specificity of the antiserum. We also examined the Vas level in the mei-P26fs1/mei-P26mfs1 ovarian extract by immunoblot and found no difference from the wild-type ovarian extract (Fig. 3B). Thus, vas is required for normal accumulation of Mei-P26, but mei-P26 is not required for Vas expression. Using real-time PCR methods, we further investigated whether the levels of mei-P26 mRNA differ from wild type in vasPH165 ovaries, but found no difference (Fig. 3C). Thus, we conclude that Vas activates mei-P26 expression at the level of translation, rather than by influencing the stability of its mRNA.
Figure 3.
Mei-P26 protein level is reduced in vas mutant ovaries. (A) Immunoblot of ovarian extracts from mei-P26fs1/mei-P26mfs1, vasPH165, and wild-type (OreR) females using anti-Mei-P26, which was preadsorbed with fixed mei-P26fs1/mei-P26mfs1 ovaries. The same blot was probed with anti-α-tubulin as a loading control. (Lane 1) mei-P26fs1/mei-P26mfs1 ovarian extract serves as a negative control. (Lane 2) Homozygous vasPH165 ovarian extract. (Lane 3) Wild-type ovarian extract. The Mei-P26 level is reduced in lane 2 as compared with lane 3 (arrow). (B) Immunoblot using anti-Vas on extracts prepared from mei-P26fs1/mei-P26mfs1 ovaries (lane 1), homozygous vasPH165 ovaries (lane 2), and wild-type ovaries (lane 3). The Vas level (arrow) is unaffected by mei-P26 mutations. Molecular weight markers are indicated in kilodaltons. (C) Real-time PCR data comparing mei-P26 RNA levels in mei-P26fs1/mei-P26mfs1 (mei-P26), vasPH165 (vas), and wild-type (wt) ovaries, showing that mei-P26 RNA levels are unaffected by vas mutation. The data presented are averaged from four independent experiments (12 reactions) for each genotype, and the error bars indicate standard deviations.
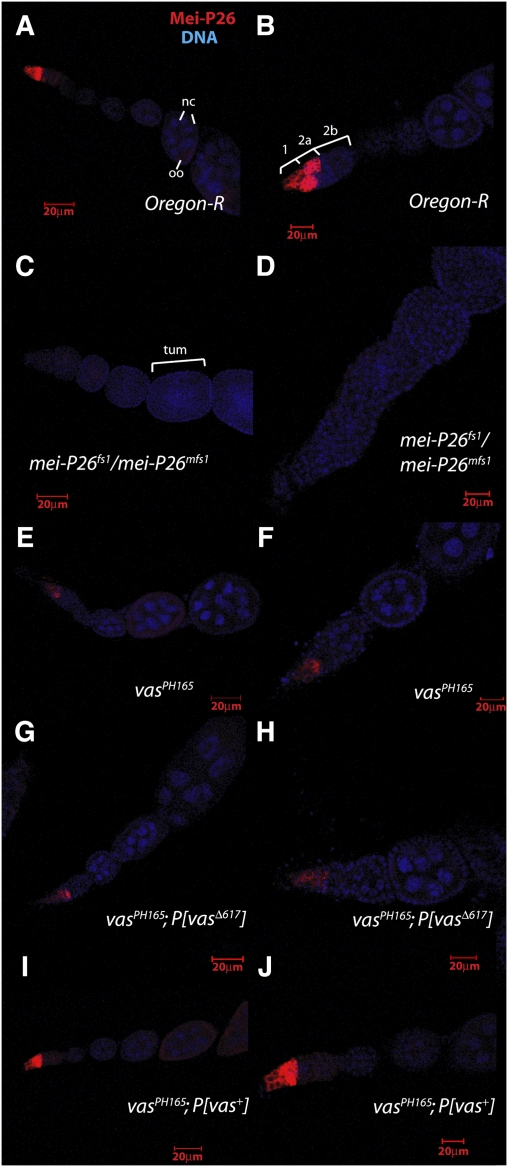
Mei-P26 is expressed at low levels in stem cells, but its level increases through transit-amplifying divisions such that it accumulates at high levels in early 16-cell germ cell cysts (Fig. 4A,B; Neumüller et al. 2008). We used mei-P26fs1/mei-P26mfs1 ovaries as negative controls and to ensure the specificity of the immunostaining reaction (Fig. 4C,D). Consistent with the immunoblotting data, Mei-P26 accumulation was greatly reduced in homozygous vasPH165 ovaries (Fig. 4E,F). This result was highly reproducible from sample to sample. We further investigated whether Mei-P26 accumulation is reduced in a site-directed vas mutant (vasΔ617) (Johnstone and Lasko 2004), which is abrogated for interaction with eIF5B. We reproducibly found that accumulation of Mei-P26 protein was greatly reduced in vasPH165; P{vasΔ617} (Fig. 4G,H), but that expression of a vas+ transgene fully rescued wild-type levels of Mei-P26 expression in vasPH165 ovaries (Fig. 4I,J). Taken together with our previous data, we conclude that Vas positively regulates mei-P26 translation via its interaction with eIF5B.
Figure 4.
Vas activity affects Mei-P26 protein accumulation. (A,B) In wild-type ovaries, Mei-P26 protein is present in stem cells, and its expression reaches a peak in 16-cell cysts. It is essentially absent in subsequent stages of oogenesis. In A, an egg chamber, which in normal development includes 15 polyploid nurse cells (nc) and an oocyte (oo), is labeled. In B, the different developmental stages of the germarium are labeled. Region 1 (1) contains the stem cells and the mitotic region in which the cystocytes divide to form two-, four-, eight-, and 16-cell cysts; region 2a (2a) contains early 16-cell cysts, which mature in region 2b (2b), becoming associated with a monolayer of precursor follicle cells. Region 2b is also the region in which nurse cells and oocytes differentiate (Spradling et al. 1997). Mei-P26 is expressed in region 1, peaking in early 16-cell cysts. (C,D) Little Mei-P26 signal is apparent in mei-P26fs1/mei-P26mfs1ovaries. These egg chambers are tumorous (tum), failing to differentiate nurse cells and oocytes. (E,F) Accumulation of Mei-P26 protein throughout its expression domain is substantially reduced in homozygous vasPH165 ovaries. (G,H) Accumulation of Mei-P26 protein is similarly reduced in vasPH165; P{vasΔ617} ovaries. (I,J) Mei-P26 protein expression in vasPH165 ovaries is rescued to wild-type levels by expression of a wild-type vas+ transgene.
Vas protein binds directly to a (U)-rich motif in the mei-P26 3′ UTR
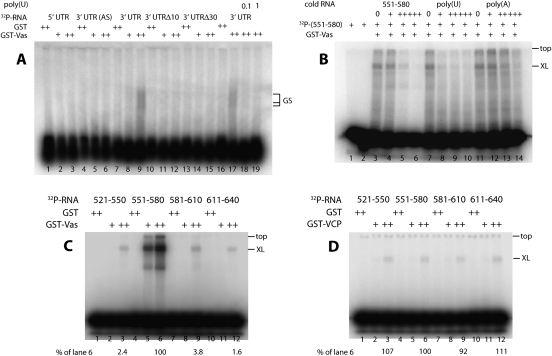
To investigate whether Vas can bind directly to mei-P26 mRNA, we carried out electrophoretic mobility shift assays (EMSAs) with purified GST-Vas that was expressed in Escherichia coli and various radiolabeled RNAs derived from the mei-P26 5′ UTR or 3′ UTR. We found that GST-Vas bound to the mei-P26 3′ UTR sense transcript, but not to the mei-P26 5′ UTR or to a mei-P26 3′ UTR antisense transcript (Fig. 5A, lanes 1–9). We further found that recovery of RNA species with retarded electrophoretic mobility depended on a particular segment of the mei-P26 3′ UTR that contains a (U)-rich motif (nucleotides 551–580, which has the following sequence: 5′-AGGUCGAGCUCCUAUUUUUUUUUUCCCCGU-3′) (Fig. 5A, lanes 10–15). We also observed that gel retardation was sensitive to the inclusion of unlabeled poly(U) (Fig. 5A, lanes 17–19).
Figure 5.
RNA-binding assays reveal a (U)-rich motif that is recognized specifically by full-length GST-Vas. (A) Gel retardation assays to investigate the RNA-binding properties of Vas in respect to mei-P26 mRNA. GST (1.5 μg) or GST-Vas (+, 2 μg; or ++, 4 μg) was incubated with 30 ng of 32P-labeled RNA probes as follows: (Lanes 1–3) Full-length mei-P26 5′ UTR RNA; (lanes 4–6) Full-length mei-P26 3′ UTR RNA (antisense strand). (Lanes 7–9,16–19) Full-length mei-P26 3′ UTR RNA (sense strand). (Lanes 10–12) mei-P26 3′ UTR RNA deleted for the (U)-rich element (nucleotides 565–574). (Lanes 13–15) mei-P26 3′ UTR RNA deleted for a larger segment containing the (U)-rich element (nucleotides 551–580). Unlabeled poly(U) was added to the samples loaded in lanes 18 and 19 (0.1 μg and 1.0 μg, respectively). RNAs were separated on a 0.7% agarose gel. Species with retarded electrophoretic mobility (GS) were observed only when GST-Vas and full-length mei-P26 3′ UTR were present, and poly(U) competitor RNA was absent. In those reactions, the molar ratio of GST:Vas to mei-P26 3′ UTR RNA was 325:1. (B) Cross-linking assays to test the ability of Vas to bind nucleotides 551–580 of the mei-P26 3′ UTR in the presence of various competitor RNAs. Lanes 1 and 2 are negative controls (no added protein and GST added, respectively). (Lanes 3–14) GST-Vas (4 μg) and 32P-labeled mei-P26 3′ UTR nucleotides 551–580 (20 ng) were UV-cross-linked in the presence of competitors as follows: (Lanes 3–6) mei-P26 3′ UTR nucleotides 551–580. (Lanes 7–10) Poly(U). (Lanes 11–14)Poly(A). Competitor RNAs were added at 0 μg (labeled 0), 0.01 μg (+), 0.1 μg (++), and 1 μg (+++). The position at which samples were loaded onto the gel is labeled “top” and the position at which the major cross-linked species migrates is labeled “XL.” (C) Cross-linking assays to compare the relative affinity of full-length GST-Vas to nucleotides 551–580 of the mei-P26 3′ UTR to its affinity to neighboring 30-nt fragments of the mei-P26 3′ UTR. GST (1.5 μg) or GST-Vas (+, 2 μg; or ++, 4 μg) was incubated with the following 32P-labeled (30-ng) RNA probes as follows: (Lanes 1–3) mei-P26 3′ UTR nucleotides 521–550. (Lanes 4–6) mei-P26 3′ UTR nucleotides 551–580. (Lanes 7–9) mei-P26 3′ UTR nucleotides 581–610. (Lanes 10–12) mei-P26 3′ UTR nucleotides 611–640. The position at which samples were loaded onto the gel is labeled “top” and the position at which the major cross-linked species migrates is labeled “XL.” The intensities of the major cross-linked species were measured with ImageQuant software, and are presented as a percentage of that measured in lane 6. In this experiment, when 4 μg of GST-Vas were present, the molar ratio of protein to RNA was 12:1. (D) Cross-linking assays as in C but using 4 μg of GST-VCP instead of the full-length GST-Vas, and using 20 ng of the RNA probes. Unlike GST-Vas, GST-VCP does not preferentially bind the 551–580 segment. In this experiment, when 4 μg of GST-VCP were present, the molar ratio of protein to RNA was 25:1. All of the transcripts used in this figure derive from a mei-P26 cDNA that contains 10 consecutive U residues in its 3′ UTR (nucleotides 565–574), but that differs from the corresponding region of the consensus genome sequence, which has only nine consecutive T residues (Celniker et al. 2002).
To further examine the Vas–mei-P26 interaction, we used a UV cross-linking assay that confirmed a specific interaction between GST-Vas and the 551–580 segment of the 3′ UTR. This interaction is sensitive to competition with unlabeled poly(U), but not to poly(A), illustrating the importance of the (U)-rich motif for Vas binding (Fig. 5B). We used this assay to compare the relative affinity of GST-Vas for the 551–580 segment versus that for neighboring 30-nt segments of the mei-P26 3′ UTR and observed a 30- to 60-fold preference for the segment containing the (U)-rich motif (Fig. 5C).
A fragment of Vas consisting of amino acids 200–623 (which we term Vasa core protein, or VCP) was cocrystallized with an oligomer of poly(U) and a nonhydrolyzable ATP analog for high-resolution structural analysis (Sengoku et al. 2006). In this way, VCP–RNA binding was shown to involve a series of hydrophilic interactions between the protein and the phosphate and 2′-hydroxy groups of the sugar-phosphate backbone, and not the nucleotide bases. This argues against the possibility of sequence-specific binding, which we nonetheless observed for full-length GST-Vas. To test whether GST-VCP behaves differently from GST-Vas in our assays, we expressed GST-VCP and tested its relative affinities for four 30-nt oligonucleotides, including the 551–580 segment. We found that, unlike GST-Vas, GST-VCP binds weakly and essentially equally to all four probes (Fig. 5D). The affinity of GST-VCP binding to the RNA probes was comparable with that of full-length GST-Vas to probes that did not contain the critical (U)-rich motif. This result strongly suggests that, while the core fragment of Vas is capable of a basal level of RNA binding, another domain located outside of amino acids 200–623 can contribute to RNA binding in a sequence-specific manner.
(U)-rich motifs are frequent in Drosophila 3′ UTRs
To gain some insight into the potential number of Vas target mRNAs, we investigated the frequency of polyuridine tracts in the 3′ UTRs of Drosophila melanogaster mRNAs. We obtained the sequences of 17,887 predicted 3′ UTRs, totaling 8,326,756 nt from FlyBase (Wilson et al. 2008), and searched for instances of seven or more consecutive U residues. Such U-motifs are quite frequent in 3′ UTRs: (U)7 or greater appeared 4030 times, or once every 2066 nt; (U)8 or greater appeared 2419 times, or once every 3422 nt; (U)9 or greater appeared 1622 times, or once every 5134 nt; and (U)10 or greater appeared 998 times, or once every 8343 nt. As the average 3′ UTR length is 465 nt, this analysis implies that a maximum of 465 out of 2066, or 22.5%, would contain a (U)7 or greater motif, although the actual number is somewhat smaller, as we identified many individual mRNAs, most with long 3′ UTRs, that contained two or more such repeats. Shorter U-motifs are frequent enough to be found in most 3′ UTRs. As it is unlikely that Vas actually interacts with such a large set of mRNAs, it is probable that additional sequence or structural elements contribute to the definition of a Vas-binding site. As Vas is present mostly in mRNP complexes in vivo, other proteins associated with it may confer additional specificity. Also, some mRNAs that contain a Vas-binding element in their 3′ UTR may never be coordinately expressed with Vas. Polyuridine repeats were not overrepresented among the 3′ UTRs of the mRNAs we coimmunoprecipitated with Vas. However, of the 31 mRNAs we recovered in our immunoprecipitations that contained 3′ UTR polyuridine repeats seven or more residues in length, seven (mei-P26, gcl, cib, pxt, Bsg25D, Vap-33-1, and TH1) are enriched in blastoderm pole cells. This 22.6% frequency of pole cell-enriched mRNAs is far higher than is found by random sampling or from large-scale gene expression surveys, suggesting that filtering our coimmunoprecipitation results for the presence of U-motifs may predict direct targets of Vas in addition to mei-P26.
We used two modeling programs—CentroidFold (Sato et al. 2009) and the SRNA program of Sfold (Ding et al. 2004)—to predict secondary structures for a 500-nt and a 200-nt region, respectively, surrounding the U-motif that is recognized by Vas. Both programs predicted that the U residues were mostly in a double-stranded base-paired region that was interrupted by several unpaired residues (Supplemental Fig. 1). We used CentroidFold to examine whether the U-motifs in the six other Vas-associated localized mRNAs that possessed them would also be predicted to be base-paired. In the gcl 3′ UTR, there is a (U)7 motif that is predicted to be entirely base-paired. The U-motifs of cib and vap-33-1 are similar to that of mei-P26 in that they are partially base-paired. The latter of these RNAs also contains a (U)6 motif that is partially base-paired. In contrast, the U-motifs of Bsg25D and TH1 are in unstructured regions, and only the first residue of a (U)7 motif in pxt is predicted to be in a base pair.
In vivo expression of transgenic GFP-Mei-P26 depends on vas activity and the mei-P26 3′ UTR-binding site
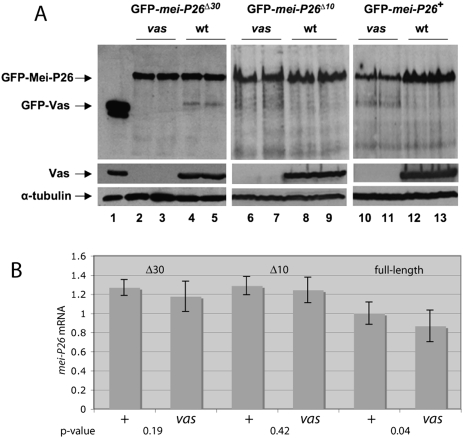
To determine the significance of the interaction between Vas and the 3′ UTR-binding site to mei-P26 expression in the ovary, we produced transgenic lines expressing GFP-tagged versions of Mei-P26, with either a full-length 3′ UTR or deletions of nucleotides 551–580 (△30) or 565–574 (△10). We introduced these transgenes into either a wild-type or vasPH165 genetic background, and monitored the effects of vas activity on their expression. As we described for endogenous Mei-P26, expression of GFP-Mei-P26 from the control construct with an intact 3′ UTR is much greater in wild-type ovaries than in comparably staged vasPH165 ovaries (Fig. 6A). Quantitation of four such experiments indicated that, when normalized for endogenous α-tubulin expression, the level of GFP-Mei-P26 from a construct with an intact 3′ UTR in vasPH165 ovaries was 51.8% ± 8.8% of that in similarly staged wild-type ovaries. This differential expression depends on the presence of the (U)-rich motif in the mei-P26 3′ UTR, however, as GFP-Mei-P26 expression is essentially equal in wild-type and vasPH165 ovaries when that element is removed (Fig. 6). Mei-P26 expression from the △30 construct was expressed in vasPH165 ovaries at 96.9% ± 5.2% of wild-type levels (n = 4), while Mei-P26 expression from the △10 construct was expressed in vasPH165 ovaries at 98.8% ± 8.9% of wild-type levels (n = 4). These differences between GFP-Mei-P26 expression from the various constructs in vasPH165 ovaries are highly significant (full-length vs. △10, P < 0.0001; full-length vs. △30, P < 0.0003; unpaired t-test). We did not observe a comparable effect of vas activity on GFP-mei-P26 mRNA levels (Fig. 6B). In vas extracts, a modest reduction in full-length GFP-mei-P26 mRNA to 86.6% ± 15.8% (P = 0.04) was observed, as well as smaller, not statistically significant, reductions in vas extracts for mRNAs of both deletion constructs. We conclude that the (U)-rich motif is essential for Vas-mediated regulation of mei-P26 in vivo, and that this regulation occurs primarily at the level of translation.
Figure 6.
The Vas-binding site in the mei-P26 3′ UTR affects GFP-Mei-P26 expression in vivo. (A) Immunoblot of ovarian extracts from various genotypes. α-GFP was used to detect GFP-Mei-P26 expressed from transgenes as indicated. (vas) Transgene is expressed in a vasPH165 genetic background; (wt) transgene is expressed is a wild-type genetic background. α-Vas was used to confirm the genotypes, and anti-α-tubulin was used as a loading control. Quantitative data from four independent experiments are presented in the Results. (Lane 1) P[GFP-vas] serves as positive control for α-GFP. (Lanes 2,3) vasPH165; P[GFP-mei-P26Δ30]. (Lanes 4,5) +; P[GFP-mei-P26Δ30]. The level of GFP-Mei-P26 is equivalent in wild-type and vas mutant backgrounds. (Lanes 6,7) vasPH165; P[GFP-mei-P26Δ10]. (Lanes 8,9) +; P[GFP-mei-P26Δ10]. The level of GFP-Mei-P26 is equivalent in wild-type and vas mutant backgrounds. (Lanes 10,11) vasPH165; P[GFP-mei-P26]. (Lanes 12,13) +; P[GFP-mei-P26]. The level of GFP-Mei-P26 is reduced in the vas mutant background as compared with wild-type, indicating that the Vas-binding site is essential for Vas-mediated activation of expression. (B) Real-time PCR data comparing mei-P26 RNA levels from the same transgenic constructs in wild-type and in vasPH165 genetic backgrounds. mRNA levels from all three constructs are slightly reduced in vasPH165 extracts, to 86.6% for full-length, 96.5% for GFP-mei-P26Δ10, and 92.8% for GFP-mei-P26Δ30. The P-values for these differences are given for each construct.
Discussion
In this study, we demonstrated that Vas regulates mei-P26 expression in vivo, and that a (U)-rich element in the mei-P26 3′ UTR interacts with Vas in vitro and is required for Vas-mediated regulation in vivo. DEAD-box helicases such as Vas were not believed previously to be sequence-specific nucleic acid-binding proteins (Sengoku et al. 2006). We showed, however, that Vas demonstrates specific binding, but that this requires domains that are distinct from the motifs shared by all DEAD-box proteins. Vas contains nine RGG repeats located between amino acids 17 and 165, within the region we implicated in binding specificity that lies outside of the canonical DEAD-box segment. While the region N-terminal to the common DEAD-box motifs is highly variable in the sequences of Vas orthologs from different species, the presence of RGG repeats within that region is conserved; for example, zebrafish Vas contains nine such motifs, and human Vas (DDX4) contains four. Two RGG repeats are present in mammalian fragile X mental retardation protein (FMRP), and have been shown to specifically recognize a G quartet structure in semaphorin 3F RNA, indicating that this motif can discriminate among target RNAs (Menon and Mihailescu 2007). RGG repeats are often present in proteins that contain other RNA-binding domains; for example, Vas contains a DEAD-box signature, while FMRP contains two hnRNPK homology (KH) domains; thus it has been proposed that they serve an auxiliary role in RNA binding (McBride and Silver 2001). We suggest that the RGG repeats of Vas play such a role by conferring specificity to its association with RNA. RGG motifs are also targets for arginine methyltransferases, and arginine methylation has been linked to modulating the RNA-binding activity of heterogeneous nuclear RNP (hnRNP) A1 (Kim et al. 1997). It is tempting to speculate that the RNA-binding activity of Vas might be similarly modulated, perhaps through the activity of the arginine methyltransferase Capsuléen, which like Vas is essential for germ cell specification (Gonsalvez et al. 2006; Anne et al. 2007).
A model has been proposed for Vas-mediated translational activation whereby Vas recruits eIF5B to target mRNAs (Carrera et al. 2000; Johnstone and Lasko 2004). This study suggests that Vas can itself discriminate among potential mRNA targets, although we cannot exclude that Vas may be recruited to other target mRNAs through indirect associations involving partner RNA-binding proteins. Precedents for regulation of translation at the step of subunit joining exist in several systems. For example, in early erythroid precursor cells, the mRNA encoding 15-lipoxygenase (r15-LOX) is translationally silenced at this step of translation initiation, dependent on a cytidine-rich 3′ UTR element termed DICE (differentiation control element) and on two RNA-binding proteins: hnRNP K and hnRNP E1 (Ostareck et al. 2001; Reimann et al. 2002; Messias et al. 2006). Phosphorylation of a specific tyrosine residue of hnRNP K by c-Src reduces its affinity for DICE, thus activating translation. Another example is provided by the ASH1 mRNA in Saccharomyces cerevisiae, which is translationally repressed before localizing to the bud cortex by Puf6p, which binds the RNA and blocks subunit joining through an interaction with eIF5B (Deng et al. 2008). As for r15-LOX, repression is alleviated by phosphorylation of the RNA-binding protein. Vas differs from these other translational regulators in that it positively regulates its targets; like hnRNP K and Puf6p, however, post-translational modification of Vas has also been linked to a reduction of its activity (Ghabrial and Schüpbach 1999; Pane et al. 2007).
Several phenotypes manifested in vas-null ovaries point toward a function for Vas in restricting cell fate during cystocyte divisions, and these could result from reduced mei-P26 expression. The relationship between Mei-P26 and Vas may be more complex, however, as both are linked to small RNA metabolism. Mei-P26 binds to AGO1, an RNase that is a core component of the RNA-induced silencing complex (RISC) that is involved in miRNA-mediated translational repression and RNA degradation pathways (Neumüller et al. 2008). Both Mei-P26 and AGO1 have been implicated in regulating GSC fate; Mei-P26 restricts growth and proliferation and promotes differentiation, while AGO1 does the reverse (Yang et al. 2007; Neumüller et al. 2008). Piwi, a key component of rasiRNA and piwi-interacting RNA (piRNA) pathways, has been implicated in stem cell self-renewal (Cox et al. 2000). Vas is associated with Piwi, Aubergine, and other components of the rasiRNA pathway (Megosh et al. 2006; Vagin et al. 2006), and has itself been linked to retrotransposon silencing (Shpiz et al. 2009). Therefore, Vas appears to be involved in both the AGO1 pathway, through its regulation of Mei-P26, and the rasiRNA pathway (Vagin et al. 2006), potentially making it a key regulator of processes mediated by small RNAs in GSCs and early-stage committed cells.
Materials and methods
Purification of Vas-binding mRNAs
We modified previous in vivo cross-linking and immunoprecipitation methods (Liu et al. 2003; Ule et al. 2003) with additional mRNA purification steps and a second Vas immunoprecipitation. Our methods are described in detail in the Supplemental Material.
Western analysis
Extracts were prepared from 3- to 5-d-old ovaries, and proteins were resolved on 7.5% SDS-PAGE gels and transferred onto nitrocellulose membranes that were blocked overnight at 4°C in PBS, 0.05% Tween 20, and 2% skim milk (PBSTM). Membranes were incubated for 1 h at room temperature with primary antibodies diluted in PBSTM, washed with PBSTM, then incubated for 1 h at room temperature with HRP-conjugated secondary antibodies (GE Healthcare) diluted 1:5000 in PBSTM. Membranes were washed with PBSTM, and proteins were detected on X-ray films using Western Lightning Plus-ECL (PerkinElmer). Rabbit anti-Mei-P26 was used at 1:1000, and the antibody was purified by preabsorption with fixed ovaries of mei-P26fs1/mei-P26mfs1 before using. Rabbit anti-Vas was used at 1:5000. Mouse anti-α-tubulin (Sigma) was used at 1:10,000. Films were scanned and quantitation of results was carried out using Quantity ONE software (Bio-Rad).
Immunohistochemistry
Ovaries were dissected from 3- to 5-d-old females in PBS and fixed in 100 μL of PBS, 1% NP-40, 600 μL of heptane, and 100 μL of 10% formaldehyde for 20 min. Samples were rinsed three times, washed three times for 10 min with PBST (PBS + 0.3% Triton X-100), and blocked in PBSTA (PBST + 1% BSA) for 1 h at room temperature. Samples were incubated with primary antibodies overnight at 4°C in PBST, and the antibody was purified by preadsorption with fixed ovaries of mei-P26fs1/mei-P26mfs1 before using. Samples were rinsed three times, washed three times for 20 min with PBST, then blocked in 1 mL of PBSTA containing 80 μL of normal goat serum (NGS) for 1 h at room temperature. Samples were incubated in the dark with fluorescent secondary antibody (preadsorbed goat anti-rabbit Alexa Fluor555, Molecular Probes), final dilution 1:1000 in PBST, and 4% NGS for 90 min at room temperature, then washed four times for 5 min and twice for 15 min in PBST in the dark. Samples were counterstained with DAPI, mounted in anti-fade reagent in glycerol/PBS from the SlowFade Antifade Kit (Molecular Probes), and examined under confocal microscopy (Zeiss LSM510). Rabbit anti-Mei-P26 was used at a dilution of 1:1000, rabbit anti-Vas was used at a dilution of 1:2000, and mAb 1B1 was used at a dilution of 1:10.
EMSAs
GST-Vas and GST were expressed in E. coli and purified by glutathione-Sepharose chromatography. mei-P26-derived transcripts were prepared with MAXIscript (Ambion). Proteins were incubated for 20 min at room temperature with 30 ng of [α-32P]UTP-labeled mei-P26 3′ UTR transcript (sense RNA or antisense RNA) in 15 μL of incubation buffer (40 mM Tris-HCl buffer at pH 7.5, 50 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 1 mM AMPPNP). After incubation, samples were separated on a 0.7% agarose gel. The gel was dried onto chromatography paper and analyzed on a Storm 840 PhosphorImager (Molecular Dynamics).
UV cross-linking assays
GST-Vas or GST was incubated for 20 min at room temperature with synthetic 5′-32P-labeled 30-nt RNAs in 15 μL of 20 mM Tris-HCI buffer (pH 8.0) containing 5 mM MgCl2, 1 mM dithiothreitol, and 1 mM AMP-PNP. For competitive UV cross-linking assays, unlabeled competitors were added. The reaction mixtures were then irradiated with a 254-nm UV lamp, 8 W (MRL-58 Multiple-Ray Lamp, UVP) for 25 min at 4°C. The distance between samples and the lamp was 2 cm. After irradiation, samples were subjected to SDS-PAGE on a 10% polyacrylamide gel, which was then dried and analyzed on a PhosphorImager as described above. RNA probes were denatured/renatured by incubating for 5 min at 95°C with gradual cooling down to room temperature before using.
Real-time PCR analysis
Total RNA was isolated from five pairs of similarly staged 1- to 3-d-old ovaries of relevant phenotypes using the RNeasy mini-kit (Qiagen). Possible trace contaminant DNA was removed by DNase I digestion. RNA concentrations were then adjusted to 200 μg/mL. Reverse transcription was performed using the DyNAmo cDNA synthesis kit (Finnzymes) in a 10-μL reaction volume with random primers and 500 ng of RNA for 45 min at 37°C. Negative controls (NRTs) omitted reverse transcriptase. Quantitative PCR was performed using a DyNAmo Flash SYBR Green kit (Finnzymes) using a C1000 Thermal Cycler (Bio-Rad) according to the manufacturer's protocol. Two reference genes (β-Tub and rp49) were used for each sample. The following primers were used: mei-P26: forward, 5′-CGTTCTCAGCGACGATGCC-3′, reverse, 5′-GCAGGAACCAGAGCTGGAG-3′, amplicon size 112 nt; GFP: forward, 5′-GAACCGCATCGAGCTGAAGG-3′, reverse, 5′-CGCGGATCTTGAAGTTCACC-3′, amplicon size 146 nt. Experiments were run in triplicate with an NRT control. PCR reactions were run in a 10-μL volume for 30 cycles. Data from each experiment were analyzed by CFX Manager software (Bio-Rad). Data from multiple experiments were pooled and analyzed using Excel (Microsoft).
mei-P26 transgenic flies
Deletions in the mei-P26 3′ UTR were generated from cDNA clone GH10646 using the Gene Tailor Site-Directed Mutagenesis System (Invitrogen), and cloned into P[w+Pvas-egfp] (Nakamura et al. 2001). Detailed cloning procedures are available upon request. Constructs were verified by DNA sequencing (Genome Quebec Innovation Center). P-element-mediated germline transformation was carried out using standard techniques (Spradling and Rubin 1982). To compare relative protein levels, ovarian extracts from 1- to 3-d-old transgenic flies were resolved on 10% SDS-PAGE gels and transferred onto nitrocellulose. Immunoblots were performed using anti-GFP antibody (1:500; Roche), anti-Vas (1:5000), and anti-α-tubulin (1:10,000).
Acknowledgments
We are grateful to Jan-Michael Kugler for helpful comments on the manuscript. This work was funded by grant R01-HD036631 to P.L. from the National Institute of Child Health and Development.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1820709.
Supplemental material is available at http://www.genesdev.org.
References
- Abdu U, Brodsky M, Schüpbach T. Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of gurken through Chk2/Mnk. Curr Biol. 2002;12:1645–1651. doi: 10.1016/s0960-9822(02)01165-x. [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuléen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jäckle H, Lasko P. Vasa mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schüpbach T. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Cho P, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: Inhibition of caudal via 5′–3′ tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Cho P, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in the Drosophila embryo. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse KN, Ferguson SB, Schüpbach T. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev Biol. 2008;313:713–724. doi: 10.1016/j.ydbio.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes & Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. Sfold Web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo P, Ephrussi A. Bruno regulates gurken during Drosophila oogenesis. Mech Dev. 2003;120:289–297. doi: 10.1016/s0925-4773(02)00454-9. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Schüpbach T. Activation of a meiotic checkpoint regulates translation of gurken during Drosophila oogenesis. Nat Cell Biol. 1999;1:354–357. doi: 10.1038/14046. [DOI] [PubMed] [Google Scholar]
- Glasscock E, Singhania A, Tanouye MA. The mei-P26 gene encodes a RING finger B-box coiled-coil-NHL protein that regulates seizure susceptibility in Drosophila. Genetics. 2005;170:1677–1689. doi: 10.1534/genetics.105.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, Dart5, is essential for germ cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Kadryova LY, Habara Y, Lee TH, Wharton RP. Translational control of Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, Papov VV, Schneiders JM, Szer W, Wilson SH, Paik WK, et al. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly–Gly–Gly–Arg–Gly–Gly–Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36:5185–5192. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- Kugler JM, Lasko P. Localization, anchoring, and translational control of oskar, gurken, bicoid, and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of Vasa protein correlates with, but is not sufficient for, pole cell development. Genes & Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Liu N, Dansereau DA, Lasko P. Fat Facets interacts with Vasa in the Drosophila pole plasm and protects it from degradation. Curr Biol. 2003;13:1905–1909. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- McBride AE, Silver PA. State of the arg: Protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of Piwi and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Menon L, Mihailescu MR. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messias AC, Harnisch C, Ostareck-Lederer A, Sattler M, Ostareck DH. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J Mol Biol. 2006;361:470–481. doi: 10.1016/j.jmb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila Cup is an eIF4E-binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–246. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155:1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Reimann I, Huth A, Thiele H, Thiele BJ. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX RNA 3′-UTR control element DICE. J Mol Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: A Web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Maternal-effect mutations altering the anterior–posterior pattern of the Drosophila embryo. Rouxs Arch Dev Biol. 1986;195:302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37:268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Spradling AC, de Cuevas M, Drummond-Barbosa D, Keyes L, Lilly M, Pepling M, Xie T. The Drosophila germarium: Stem cells, germ line cysts, and oocytes. Cold Spring Harb Symp Quant Biol. 1997;62:25–34. [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for Gurken accumulation in the oocyte, and is involved in oocyte determination and germ line cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA. rasiRNAs, DNA damage, and embryonic axis specification. Cold Spring Harb Symp Quant Biol. 2006;71:171–180. doi: 10.1101/sqb.2006.71.066. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends upon regulation of gurken by Vasa. Development. 1998;125:1723–1732. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB FlyBase Consortium. FlyBase: Integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P, Chen D. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Zaccai M, Lipshitz HD. Differential distributions of two adducin-like protein isoforms in the Drosophila ovary and early embryo. Zygote. 1996;4:159–166. doi: 10.1017/s096719940000304x. [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci. 2004;101:14800–14805. doi: 10.1073/pnas.0406451101. [DOI] [PMC free article] [PubMed] [Google Scholar]