Abstract
Medulloblastoma is the most common solid malignancy of childhood, with treatment side effects reducing survivors’ quality of life and lethality being associated with tumor recurrence. Activation of the Sonic hedgehog (Shh) signaling pathway is implicated in human medulloblastomas. Cerebellar granule neuron precursors (CGNPs) depend on signaling by the morphogen Shh for expansion during development, and have been suggested as a cell of origin for certain medulloblastomas. Mechanisms contributing to Shh pathway-mediated proliferation and transformation remain poorly understood. We investigated interactions between Shh signaling and the recently described tumor-suppressive Hippo pathway in the developing brain and medulloblastomas. We report up-regulation of the oncogenic transcriptional coactivator yes-associated protein 1 (YAP1), which is negatively regulated by the Hippo pathway, in human medulloblastomas with aberrant Shh signaling. Consistent with conserved mechanisms between brain tumorigenesis and development, Shh induces YAP1 expression in CGNPs. Shh also promotes YAP1 nuclear localization in CGNPs, and YAP1 can drive CGNP proliferation. Furthermore, YAP1 is found in cells of the perivascular niche, where proposed tumor-repopulating cells reside. Post-irradiation, YAP1 was found in newly growing tumor cells. These findings implicate YAP1 as a new Shh effector that may be targeted by medulloblastoma therapies aimed at eliminating medulloblastoma recurrence.
Keywords: Hippo, Sonic hedgehog, TEAD1, YAP1, cerebellum, medulloblastoma
Medulloblastoma is the most common malignant solid tumor in children. These tumors arise in the developing cerebellum, a region of the brain that undergoes rapid expansion after birth, during the first years in humans and the first 2 wk in mice. Current treatments for medulloblastoma include radiation, surgery, and chemotherapy, all of which are associated with devastating physical and mental side effects in long-term survivors, including life-long cognitive and psychological damage (Packer et al. 1999). Attempts to reduce levels of radiation to reduce side effects are associated with tumor recurrence. Recently, cells within medulloblastomas that survive radiation treatment and can repopulate the tumors post-radiation have been identified (Calabrese et al. 2007; Hambardzumyan et al. 2008). These cells reside in regions adjacent to blood vessels, and their post-radiation survival is promoted by PI3 kinase pathway activity (Hambardzumyan et al. 2008). Ideal medulloblastoma therapies will attack the tumors and eliminate the tumor reinitiating cells but spare the rest of the brain. Unfortunately, the poor understanding of molecular events leading to medulloblastoma formation, maintenance, and recurrence has hindered the advancement of treatment options.
Cerebellar granule neural precursors (CGNPs) are proposed cells of origin for certain classes of medulloblastoma (Provias and Becker 1996). After birth, CGNPs undergo a rapid expansion phase in the cerebellar external granule layer (EGL). CGNPs then exit the cell cycle and migrate through the underlying layer of Purkinje neurons toward the internal granule layer (IGL), where they complete their differentiation program (Hatten and Heintz 1995). CGNP proliferation is dependent on signaling by Sonic hedgehog (Shh), secreted by Purkinje neurons (Dahmane and Ruiz i Altaba 1999).
Aberrant activation of the Shh signaling pathway is implicated in the formation of medulloblastomas (Raffel et al. 1997; Reifenberger et al. 1998). This can be phenocopied in mice, in that inducing increased Shh pathway activity causes medulloblastomas (Wetmore 2003; Fults 2005). Cell division is a complex process that requires integration of many intracellular pathways. We showed previously that Shh signaling can cooperate with insulin-like growth factor (IGF) signaling (Kenney et al. 2004), which promotes cell survival and growth through its effects on Akt and mammalian target of rapamycin (mTOR), respectively (Foulstone et al. 2005; Kurmasheva and Houghton 2006). Shh:IGF pathway interactions also promote proliferation through activation of insulin receptor substrate 1 (IRS1) in CGNPs (Parathath et al. 2008). Consistent with cooperative mechanisms enhancing Shh-mediated transformation, the incidence of Shh-mediated medulloblastomas in mice can be increased by the use of radiation, loss of p53, or concurrent activation of proteins that promote cell survival and proliferation, such as Bcl2, IGF, and N-myc itself (Fults 2005; Marino 2005).
Here, we investigated whether Shh mitogenic signaling also interacts with the recently described tumor-suppressive Hippo pathway in CGNPs and medulloblastoma. This pathway restricts organ size increase by inhibiting the activity of the transcriptional cofactor yes-associated protein 1 (YAP1); YAP1 itself can promote proliferation and transformation. YAP1 is a WW domain-containing transcriptional coactivator (Yagi et al. 1999) that has been shown recently to cooperate with myc in a mouse model of hepatocellular carcinoma (Zender et al. 2006). The name WW derives from the presence of two signature tryptophan residues that are spaced 20–23 amino acids apart. YAP1 overexpression in human mammary epithelial cells leads to their malignant transformation (Overholtzer et al. 2006). The YAP1 gene is located in the 11q22 amplicon, which is frequently observed in different human cancers, including glioblastomas, squamous cell carcinoma, and pancreatic, oral, cervical, ovarian, and lung cancers, among others (Weber et al. 1996; Imoto et al. 2002; Dai et al. 2003; Baldwin et al. 2005; Bashyam et al. 2005; Hermsen et al. 2005; Lambros et al. 2005).
The components and the function of the Hippo pathway are well conserved in mammals (Zhao et al. 2007). YAP1 interacts with and regulates the activity of several transcription factors, including RUNX2, SMAD7, p73, p53BP2, and the TEA domain transcription factor (TEAD) family members (Saucedo and Edgar 2007). When YAP1 is phosphorylated by the Lats1 tumor suppressor, it translocates to the cytoplasm, where it interacts with 14–3–3 proteins and is thought to be inactive. Recently, Cao et al. (2008) identified a role for YAP1 in regulating chick neural tube progenitor number through interactions with TEAD. However, although a role for Shh in neural precursor proliferation and patterning in the neural tube is well established (Ulloa and Briscoe 2007), a relationship between the Hippo and Shh pathways has not yet been shown.
In this study, we demonstrate that YAP1 and its transcriptional partner, TEAD1, are highly expressed in Shh-driven medulloblastomas in both humans and mice. We also report, for the first time, amplification of YAP1 in a subset of human medulloblastomas—specifically, SHH-associated medulloblastomas. Moreover, we show that YAP1 expression is up-regulated by the Shh pathway in proliferating CGNPs, that Shh signaling regulates YAP1 nuclear localization through its binding to IRS1, and that YAP1 activity promotes CGNP proliferation, at least in part through interactions with TEAD1. In mouse medulloblastomas, YAP1 protein localized to the cells occupying the perivascular niche (PVN) that have been proposed to have cancer stem cell properties. Indeed, YAP1-positive cells remained alive and disseminated through the tumor after the tumor bulk cells were eradicated by radiation. Our findings mark YAP1 as a mediator of normal proliferation in the developing cerebellum, and as a potential target for medulloblastoma therapies aimed at eliminating tumor-reinitiating cells.
Results
YAP1 is overexpressed and amplified in human medulloblastoma
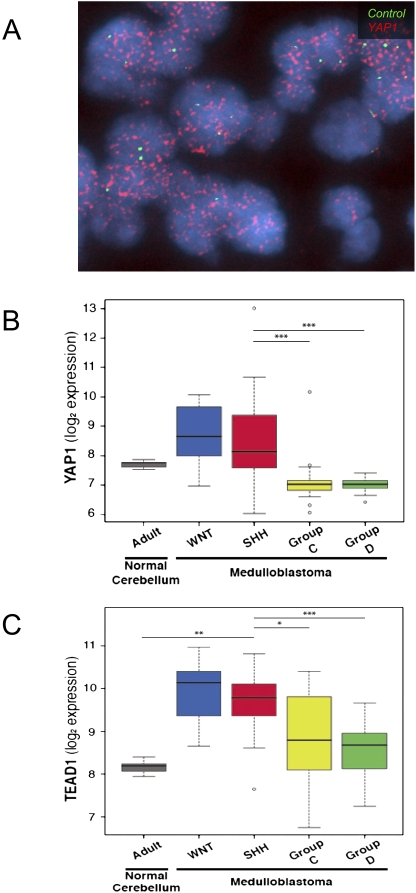
A role for YAP1 in medulloblastoma, for which cerebellar neural precursor cells are a postulated cell of origin, has not been determined. To determine whether YAP1 may be involved in human medulloblastoma, we performed interphase fluorescence in situ hybridization (FISH) on a human medulloblastoma tissue microarray comprised of 67 medulloblastomas. We observed high-copy amplification of YAP1 but not of a centromeric control probe in two tumors; protein analysis indicates that the representative tumor shown has high levels of YAP1 protein. Moreover, when we analyzed results from a recent large-copy-number study of the medulloblastoma genome (Northcott et al. 2009b), we found a single medulloblastoma with a high-copy-number amplification of YAP1 on chromosome 11q22 (Fig. 1A; data not shown); this medulloblastoma belonged to the SHH subset of tumors as determined by its gene expression pattern. Next, we carried out gene expression analysis of a medulloblastoma collection comprising >200 samples. Examination of YAP1 expression revealed that it is highly overexpressed in both SHH- and WNT-dependent medulloblastomas as compared with other normal cerebellar controls (Fig. 1B; Northcott et al. 2009a). YAP1 is specifically up-regulated in SHH and WNT medulloblastoma subgroups as compared with Group C and Group D medulloblastomas (comprised largely of classical medulloblastomas). Similarly, TEAD1, the major transcriptional partner of YAP1 (Zhao et al. 2008), is overexpressed in both SHH- and WNT-dependent medulloblastomas as compared with normal controls and Group C and Group D medulloblastomas (Fig. 1C). Similar results were observed when we analyzed a publicly available smaller database (Supplemental Fig. 1; Kool et al. 2008). Taken together, these data comprise the first observations of YAP1 up-regulation/amplification in human medulloblastomas.
Figure 1.
YAP1 is amplified in a subset of human medulloblastomas, and YAP1 and TEAD1 are overexpressed in human Shh- and Wnt-driven medulloblastomas. (A) Interphase FISH with a YAP1 probe (red signal) revealed a double-minute pattern of hybridization in two out of 67 (3%) human medulloblastomas. A control probe targeting 11p11.2 (green signal) showed a normal complement. (B) Box plot showing YAP1 mRNA expression obtained from an exon array profiling of 110 human medulloblastomas and 14 cerebella. YAP1 is highly expressed in SHH- and WNT-driven medulloblastomas. These plots are a useful means of displaying differences between populations (i.e., medulloblastoma subgroups) as they depict groups of numerical data (in this case, signal intensity/expression level for the respective genes in adult cerebellum and medulloblastoma subgroups) through their five-number summaries: the smallest observation (sample minimum = lower line), lower quartile (Q1 = bottom of box), median (Q2 = line in box), upper quartile (Q3 = top of box), and largest observation (sample maximum = upper line). These plots are also able to identify any observations that may represent outliers (circles outside the boxes). Note the outlier above the YAP1 SHH plot is a medulloblastoma with genomic amplification of YAP1 and concordant YAP1 expression. (C) Box plot showing TEAD1 mRNA expression in the same sample series. Note high levels of expression in WNT- and SHH-associated medulloblastomas. To statistically compare the expression of YAP1 and TEAD1 in SHH-driven medulloblastomas to their relative expression in the individual subgroups (and to the normal adult cerebellum), we performed the Wilcoxon rank-sum (Mann-Whitney) test. Statistically significant differences are indicated as (*) P < 0.01; (**) P < 0.001; (***) P < 0.0001. These results show that YAP1 and TEAD1 expression is significantly higher in SHH-driven (and WNT-driven) medulloblastoma than Group C and Group D tumors. Similar results of significance were obtained when comparing YAP1 and TEAD1 expression in WNT medulloblastoma to either adult cerebellum or GroupC/D tumors.
Shh up-regulates YAP1 in primary CGNP cultures
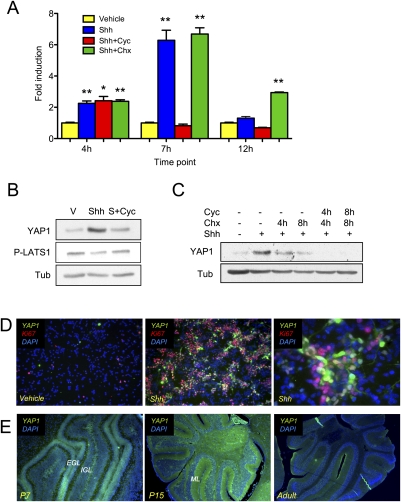
SHH pathway-activated medulloblastomas are thought to arise from CGNPs in the developing cerebellum that depend on Shh signaling for their expansion during development. Therefore, we wished to determine whether the association of YAP1 with Shh signaling was conserved between cancer and developmental biology. We carried out quantitative RT–PCR analysis of mRNA extracts from primary CGNP cultures that were maintained without serum and that were treated with purified Shh protein, which sustains their proliferation, or with Shh vehicle, in whose presence these cells undergo differentiation. As shown in Figure 2A, Shh treatment resulted in rapidly increased levels of YAP1 mRNA. This increase was prevented in the presence of the Smoothened inhibitor cyclopamine, but not when cells were treated with the protein synthesis inhibitor cycloheximide, indicating that new protein synthesis is not required to up-regulate YAP1 expression. The increase in YAP1 mRNA levels was not maintained over time.
Figure 2.
YAP1 mRNA and protein expression are up-regulated by Shh. (A) YAP1 mRNA expression in CGNPs treated as indicated was analyzed by real-time PCR and fold change is represented. There is a transient increase in YAP1 mRNA expression in Shh-treated cells with a maximum increase at 7 h. Treatment with cyclopamine (cyc) blocks YAP1 mRNA up-regulation, while treatment with cycloheximide (chx) does not. Statistically significant differences are indicated as (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) CGNPs were cultured in vitro for 48 h in the presence or absence of Shh, then treated with cyclopamine for 12 h when indicated. Shh treatment leads to YAP1 protein accumulation and reduced LATS1 phosphorylation. Both events are prevented in the presence of cyclopamine. (C) CGNPs were cultured in vitro for 48 h in the presence or absence of Shh ± cyclopamine and/or cycloheximide. Treatment with cyclopamine accelerates YAP1 protein degradation. (D) CGNPs cultured in the absence or presence of Shh were fixed and immunostained for YAP1 and Ki67. (Left) CGNPs expressing YAP1 are also Ki67 positive. (E) Cerebella of SW129 mice were immunostained for YAP1 at three developmental stages. (Left) At P7, YAP1 is expressed mainly in the EGL. (Middle) At P15, YAP1 is present in granule cells in the IGL, and in their processes in the molecular layer. In adult cerebella, we also detect YAP1 protein distributed sparsely throughout the IGL.
When we analyzed CGNP protein lysates by Western blotting after 48 h of Shh treatment, we detected high levels of YAP1 protein (Fig. 2B), together with a decrease in phospho-LATS (P-LATS), the kinase that phosphorylates and inactivates YAP1. To gain further insight into the mechanism through which Shh regulates YAP1, we treated CGNPs with cyclopamine for 12 h. This attenuated the Shh-induced increase in YAP1 protein and restored P-LATS1 levels (Fig. 2B). To determine whether Shh signaling regulates YAP1 stability, we carried out cycloheximide chases in the presence and absence of cyclopamine. As shown in Figure 2C, in the presence of Shh, YAP1 protein has a half-life of ∼4 h. In the presence of cyclopamine, YAP1 degradation occurs more rapidly, indicating a role for the Shh pathway in stabilizing YAP1 protein. Our results indicate that Shh mediates a transient increase in YAP1 expression and a sustained stabilization of YAP1 protein.
In the Shh-treated CGNPs, YAP1 was in cells undergoing proliferation, as determined by colocalization with the proliferation marker Ki67 (Fig. 2D; Supplemental Fig. 2B). During postnatal cerebellar development, CGNPs proliferate in the EGL. We carried out immunofluorescent staining of mouse cerebellar sections at three developmental stages: postnatal day 7 (P7), when CGNP proliferation is at its peak; P15, when proliferation is winding down; and in adulthood, when cerebellar development is complete. As shown in Figure 2E, YAP1 antibodies marked the EGL during the peak of CGNP proliferation. At P15, YAP1 was present in granule cells in the IGL, and in their processes in the molecular layer. Interestingly, we also detected YAP1 protein sparsely distributed throughout the IGL in adult mouse cerebella. Staining is specific, as determined by isotype control tests (Supplemental Fig. 3A). These results indicate that YAP1 protein is associated with CGNPs undergoing Shh-dependent proliferation in vitro and in vivo, and that YAP1 may play roles in later stages of cerebellar development as well.
Interactions with IRS1 regulate YAP1 localization
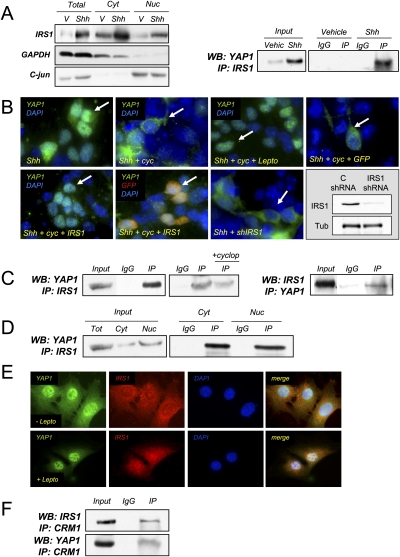
Recently, we demonstrated that Shh signaling promotes stabilization of IRS1, and that IRS1 is present in the nucleus of proliferating CGNPs (Fig. 3A, left; Parathath et al. 2008). Since we also detected YAP1 in the nucleus of CGNPs (Fig. 2D), and previous studies have implicated IRS1 in transcriptional regulation of proliferation-associated genes (Wu et al. 2008), we asked whether these two proteins interact. When we immunoprecipitated IRS1 from Shh-treated CGNPs, we detected YAP1 by Western blotting (Fig. 3A, right). Immunofluorescence analysis showed that, in the presence of Shh alone, YAP1 was present in the nucleus (Fig. 3B, top left). Interestingly, cyclopamine treatment caused YAP1 to leave the nucleus (Fig. 3B, top panel, second from left). YAP1 cytoplasmic localization was prevented by the addition of leptomycin, which inhibits the nuclear export protein CRM1 (Moroianu 1998). A role for IRS1 in promoting YAP1 nuclear localization is supported by our observation that retrovirus-mediated IRS1 expression retained YAP1 in the nucleus even in the presence of cyclopamine (Fig. 3B, bottom panel, left), while overexpression of GFP alone did not (Fig. 3B, top right). Cells that were infected with IRS1 possess nuclear YAP1 (Fig. 3B, bottom panel, second from left; Supplemental Fig. 3B). Conversely, shRNA lentivirus-mediated IRS1 knockdown prevented YAP1 nuclear accumulation (Fig. 3B, bottom panel, second from right). The knockdown efficiency was close to 80% (Fig. 3B, bottom right).
Figure 3.
IRS1 interacts with YAP1 and regulates its nuclear accumulation. (A) Subcellular fractionation of CGNPs cultured in the presence or absence of Shh. (Left) The greatest accumulation of IRS1 protein after Shh treatment takes place in the nucleus. (Right) Immunoprecipitation of IRS1 in Shh-treated CGNPs brings down YAP1. (B, top panel, left) Immunostaining shows that YAP1 is mainly nuclear in Shh-treated CGNPs. (Top panel, second from left) In the presence of cyclopamine, YAP1 is excluded from the nucleus. (Top panel, second from right) In the presence of cyclopamine and leptomycin, YAP1 accumulates in the nucleus. Overexpressing IRS1 in the presence of cyclopamine leads to YAP1 accumulation in the nucleus (bottom panel, left), while overexpressing control viruses (GFP) does not (top panel, right). (Bottom panel, second from left) Cells that get infected with IRS1 (GFP reporter) viruses have nuclear YAP1. (Bottom panel, second from right) IRS1 knockdown prevents YAP1 from accumulating in the nucleus. (Bottom panel, right) The knockdown efficiency is shown by Western blot. (C) Immunoprecipitation of IRS1 in the Pzp53 medulloblastoma cell line coprecipitates YAP1. (Left) The YAP1:IRS1 interaction was decreased in the presence of cyclopamine. (Right) IRS1 was also detected in YAP1 immunoprecipitates. (D) Subcellular fractionation of Pzp53 cells and subsequent IRS1 immunoprecipitation. YAP1 was detected in both the nuclear and cytoplasmic precipitates. (E) Pzp53med cells were treated with leptomycin, fixed, and immunostained for YAP1 and IRS1. (Top row) In untreated cells, YAP1 and IRS1 are both nuclear and cytoplasmic. (Bottom row) In leptomycin-treated cells, there is an accumulation of both proteins in the nucleus. (F) Immunoprecipitation of CRM1 in Pzp53 cells. Both YAP1 and IRS1 were detected in the immunoprecipitate.
To further investigate whether YAP1:IRS1 interactions play roles in YAP1 nuclear localization, we turned to Pzp53med cells, a cell line derived from a Ptc+/−/p53−/− mouse medulloblastoma (Berman et al. 2002) that has been used to model Shh-mediated proliferation events (Corcoran and Scott 2001). As shown in Figure 3C, we detected YAP1 in the IRS1 immunoprecipitate from Pzp53med cells. When we carried out the reciprocal immunoprecipitation, we detected IRS1 in YAP1 immunoprecipitates. The interaction between IRS1 and YAP1 was reduced in the presence of cyclopamine. When we carried out subcellular fractionation, we found that IRS1 and YAP1 interacted in both the cytoplasm and the nucleus (Fig. 3D), suggesting that IRS1 may interact with both nuclear and cytoplasmic (phosphorylated) YAP1. Indeed, in IRS1 immunoprecipitates, we can detect phosphorylated YAP1 (Supplemental Fig. 4A). Consistent with a role for CRM1 in mediating IRS1 and YAP1 export to the cytoplasm, when we treated the Pzp53med cells with leptomycin, we observed an accumulation of both proteins in the nuclear fraction (Fig. 3E). Moreover, when we immunoprecipitated CRM1, the protein inhibited by leptomycin, we were able to detect both IRS1 and YAP1 (Fig. 3F). Taken together, these results suggest a model in which Shh stabilizes IRS1 (Parathath et al. 2008) and induces YAP1, and that IRS1:YAP1 interactions regulate their subcellular localization.
YAP1 interacts with TEAD1 in CGNPs and medulloblastoma cells
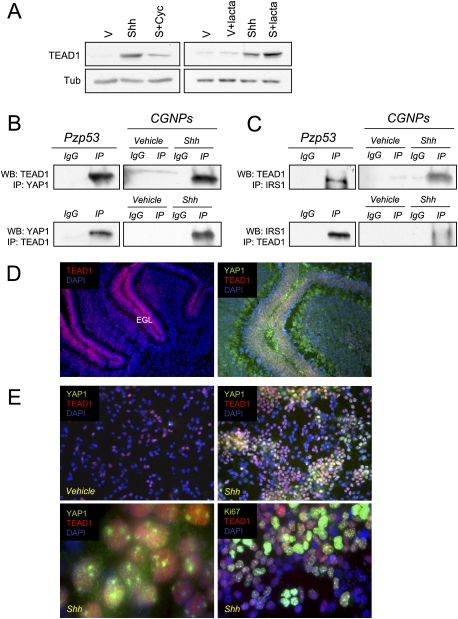
It has been shown recently that YAP1 interactions with the TEAD family of transcription factors are important for maintaining the neural progenitor state of cells in the developing chick spinal cord (Cao et al. 2008). Since Shh inhibits differentiation of CGNPs (Wechsler-Reya and Scott 1999), we wished to determine whether the YAP1:TEAD interaction was conserved in mammalian cerebellar neural progenitors. We first asked whether Shh treatment affected expression of TEAD1, but we found no effects of Shh treatment on TEAD1 mRNA levels in CGNPs (data not shown). Instead, as we found with YAP1, Shh promoted the accumulation of TEAD1 protein. There is no accumulation of TEAD1 in vehicle-treated cells in the presence of the proteasome inhibitor lactacystin, indicating that, even though the mRNA may be present at low levels, it is not being translated in the absence of Shh. However, in the presence of lactacystin in Shh-treated cells, there is an accumulation of TEAD1 (Fig. 4A).
Figure 4.
TEAD1 interacts with YAP1 in CGNPs. (A) CGNPs were cultured for 48 h in the presence or absence of Shh and treated with cyclopamine (12 h) or lactacystin (6 h) as indicated. (Left) In the presence of Shh, there is an accumulation of TEAD1 protein that is blocked by cyclopamine. (Right) In the presence of lactacystin and Shh, there is an accumulation of TEAD1 compared with Shh alone. (B) Immunoprecipitation of YAP1 and TEAD1 in Pzp53 cells and CGNPs. TEAD1 was detected in YAP1 precipitates in both Pzp53 and Shh-treated CGNPs (top panel) and vice versa (bottom panel). (C) Immunoprecipitation of TEAD1 and IRS1 in Pzp53 cells and CGNPs. TEAD1 was detected in IRS1 immunoprecipitates in Pzp53 and Shh-treated CGNPs (top panel) and vice versa (bottom panel). (D) Cerebella of SW129 P7 mice were immunostained for TEAD1. (Left) TEAD1 is found in the EGL. (Right) TEAD1 and YAP1 are coexpressed in the EGL of the cerebellum. (E) CGNPs were cultured for 48 h and immunostained for TEAD1, YAP1, and Ki67. (Top left) In the absence of Shh, TEAD1 protein expression is very low. (Top right) In the presence of Shh, TEAD1 accumulates. (Bottom left) YAP1 and TEAD1 are localized in the nucleus. (Bottom right) TEAD1 is coexpressed with Ki67.
As shown in Figure 4B, when we immunoprecipitated endogenous YAP1, we could detect TEAD1 by Western blotting, and vice versa. Moreover, TEAD1 and IRS1 also interact, based on coimmunoprecipitation experiments carried out in Pzp53med cells and CGNPs (Fig. 4C). In the developing cerebellum, TEAD1 was found in the EGL (Fig. 4D, left) where it colocalizes with YAP1 (Fig. 4D, right). Consistent with our Western blot analyses, immunofluorescent staining revealed up-regulation of YAP1 and TEAD1 in Shh-treated CGNPs (Fig. 4E, top panels). In primary CGNP cultures treated with Shh, we found both YAP1 and TEAD1 in the nucleus (Fig. 4E, bottom left panel). A high percentage of cells expressing TEAD1 also express YAP1. All YAP1 and many TEAD1-positive cells express the proliferation marker Ki67 as well (Fig. 4E, bottom right; Supplemental Fig. 2B). Taken together, these results suggest that YAP1 and TEAD1 function in a complex in Shh-treated proliferating CGNPs, consistent with the study by the Gage group (Cao et al. 2008). However, we cannot rule out the possibility that TEAD1 has additional interacting partners in CGNPs. Likewise, YAP1 may function in additional complexes in Shh-stimulated CGNPs. Our future studies will identify novel YAP1 interactors in cerebellar precursors.
YAP1 is required to sustain CGNP proliferation
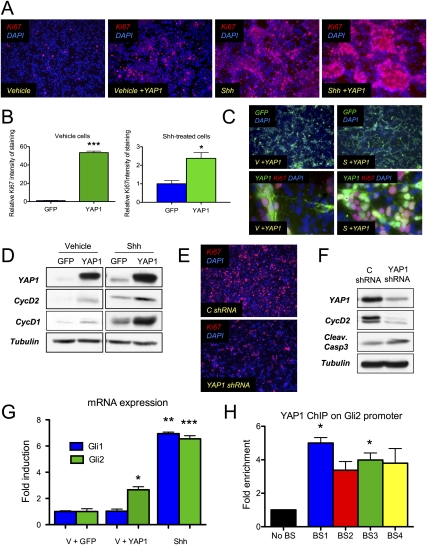
Shh is the obligate mitogen for CGNPs; when it is withdrawn from their culture medium, CGNPs leave the cell cycle within 6 h (Kenney and Rowitch 2000) and begin to differentiate. Downstream effectors of Shh mitogenic signaling include Gli1, Gli2, N-myc, and IRS1. It has been shown that ectopic expression of these genes can drive CGNP proliferation even in the absence of Shh (Kenney et al. 2003; Oliver et al. 2003; Parathath et al. 2008). We wished to determine whether YAP1, too, could sustain CGNP proliferation and/or synergize with exogenous Shh. We used retroviral transduction to express YAP1 in CGNPs that had been pretreated with Shh to maintain their proliferative state, then maintained them in culture with or without Shh for 24 h. Ectopic YAP1 expression significantly increased proliferation in cells grown without Shh (Fig. 5A,B), and YAP1 overexpression also elicited a 2.5-fold increase in proliferation in the presence of Shh. However, in cultures left without Shh after infection, which had a similar infection efficiency as those grown in Shh post-infection (Fig. 5C, top panel), proliferation was less than in Shh-treated cultures, indicating that YAP1 alone is not sufficient to completely recapitulate CGNP mitogenic response to Shh, perhaps due to the requirement for Shh signaling to up-regulate YAP1’s binding partner TEAD1, which is nearly absent from vehicle-treated cells (Fig. 3). In both conditions, the majority of cells expressing Ki67 are the ones expressing YAP1 (Fig. 5C, bottom). As shown in Figure 5D, we achieved high levels of YAP1 expression, and this was associated with increased levels of cyclin D1 and cyclin D2—markers for CGNP proliferation.
Figure 5.
YAP1 overexpression induces proliferation of CGNPs. (A) CGNPs were transduced with YAP1-expressing retroviruses. Cells were immunostained for Ki67. (Left) In the absence of Shh, few cells proliferate. When YAP1 is overexpressed in the absence of Shh, proliferation increases (second from the left), although not to the same extent as in the presence of Shh (second from the right). (Right) Overexpression of YAP1 in the presence of Shh leads to increased proliferation. (B) Automated quantification of Ki67 staining in CGNPs transduced with GFP or YAP1 retroviruses. Three different fields were considered in each case. Statistically significant differences are indicated as (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (C, top panel) GFP immunostaining shows similar infection efficiency in Shh-treated cells and cells from which Shh was withdrawn after infection. (Bottom panel) YAP1 + Ki67 immunostaining shows that the majority of cells expressing Ki67 are the ones expressing YAP1. (D) Western blot showing the increase in YAP1 protein expression after viral transduction. YAP1 overexpression leads to cyclin D1 and cyclin D2 up-regulation, reflecting an increase in proliferation. (E) Ki67 immunostaining in CGNPs infected with YAP1 shRNAs shows a decrease in proliferation compared with control shRNA-infected cells. (F) Western blot showing YAP1 protein levels were reduced by 65% in cells infected with YAP1 shRNA lentiviruses. Cyclin D2 levels were dramatically decreased with only a subtle increase in cleaved caspase 3. (G) CGNPs were transduced with YAP1-expressing retroviruses. After 24 h, Gli2 mRNA expression was analyzed by real-time PCR. YAP1 overexpression induces a statistically significant induction in Gli2 mRNA levels in the absence of Shh. (*) P < 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001. (H) ChIP analysis was carried out to test the presence of YAP1 on the Gli2 promoter. Four regions containing TEAD1-binding sites were assessed. Fold enrichments normalized to the level observed at the control region are shown. Statistically significant differences are indicated as (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
YAP1 is not only sufficient to increase CGNP proliferation but is also required to sustain their proliferation, since knocking down YAP1 expression in the presence of Shh leads to a dramatic decrease in proliferation with only a subtle increase in apoptosis (Fig. 5E,F). Our observation that YAP1 is required to sustain Shh-induced CGNP proliferation is in agreement with previous studies that have shown that YAP1 is limiting for proliferation (Buttitta and Edgar 2007; Pan 2007; Zhao et al. 2007; Zeng and Hong 2008). A database search (http://www.switchdb.com/motifs) revealed four TEAD1-binding sites in the promotor of the Shh effector Gli2 (Supplemental Fig. 4B). Consistent with YAP1-mediated Gli2 induction contributing to YAP1’s proliferative effects in CGNPs, we observed increased expression of Gli2 in vehicle-treated CGNPs infected with YAP1 retroviruses. In the presence of Shh, Gli2 expression was already at maximal levels (Fig. 5G). In order to confirm a role for YAP1/TEAD1 in regulating the Gli2 promotor, we conducted a chromatin immunoprecipitation (ChIP) assay to scan for YAP1 binding in the regions that we found to contain putative TEAD1-binding sites. As shown in Figure 5H, we found statistically significant evidence that YAP1 binds to two out of the four TEAD1-binding sites that we analyzed. These results suggest that one mechanism through which YAP1 could drive CGNP proliferation is induction of Gli2, but the inability of YAP1 to fully recapitulate the Shh proliferative response indicates roles for other transcription factors in regulating expression of Gli2 and other Shh mitogenic effectors.
YAP1 is up-regulated in mouse Shh-induced medulloblastomas, where it localizes to the tumor cells in the PVN
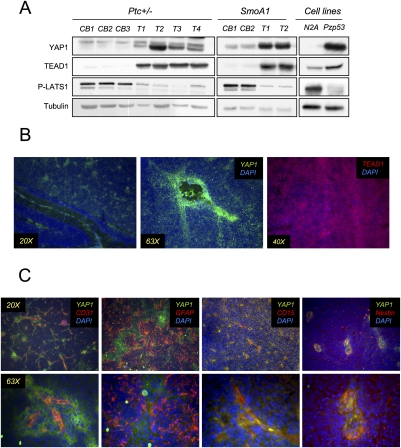
To determine whether the proliferative function of YAP1 and TEAD1 in Shh-stimulated CGNPs might be conserved in Shh-mediated tumorigenesis, we analyzed their levels in medulloblastomas harvested from mice heterozygous for the tumor suppressor Ptc, an inhibitory component of the Shh receptor complex, in comparison with adjacent, non-tumor-containing cerebellar tissue. We also analyzed medulloblastomas arising in NeuroD2-SmoA1 transgenic mice, which express a constitutively active mutant allele of Smo (Hallahan et al. 2004), the positive regulator of the Shh signaling pathway. As shown in Figure 6A, both Shh-induced medulloblastoma models had high levels of YAP1 and TEAD1 protein. Similarly, YAP1 and TEAD1 were detected in Pzp53med lysates, but not in lysates from mouse N2A neuroblastoma cells, indicating that YAP1 and TEAD1 up-regulation is linked to Shh signaling and cerebellar neural precursors, and is not a general marker of proliferation. Interestingly, we also observed that medulloblastomas exhibited reduced levels of LATS1 activity in comparison with non-tumor-bearing cerebella. LATS1 phosphorylation of YAP1 causes its cytoplasmic localization and thus reduces its transcriptional coactivation capacity.
Figure 6.
YAP1 and TEAD1 are highly expressed in mouse medulloblastomas. (A) Western blot showing YAP1 and TEAD1 levels in medulloblastomas from Patched heterozygous mice (left) and NeuroD2-SmoA1 transgenic animals (middle). (Right) YAP1 and TEAD1 are also present at high levels in the Pzp53 medulloblastoma cell line compared with the N2A neuroblastoma cell line. PLATS1 was decreased in tumors and in the medulloblastoma cell line. (B) Medulloblastomas obtained from NeuroD2-SmoA1 transgenic mice were immunostained for YAP1 and TEAD1. (Left, middle) Although the expression of YAP1 is high throughout the tumor, it is especially strong around the blood vessels. (Right) TEAD1 protein is found throughout the tumor. (C) YAP1 was costained for different markers in medulloblastomas. (Left) Costaining with CD31 shows YAP1 in the PVN. (Second from left) YAP1 is not found in perivascular astrocytes, as determined by GFAP costaining. YAP1 is colocalized with CD15 (second from right) and with nestin (right).
NeuroD2-SmoA1 and Ptc+/− medulloblastomas contain heterogeneous cell types, including tumor cells, fibroblasts, vasculature cells, and entrapped or invading glial cells. The tumor cells themselves vary in their degree of differentiation and level of proliferation. To determine which cells express YAP1, we carried out immunofluorescent staining for YAP1 in conjunction with cell type markers. To our surprise, we found that, although YAP1 was diffusely expressed throughout the medulloblastomas, its expression was strikingly high in regions associated with blood vessels (Fig. 6B). In contrast, TEAD1 was expressed diffusely throughout the tumor bulk. We confirmed the YAP1-expressing regions as perivascular by immunostaining for the endothelial cell marker CD31 (Newman and Albelda 1992). The YAP1 signal did not reflect its expression in glial cells, as YAP1 immunostaining did not overlap with staining for the glial cell marker GFAP (Fig. 6C; Supplemental Fig. 5A). Cells residing in the PVN of medulloblastomas have been reported to have tumor stem cell properties, and they are positive for stem and progenitor cell markers such as nestin (Calabrese et al. 2007; Hambardzumyan et al. 2008). Recently, CD15 has also been identified as a marker of cells capable of tumor propagation (Read et al. 2009). As shown in Figure 6C and Supplemental Figure 4A, we determined that the highly YAP1-positive cells in the medulloblastomas might be these so-called tumor stem cells, as they express CD15 and nestin. YAP1 perivascular staining is specific, as determined by isotype control tests (Supplemental Fig. 5B).
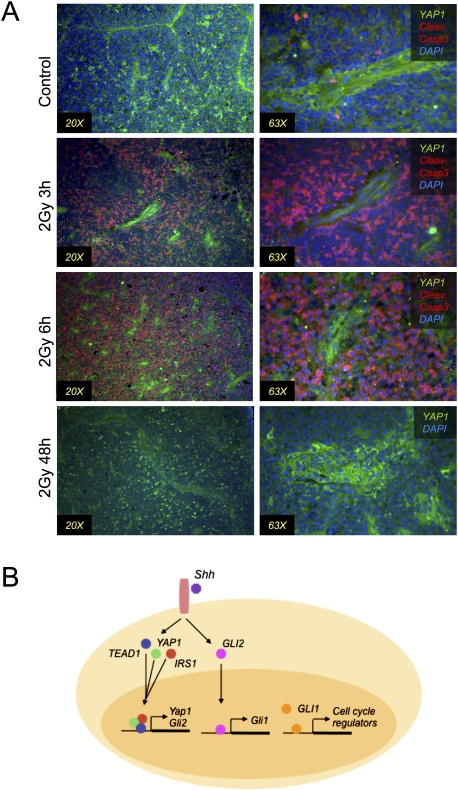
The Holland group (Hambardzumyan et al. 2008) has elegantly demonstrated that medulloblastoma tumor cells occupying the PVN survive irradiation at levels that kill the tumor bulk, and that these cells resume proliferation post-irradiation and therefore cause the regrowth of the tumors. Since YAP1 has been implicated in both survival and proliferation, we wished to determine whether its expression might be associated with tumor repopulation. We irradiated mice bearing Ptc+/− medulloblastomas, then allowed the animals to survive for 3, 6, or 48 h, before carrying out immunostaining for YAP1 and a marker of cell death (cleaved caspase 3). As shown in Figure 7A, in nonirradiated control tumors, there is little detectable cleaved caspase 3. At 3 and 6 h post-irradiation, the majority of cells in the tumor bulk were positive for cleaved caspase 3, but the YAP1-positive cells in the PVN did not have cleaved caspase 3, indicating that the YAP1-expressing cells escaped irradiation-induced cell death. By 48 h after irradiation, when it has been shown that tumor-repopulating cells are proliferating, we detected strongly YAP1-positive cells in the tumor bulk as well as the PVN (Fig. 7A). Our findings thus implicate YAP1 in survival and, potentially, tumor recurrence after medulloblastoma irradiation.
Figure 7.
YAP1 is present in perivascular cells that are resistant to radiation. (A) Immunostaining for the indicated proteins in medulloblastomas from mice irradiated with 2 Gy γ radiation. Three hours and 6 h after irradiation, most of the cells undergo apoptosis, as shown by cleaved caspase 3 staining. Cells with YAP1 are resistant to radiation. At 48 h post-irradiation, YAP1+ cells appear throughout the tumor bulk. (B) Model showing that the Shh pathway leads to YAP1 expression, protein stabilization, and nuclear accumulation. TEAD1 and IRS1 are stabilized by Shh, and IRS1 also translocates to the nucleus. YAP1, TEAD1, and IRS1 interact with each other and might regulate gene expression together. The YAP1:TEAD1 complex regulates expression of Gli2, which translocates to the nucleus downstream from activated Smoothened, where it regulates Gli1 transcription, which in turn regulates the expression of cell cycle regulators.
Discussion
Proliferation of cerebellar neural precursors, postulated medulloblastoma cells of origin, requires Shh pathway activation, and aberrant activation of this pathway is implicated in medulloblastomas. Thus, elucidating links between the hedgehog signaling pathway and mechanisms regulating cell cycle progression will yield insight into both developmental neurobiology and medulloblastoma etiology. We investigated potential interactions between the Shh pathway and the tumor-suppressive Hippo pathway, whose negatively regulated target, YAP1, has been shown recently to be required for neural progenitor cell maintenance in the chick neural tube (Cao et al. 2008) and for expansion of undifferentiated progenitor cells in the intestines (Camargo et al. 2007), and is known to have oncogenic roles in other systems (Overholtzer et al. 2006; Zender et al. 2006). We identified YAP1 as being either amplified or up-regulated in human Shh-associated medulloblastomas. Shh signaling promoted YAP1 mRNA up-regulation, protein accumulation, and nuclear localization in proliferating CGNPs. YAP1 ectopic expression was sufficient to drive Shh-independent proliferation, while YAP1 knockdown dramatically reduced CGNP proliferation. The presence of potential Gli- and N-myc-binding sites in the YAP1 promotor (Supplemental Fig. 2A) suggests that these transcription factors may regulate its expression, a topic for future analysis. Finally, we localized YAP1 to cells that comprise the tumor-repopulating compartment in mouse Shh-induced medulloblastomas.
YAP1 itself does not bind DNA, but activates transcription factors to which it binds, including p73, RUNX, and TEAD family members (Pan 2007). TEAD1 has been shown to play roles in neural progenitor population maintenance (Cao et al. 2008), and it has been shown that TEAD1 is the major YAP1 partner in breast cancer cell lines (Zhao et al. 2008). We observed Shh-dependent up-regulation of TEAD1 protein levels in CGNPs. Moreover, we detected the presence of YAP1:TEAD1 complexes in Shh-treated CGNPs and in Pzp53med cells, which were derived from a mouse Ptc+/−/p53−/− medulloblastoma.
One mechanism through which YAP1 promotes CGNP proliferation might be through inducing expression of Gli2 (Fig. 7B; Sasaki et al. 1999). When we carried out a database search for TEAD1-binding sites, we identified such sites in the Gli2 promotor (Supplemental Fig. 4B). Gli2 can then go on to activate Gli1 and other downstream mediators of Shh-induced proliferation. We also found TEAD1-binding sites in the YAP1 promotor, raising the possibility of a positive feedback loop in which TEAD1:YAP1 complexes not only regulate other mitogenic transcriptional regulators, but also ensure their own maintenance. Indeed, one function of YAP1 is to retain TEAD1 in the nucleus, where it can regulate target gene expression (Ota and Sasaki 2008). Such a model is in keeping with our observation that YAP1 overexpression alone was not sufficient to maintain full proliferation in Shh-deprived CGNPs, because under our experimental conditions, TEAD1 was not present due to the lack of Shh to promote its expression and accumulation.
Our data support a role for Shh-mediated YAP1 and TEAD1 induction and interaction in CGNP proliferation. In addition, we identified a potential role for interactions between YAP1 and IRS1 in regulating YAP1 nuclear localization: YAP1 and IRS1 were present in the same complex in Shh-treated CGNPs and inhibition of Smoothened caused redistribution of YAP1 to the cytoplasm, prevented by ectopic expression of IRS1. Moreover, we also detected CRM1 in complex with IRS1 and YAP1. CRM1, also known as Exportin, is the protein inhibited by leptomycin, and we found that leptomycin prevented YAP1 and IRS1 nuclear export, suggesting a role for CRM1 in this process. We showed previously that Shh stabilizes IRS1 (Parathath et al. 2008), which may potentiate IGF-mediated mitogenic signals. Our new results indicate a second function for IRS1 in Shh-mediated proliferation, promoting YAP1 nuclear localization.
Indicating conserved roles between development and medulloblastoma, we observed that Shh-induced mouse medulloblastomas possessed high levels of YAP1 and its transcriptional partner, TEAD1. Excitingly, in these tumors, YAP1 localized to the PVN, adjacent to endothelial cells. A role for endothelial cells in providing a microenviroment supportive of maintaining a cancer “stem” or “initiating” cell has been reported by several groups (Calabrese et al. 2007; Yang and Wechsler-Reya 2007). Indeed, recent studies have shown that the PVN contains a reservoir of cells capable of tumor repopulation after irradiation kills the tumor bulk (Hambardzumyan et al. 2006). In our studies, these cells expressed the highest levels of YAP1. Patients with medulloblastoma that recurs after radiation therapy have reduced survival outcome, death from a second malignant tumor more frequent than death from the original tumor (Jenkin et al. 1995). Understanding the cell biological basis for the ability of the tumor-repopulating cells to survive lethal irradiation and mount a post-irradiation proliferative response is essential for developing therapeutic strategies to eliminate this population of cells and thereby prevent medulloblastoma recurrence; our studies suggest a possible role for YAP1 in this process.
In summary, we identified the transcriptional coactivator YAP1 as a target of Shh mitogenic signaling in the developing cerebellum, whose expression is also up-regulated in Shh-associated medulloblastomas. The ability of YAP1 to drive CGNP proliferation in the absence of Shh, and its presence in medulloblastoma tumor-repopulating cells, suggests that YAP1 itself could make a good target for new medulloblastoma treatments; alternative ways to up-regulate the Hippo pathway that suppresses YAP1 activity should also be explored. A natural mechanism for regulating YAP1 activity is its phosphorylation downstream from LATS1, which promotes its cytoplasmic relocalization. Indeed, our observations that Shh promotes YAP1 nuclear localization and that Shh-induced mouse medulloblastomas have reduced levels of phosphorylated LATS1 indicate that ways to promote YAP1 cytoplasmic translocation, by manipulating the nuclear export machinery or activating Hippo signaling, may be a means to reduce YAP1’s proproliferative effects. In future studies, we will determine precisely how the Shh pathway affects YAP1 nuclear import and export, whether YAP1 can drive medulloblastoma formation on its own, and what function is carried out by YAP1 in medulloblastoma PVN cells.
Materials and methods
Animal studies
Harvest of neural precursors from neonatal mice, preparation of cerebella and tumor tissue from wild-type and mutant mice for histological analysis, and irradiation of tumor-bearing mice were carried out in compliance with the Memorial Sloan-Kettering Institutional Animal Care and Use Committee guidelines. NeuroD2-SmoA1 mice were provided by Jim Olson (Fred Hutchinson Cancer Research Center). Patched+/− mice were provided by Kathryn Anderson (Memorial Sloan-Kettering Cancer Center).
Culture of CGNPs
CGNP cultures were generated as described previously (Kenney and Rowitch 2000). Cells were plated on individual poly-DL-ornithine (Sigma) precoated plates or precoated glass coverslips. Where indicated, Shh (R&D Systems) was used at a concentration of 3 μg/mL, lactacystin (Calbiochem) was used at 10 μM, leptomycin (Sigma) was used at 1 nM, cycloheximide (Sigma) was used at 10 μg/mL, and Cyclopamine (R&D Systems) was used at 1 μg/mL.
Retrovirus production and infection
The YAP1 cassette was cloned from pBabe-YAP1 (Addgene) into the retroviral vector pWzl-IRES-GFP. 293 EBNA (Invitrogen) packaging cells were cotransfected with gag-pol and VSVg packaging plasmids plus pWzl-YAP1-IRES-GFP, pIG-IRS1-IRES-GFP (Parathath et al. 2008) or control pWzl-eGFP, using Fugene 6 transfection reagent (Roche). The media was changed 12 h after transfection and supernatants (8 mL) were harvested at 24 and 48 h, filtered through 0.45-μm syringe filters, and pooled. For infection, Shh-treated CGNPs were exposed to the viral supernatants for 3 h. Viral supernatant was then removed and replaced with fresh medium ± Shh as indicated. Cells were cultured for 48 h post-infection.
shRNA lentiviruses
293T packaging cells were cotransfected with VSVg and Delta 8.9 packaging plasmids plus Mission shRNA lentiviral plasmids (Sigma) targeting YAP1 (TRCN0000095864, TRCN0000095865, TRCN0000095866, TRCN0000095867, and TRCN0000095868) and IRS1 (TRCN0000105880, TRCN0000105881, TRCN0000105882, TRCN0000105883, and TRCN0000105884). eGFP shRNA was used as control vector (SHC005V). Specificity of knockdown for each construct was confirmed as described previously (Parathath et al. 2008). Viral supernatants for each construct were pooled, such that cells would be infected with five different lentivirsues targeting YAP1 or IRS1. Shh-treated CGNPs were exposed to the viruses for 3 h, then treated with fresh medium ± Shh and culture for 36 h.
RNA extraction and real-time PCR
Total RNA from CGNPs, cell lines, or tissues was extracted and purified using the MiRvana kit (Ambion). cDNA was prepared from 1 μg of total RNA by using iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems). RNA expression data were acquired and analyzed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Average results and standard errors are presented.
Primer sequences used were as follows: GAPDH, 5′-TGGAAGGACTCATGACCACA-3′; GAPDHR, 5′-TTCAGCTCAGGGATGACCTT-3′; YAP1F, 5′-CAGGAATTATTTCGGCAGGA-3′; YAP1R, 5′-CATCCTGCTCCAGTGTAGGC-3′; TEAD1F, 5′-CTCAGGACGGGAAAGACAAG-3′; TEAD1R, 5′-TTCCTTCTGGCAAGAACCTG-3′; Gli2F, 5′-GCAGACTGCACCAAGGAGTA-3′; Gli2R, 5′-CGTGGATGTGTTCATTGTTGA-3′; Gli1F, 5′-TGGACAAGTGCAGGTAAAACC-3′; Gli1R, 5′-AATCCGGTGGAGTCAGACC-3′.
Protein preparation, immunoprecipitation, subcellular fractionation, and immunoblotting
For immunoblot analysis, cells were washed once in PBS and protein extracts were prepared as described previously (Kenney and Rowitch 2000). Subcellular fractionation was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) following the manufacturer's instructions. Protein content was determined by using the Bio-Rad protein assay. Fifty micrograms of each sample were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on 8% polyacrylamide gels and then transferred in 20% methanol buffer at 4°C to Immobilon polyvinylidene difluoride (Millipore) membranes. Membranes were blocked in 5% milk or 3% bovine serum albumin, and immunoblotting was carried out according to standard methods. Antibodies used for Western blotting were YAP1 (Abcam and Cell Signaling), phospho-YAP1 (Cell Signaling), TEAD1 (BD Transduction Laboratories), P-LATS1 (Cell Signaling), IRS1 (Cell Signaling), Cyclin D2 (Santa Cruz Biotechnologies), GAPDH (Cell Signaling), c-jun (Calbiochem), and β-tubulin (Sigma). Donkey anti-mouse HRP-linked secondary was from Jackson Research Laboratories, and goat anti-rabbit was from Thermo Scientific. Peroxidase activity was detected using Amershams's ECL reagents and exposing membranes to Kodak Biomax film.
For immunoprecipitation studies, 1 mg of protein extract was used in each case. Ten micrograms of antibody were incubated with protein A-sepharose for 2 h. Protein extracts were precleared with protein A-sepharose for 2 h, and then incubated with the antibody plus protein A-sepharose overnight. The precipitate was washed four times and proteins were eluted with 0.2 M glycine. Antibodies used for immunoprecipitation were IRS1 (Cell Signaling), YAP1 (Abcam), TEAD1 (BD Transduction Laboratories), CRM1 (Santa Cruz Biotechnologies), and mouse and rabbit IgG (Upstate Biotechnologies).
ChIP
TEAD1-binding sites found in 5 kb upstream of Gli2 gene were considered. ChIP was carried out with the ChIP assay kit (Millipore) according to the manufacturer's instructions. Chromatin was isolated from Pzp53 medulloblastoma cells and precipitated with YAP1 antibody (Santa Cruz Biotechnologies), control Histone 3 antibody (Abcam), and IgG isotype control (Upstate Biotechnologies). Primer sequences used for quantitative PCR measurement of immunoprecipitated promotor fragments were as follows: Binding site 1 (BS1) forward, 5′-AGCACGTAGCGCAGTAGACA-3′; BS1 reverse, 5′-GAAGGCAGGATTCCCTGTTA-3′; BS2 forward, 5′-CCTCTCCCTAAACTCCCACA-3′; BS2 reverse, 5′-TTGGCCATTTTGTCTCCTCT-3′; BS3 forward, 5′-CCCAGTGACAGACCTTTTCC-3′; BS3 reverse, 5′-AGTCTCATCCACTGCAATGCT-3′; BS4 forward, 5′-GGCTCGAAAGAGATGTGACC-3′; BS4 reverse, 5′-ATGCCTGAGGACGCTTAGAA-3′; No BS forward, 5′-CTGAGGCAGTCGAAGGAGAG-3′; No BS reverse, 5′-ACACTGGCTGCCAAAATGTA-3′. No Binding site primers were selected 10 kb upstream of Gli2 gene, in a region that does not contain TEAD1-binding sites. Threshold cycles (Ct) were determined for both immunoprecipitated DNA and a known amount of DNA from the input sample for different primer pairs. Fold enrichments were calculated by determining the immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and were normalized to the level observed at the control region with no binding sites.
Immunofluorescence
Frozen sections (10 μm) were dried and then boiled in 0.01 M citric acid for 15 min for antigen retrieval. For paraffin-embedded sections, tissues were first dewaxed and rehydrated prior to antigen retrieval. CGNPs were grown on poly-DL-ornithine-coated glass coverslips as described previously (Parathath et al. 2008). The cells were fixed with 4% paraformaldehyde for 20 min. Sections and cells were analyzed by immunoflorescence according to standard methods. Antibodies used for immunofluerescence were Ki67 (Vector Laboratories), IRS1 (Upstate Biotechnologies), YAP1 (Abcam and Cell Signaling), TEAD1 (BD Transduction Laboratories), CD31 (BD Transduction Laboratories), GFAP (Cell Signaling), CD15 (Abcam), Nestin (Covance), Cleaved Caspase 3 (Cell Signaling), and mouse and rabbit IgG (Upstate Biotechnologies).
Image capturing
Staining of cultured primary cells and tissue sections was visualized with a Leica DM5000B microscope and images were taken using Leica FW400 software. For quantification, TIFF images of four random fields were taken for each experimental group using the 20× objective, and average pixel intensities were measured using Volocity software.
Human tumor collection and expression analysis
Exon array profiling and data analysis with tumor subgrouping were performed as published (Northcott et al. 2009a).
Interphase FISH
Dual-color interphase FISH was performed on 6- to 8-μm FFPE tissue sections from archived samples of 67 human medulloblastomas (St. Jude Children's Research Hospital). Probes were derived from BAC clones (Invitrogen) and labeled with either FITC or rhodamine fluorochromes. BAC clones RP11-90M3 (YAP1, 11q22) and RP11-1012N20 (11p11.2) were chosen for probe construction. All probe mixtures were diluted 1:50 in DenHyb buffer (Insitus Biotechnologies) and codenatured with the target cells on a slide moat for 12 min at 90°C. Slides were incubated overnight at 37°C on a slide moat and then washed in 4 M urea/2× SSC for 2 min at 25°C. Nuclei were counterstained with DAPI (200 ng/mL; Vector Laboratories) for viewing on a Nikon Eclipse E800 fluorescence microscope equipped with a 100-W mercury lamp; FITC, Rhodamine, and DAPI filters; 100× PlanApo (1.40) oil objective; and a COHU CCD camera. Images were captured and processed with an exposure time ranging from 0.5 to 1.5 sec for each fluorochrome using Cytovision version 3.6 software.
Acknowledgments
We are grateful to Dolores Hambardzumyan, Rebecca Bish, Massimo Squatrito, Eric Holland, Claudio Alarcon, Alexia-Ileana Zaromytidou, Joan Massague, and Mike Overholtzer for providing advice and reagents. A.F.-L receives fellowship support from the Spanish Ministry of Education. These studies were supported by funding from the NIH (NINDS R01NS061070) and Alex's Lemonade Stand Foundation to A.M.K.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1824509.
Supplemental material is available at http://www.genesdev.org.
References
- Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65:7561–7567. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Buttitta LA, Edgar BA. How size is controlled: From Hippos to Yorkies. Nat Cell Biol. 2007;9:1225–1227. doi: 10.1038/ncb1107-1225. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes & Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Scott MP. A mouse model for medulloblastoma and basal cell nevus syndrome. J Neurooncol. 2001;53:307–318. doi: 10.1023/a:1012260318979. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, Church D, Hassan AB. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- Fults DW. Modeling medulloblastoma with genetically engineered mice. Neurosurg Focus. 2005;19:E7. doi: 10.3171/foc.2005.19.5.8. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Squatrito M, Holland EC. Radiation resistance and stem-like cells in brain tumors. Cancer Cell. 2006;10:454–456. doi: 10.1016/j.ccr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & Dev. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hermsen M, Alonso Guervos M, Meijer G, van Diest P, Suarez Nieto C, Marcos CA, Sampedro A. Chromosomal changes in relation to clinical outcome in larynx and pharynx squamous cell carcinoma. Cell Oncol. 2005;27:191–198. doi: 10.1155/2005/407216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I, Tsuda H, Hirasawa A, Miura M, Sakamoto M, Hirohashi S, Inazawa J. Expression of cIAP1, a target for 11q22 amplification, correlates with resistance of cervical cancers to radiotherapy. Cancer Res. 2002;62:4860–4866. [PubMed] [Google Scholar]
- Jenkin D, Greenberg M, Hoffman H, Hendrick B, Humphreys R, Vatter A. Brain tumors in children: Long-term survival after radiation treatment. Int J Radiat Oncol Biol Phys. 1995;31:445–451. doi: 10.1016/0360-3016(94)00393-Y. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmasheva RT, Houghton PJ. IGF-I mediated survival pathways in normal and malignant cells. Biochim Biophys Acta. 2006;1766:1–22. doi: 10.1016/j.bbcan.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Lambros MB, Fiegler H, Jones A, Gorman P, Roylance RR, Carter NP, Tomlinson IP. Analysis of ovarian cancer cell lines using array-based comparative genomic hybridization. J Pathol. 2005;205:29–40. doi: 10.1002/path.1681. [DOI] [PubMed] [Google Scholar]
- Marino S. Medulloblastoma: Developmental mechanisms out of control. Trends Mol Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Moroianu J. Distinct nuclear import and export pathways mediated by members of the karyopherin β family. J Cell Biochem. 1998;70:231–239. [PubMed] [Google Scholar]
- Newman PJ, Albelda SM. Cellular and molecular aspects of PECAM-1. Nouv Rev Fr Hematol. 1992;34:S9–S13. [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009a;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009b;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer RJ, Cogen P, Vezina G, Rorke LB. Medulloblastoma: Clinical and biologic aspects. Neuro-oncol. 1999;1:232–250. doi: 10.1215/15228517-1-3-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Parathath SR, Mainwaring LA, Fernandez LA, Campbell DO, Kenney AM. Insulin receptor substrate 1 is an effector of sonic hedgehog mitogenic signaling in cerebellar neural precursors. Development. 2008;135:3291–3300. doi: 10.1242/dev.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provias JP, Becker LE. Cellular and molecular pathology of medulloblastoma. J Neurooncol. 1996;29:35–43. doi: 10.1007/BF00165516. [DOI] [PubMed] [Google Scholar]
- Raffel C, Jenkins RB, Frederick L, Hebrink D, Alderete B, Fults DW, James CD. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57:842–845. [PubMed] [Google Scholar]
- Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger J, Wolter M, Weber RG, Megahed M, Ruzicka T, Lichter P, Reifenberger G. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain:Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- Weber RG, Sommer C, Albert FK, Kiessling M, Cremer T. Clinically distinct subgroups of glioblastoma multiforme studied by comparative genomic hybridization. Lab Invest. 1996;74:108–119. [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wetmore C. Sonic hedgehog in normal and neoplastic proliferation: Insight gained from human tumors and animal models. Curr Opin Genet Dev. 2003;13:34–42. doi: 10.1016/s0959-437x(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Wu A, Chen J, Baserga R. Nuclear insulin receptor substrate-1 activates promoters of cell cycle progression genes. Oncogene. 2008;27:397–403. doi: 10.1038/sj.onc.1210636. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Wechsler-Reya RJ. Hit ’em where they live: Targeting the cancer stem cell niche. Cancer Cell. 2007;11:3–5. doi: 10.1016/j.ccr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]