Summary
Background
Candida species are successful microbial pathogens originally thought to be asexual, but several are now recognized as sexual or parasexual. Candida albicans, the most common fungus infecting humans, is an obligate diploid with a parasexual cycle involving mating, recombination, and genome reduction but with no recognized meiosis. Others (Candida lusitaniae, Candida guilliermondii) are haploid and their mating produces spores, suggestive of complete meiotic sexual cycles. However, comparative genomic analysis reveals these species lack multiple key meiotic components, including the recombinase Dmc1 and co-factors (Mei5/Sae3), synaptonemal complex proteins (Zip1-Zip4/Hop1), and the crossover interference pathway (Msh4/5).
Results
Here we elucidated the structure and functions of the mating-type locus (MAT) and established C. lusitaniae undergoes meiosis during its sexual cycle. The MAT-encoded a2 HMG and α1 α-domain factors specify a- and α-cell identity, whereas the a1 homeodomain protein drives meiosis and sporulation and functions without its canonical heterodimeric partner, α2. Despite the apparent loss of meiotic genes, C. lusitaniae undergoes meiosis during sexual reproduction involving diploid intermediates, frequent SPO11-dependent recombination, and whole genome reduction generating haploid progeny. The majority of meiotic progeny are euploid, but approximately one-third are diploid/aneuploid and likely arise via precocious sister chromatid segregation and meiotic nondisjunction.
Conclusions
In summary, the cell identity and meiotic pathways have been substantially rewired, and meiotic generation of both recombinant and aneuploidy progeny may expand genetic diversity. These findings inform our understanding of sexual reproduction in pathogenic microbes and the evolutionary plasticity of the meiotic machinery, with implications for the sexual nature of Candida albicans and the generation and consequences of aneuploidy in biology and medicine.
Introduction
Sexual reproduction is pervasive throughout biology and despite associated costs, serves to enable genetic reassortment and thereby evolution. The Fungal Kingdom provides a unique vantage point from which to explore mechanisms and transitions in modes of sexual reproduction. Here we capitalize upon recent comparative genomics resources to examine the evolutionary trajectory of sex determination and sexual reproduction in a clade of emerging yeast pathogens that infect humans.
Candida spp. commonly infect humans to cause both mucocutaneous and life-threatening systemic infections. The majority of Candida species define a monophyletic clade whose evolution was punctuated by a genetic code reconfiguration ~100 mya. Historically, all Candida species were thought to be asexual, yet several species are now known to mate and some even produce spores. The most common pathogen, C. albicans, mates and undergoes a parasexual cycle but meiosis and sporulation have never been observed, possibly consistent with a loss of meiotic machinery [1-6]. The emerging pathogen C. lusitaniae has a defined sexual cycle presumed to include meiosis based on its ability to produce spores [7-12]. To gain insight into parasexual, sexual, or asexual life cycles, multiple members of the genus Candida were sequenced at the Broad Institute [13, 14]. In contrast to the hypothesis that sexual species might retain meiotic homologs lost in parasexual species, the sexual species lack the same meiotic homologs as C. albicans and, in addition, have lost dozens of additional key meiotic genes [15]. This finding raised the question of whether meiosis occurs during sexual reproduction of any of these species.
We report here a characterization of the mating-type (MAT) locus of C. guilliermondii and C. lusitaniae, and demonstrate that although the organization of MAT is similar to C. albicans transcriptional rewiring has occurred. Additionally, despite the apparent loss of meiotic genes, C. lusitaniae undergoes a meiotic sexual cycle with Spo11-dependent recombination. Interestingly, although the majority of progeny produced are euploid, approximately 1/3 are aneuploid and could have important roles in the generation of genetic diversity within the population [16].
Results
Characterization of the Mating-type Locus of C. lusitaniae and C. guilliermondii
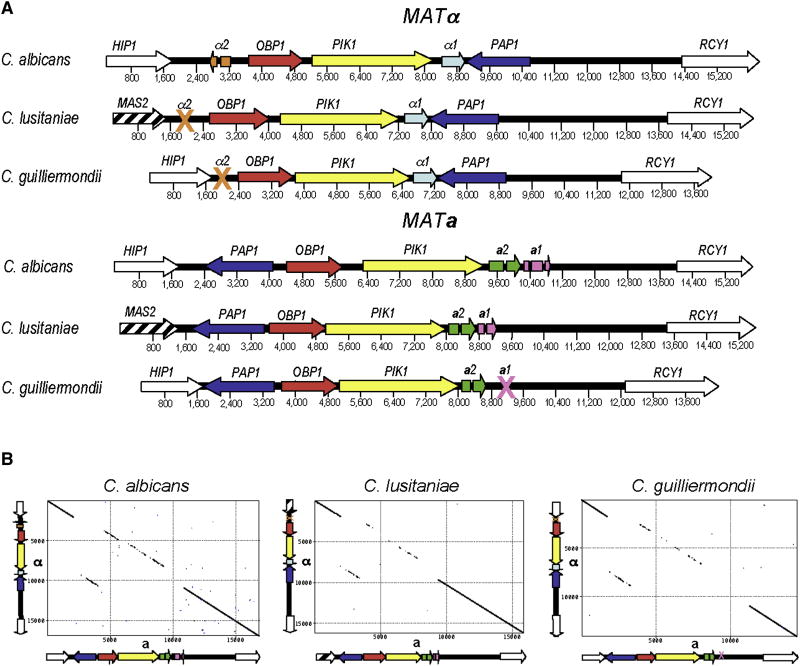
First, the mating-type (MAT) locus of C. lusitaniae and C. guilliermondii was identified. In fungi, the MAT locus is a specialized region of the genome that confers sexual identity, specifying cells as α, a, or α/a. Both species are heterothallic (outcrossing) with two opposite mating-types. MAT was defined by BLAST searches of the C. lusitaniae and C. guilliermondii genomes [13, 14] using genes of the C. albicans MTL locus encoding the α-domain factor α1, polyA polymerase Pap1α, phosphatidyl inositol-4 kinase Pik1α, and oxysterol binding protein Obp1α. The C. lusitaniae sequenced strain is MATα and homologs of four of five genes corresponding to the C. albicans MTLα locus were identified, organized into a contiguous syntenic locus spanning 7741 bp (Fig. 1). Notably, the α2 gene, which encodes a homeodomain cell identity factor, was missing. The C. guilliermondii sequenced strain contains a 9534 bp MAT a allele with four genes that share identity and synteny with four of the five C. albicans MTL a genes (a2 HMG domain factor, PAP1 a, OBP1 a,PIK1 a) (Fig. 1). Notably, the a1 gene, which also encodes a homeodomain factor, was missing. Synteny between MAT and flanking regions is largely conserved, except on the 5’ C. lusitaniae MAT border where a translocation occurred (Fig. 1). The PAP1, OBP1, and PIK1 genes are conserved and syntenic in the a and α MAT alleles of all three species, indicating they were acquired prior to their shared last common ancestor, and this organization has been preserved through considerable evolutionary divergence (see supplemental discussion).
Figure 1. Mating type (MAT) locus of C. lusitaniae and C. guilliermondii are syntenic with C. albicans.
a, Alignment of the MAT loci from the sexual species C. lusitaniae and C. guilliermondii with that of the parasexual species C. albicans. The colored genes are contained within the MAT locus (PAP1, OBP1, PIK1,a1, a2, and α1), while the white/striped genes (HIP1, MAS2, and RCY1) flank the locus. The intron and exon structure of the C. guilliermondii and C. lusitaniae a1 and a2 genes are shown. Crosses denote the loss of either the a1 or α2 transcription factor genes. The C. lusitaniae MATα allele is 7741bp and the MAT a allele is 7389 bp. The C. guilliermondii MATα allele is 9228 bp and the MAT a allele is 9534 bp. An independent C. guilliermondii MAT a allele was sequenced and found to be identical in gene content to the Broad sequence strain. b, Pustell DNA scoring matrices (created with MacVector®) demonstrating the gene rearrangement and sequence divergence between the MAT locus alleles of a single species. Gene identity flanking the locus is 97 – 99%, while genes within the locus share 60 – 80% identity. The MATα allele is plotted on the y-axis and the MAT a allele is plotted on the x-axis for each species.
The opposite MAT alleles were isolated from fertile strains and sequenced (Fig. 1). Interestingly, the MATα alleles both lack the α2 homeodomain gene. C. guilliermondii MAT a also lacks the a1 homeodomain gene whereas this gene is retained and expressed from C. lusitaniae MAT a based on RACE analysis. In S. cerevisiae and C. albicans a1 and α2 form a heterodimer that represses haploid specific genes, and in S. cerevisiae a1/α2 also promotes meiotic entry [5]. The absence of one or both components of this canonical heterodimeric cell identity determinant indicates transcriptional circuits controlling meiotic entry and haploid-specific gene repression were either lost or reconfigured as these sexual species diverged from the last common ancestor, possibly reflecting a transient nature of their diploid state or loss or altered meiosis.
The transcription factors α1 and a2 are required for mating and cell-fate identity, while a1 is required for sporulation
C. lusitaniae a and α cells mate when co-cultured and nutrient limited. Mating involves pheromone production and sensing to trigger conjugation tube formation, cell and nuclear fusion, and the formation of spore-containing asci [7, 8, 12] (Supplemental Fig. S1). Functional roles of the a2, α1, and a1 transcription factors were assigned by genetic and phenotypic analyses of independent disruption mutants isolated for each gene. Mating was assessed by microscopy, assays that monitor complementation and segregation of mutations during mating, and gene expression studies.
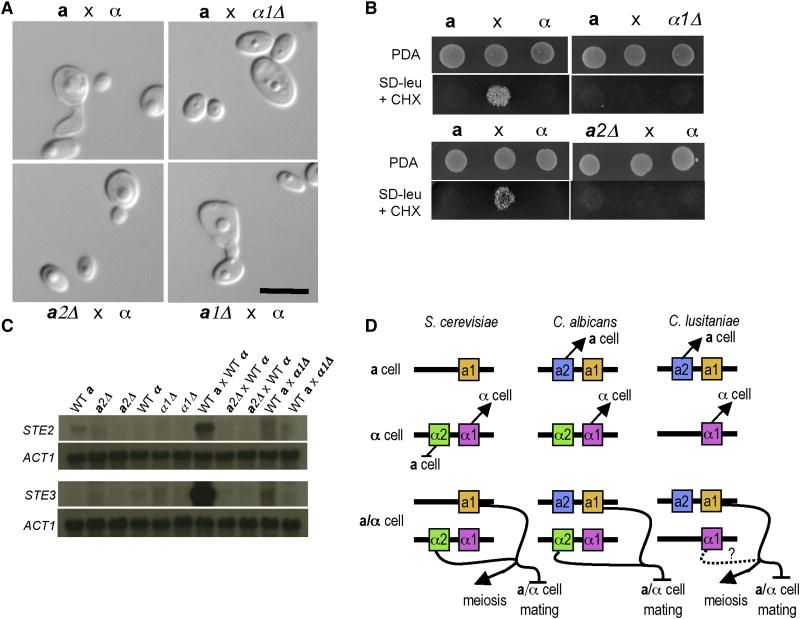
Disruption of the α1 or a2 genes abolished mating, and no mating structures (conjugation tubes, zygotes, asci, or spores) were observed (Fig. 2a). In patch and quantitative mating assays wild-type a × α crosses yielded abundant haploid recombinant products (~14%), whereas the efficiency was dramatically reduced (>400,000-fold) in a2 or α1 mutant crosses (Fig. 2b). Moreover, expression of the STE2 gene (encoding the α-pheromone receptor) is weakly induced by nutrient limitation in an a2-dependent fashion, and co-incubation with an opposite mating-type partner leads to marked induction of STE2 and STE3 in wild-type cells, but not in α1 (STE3) or a2 mutant cells (STE2) (Fig. 2c, and Supplemental Fig. S2). Therefore, a2 and α1 control cell-type specific gene expression and specify a- or α-cell fate, analogous to C. albicans (Fig. 2d). In contrast, a1 mutants mated normally to produce diploid cell-cell fusion products, but failed to sporulate (Supplemental Fig. S3). Interestingly, both a1 and an α-specific component are necessary for sporulation: diploid a/α strains sporulate whereas a/a, or α/α diploid strains do not (Supplemental Fig. S4, data not shown). This reveals a novel, α2-independent role for a1 in C. lusitaniae, in contrast to S. cerevisiae in which a1 only functions with α2 [5].
Figure 2. α1 and a2 are required for mating and control of cell identity.
a, Representative images of C. lusitaniae matings at 24 hours showing a conjugated cell pair in the wild-type mating. In crosses with α1 or a2 mutants, cells continue to bud as though an opposite mating type partner were not present, and no conjugating cells or zygotes were observed. Scale bar, 10 μm. b, Plate matings with each parent alone flanking a mating mixture in the center. After 96 hours, the matings were replica plated to media to select for recombinant progeny. No recombinant progeny were formed in either mutant mating. c, Northern blot showing the expression of the C. lusitaniae STE3 homolog (a pheromone receptor) and STE2 homolog (α pheromone receptor). All strains were grown on dilute PDA for 24 hours prior to harvesting and RNA purification. Two independent a2Δ and α1Δ mutants are shown. The expression of both pheromone receptors is induced during mating, however minimal induction is seen in a2Δ × WTα or WTa × α1Δ matings. d, Model for the control of haploid specific genes and cell identity. In both C. albicans and C. lusitaniae a2 controls a cell identity, the default cell-state in S. cerevisiae. In all three species, α1 controls alpha cell identity, and in S. cerevisiae α2 functions to repress a cell specific genes. In both S. cerevisiae and C. albicans the a1/α2 heterodimer functions to repress haploid specific genes and additionally to promote entry into meiosis in S. cerevisiae. In C. lusitaniae, a1 is necessary for sporulation and meiosis, but not conjugation and lacks the canonical partner α2. Heterozygosity at the MAT locus is necessary for sporulation, indicating a MATα component, such as α1, also promotes sporulation and meiosis. Panel D adapted from Tsong et al.[30].
Despite lacking key meiotic genes, C. lusitaniae possesses a meiotic cycle
Additional analyses completed during the Broad Institute Candida species comparative genomics project raised further questions regarding whether meiosis could occur in C. lusitaniae. C. albicans is thought to lack several important meiotic genes, and this could explain why C. albicans undergoes a parasexual cycle with no recognized meiosis [1, 17, 18]. However, comparative genomic analysis revealed the sexual species C. lusitaniae and C. guilliermondii also lack the same meiotic gene homologs missing in C. albicans and, in addition, also lack clear homologs of numerous additional key meiotic components [15]. The presumption that C. lusitaniae undergoes meiosis has been based on the ability of this species to mate and form spores. Marker assortment observed in previous genetic crosses could have resulted from independent chromosomal segregation rather than meiotic recombination, as these loci lie on different chromosomes [12] . Additionally, C. lusitaniae generally produces asci with only two rather than four spores (dyads) [9]. Mutations or altered media conditions leading to dyad production in of S. cerevisiae are known [19, 20], however meiosis typically yields four products.
To establish whether C. lusitaniae undergoes meiosis, a restriction fragment length polymorphism (RFLP) map was developed for three of the eight chromosomes (1, 5, and 6 (MAT)) and used to determine recombination frequencies following genetic crosses. An F1 progeny set was generated by crossing α leu2 chxR and a ura3 strains on potato dextrose agar, and 94 progeny were selected on YNB media that supports growth of progeny, but not of either parental strain. Isolated DNA was subject to RFLP analysis. The majority of the progeny (67/94, or ~70%) mated as a or α and inherited only one parental allele at each locus. A high level of recombination was apparent (Table 1). Based upon recombination frequencies calculated for each region, the average centiMorgan values (1 cM= 1% recombination) were 4 to 16 kb/cM (Chr 1: 8.85 - 15.7 kb/cM, Chr 5: 3.89 - 4.78 kb/cM, and Chr 6: 3.92 - 7.63 kb/cM), well within the range of known meiotic recombination frequencies for other fungi, including S. cerevisiae (~2.7 kb/cM), Cryptococcus neoformans (~13.2 kb/cM), and Aspergillus nidulans (~43 kb/cM). Thus, recombination occurs frequently during C. lusitaniae sexual reproduction, consistent with meiosis. Analysis of a second F1 progeny set yielded similar results (data not shown).
Table 1.
RFLP typing reveals frequent recombination in F1 progeny
| JLR610CL16 | F1 Progeny | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (P1) | (P2) | |||||||||||||||||
| 1 | 3 | 6 | 11 | 12 | 30 | 55 | 58 | 19 | 7 | 5 | 33 | |||||||
| H | H | H | H | H | H | H | H | H | H | H | H | D | D | |||||
| CHRa | Distb | chxR | chxS | chxR | chxR | chxR | chxR | chxR | chxS | chxS | chxS | chxR | chxR | chxS | chxS | % Recf | kb / cMg | % spo11h |
| 1 | 400 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 |  |
|||
| 1 | 500 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | ||||
| 1 | 600 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 2 | ||||
| 1 | 1106c | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | ||||
| 5 | 500 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 2 |  |
|||
| 5 | 600 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2 | ||||
| 5 | 629d | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ||||
| 6 | 13 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 2 | 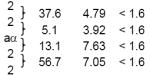 |
|||
| 6 | 193 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | ||||
| 6 | MATe | α | a | a | a | a | a | α | α | a | α | aα | a | α | ||||
| 6 | 303c | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 1 | ||||
| 6 | 703 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | ||||
Parental strains JLR610 (P1) MATα leu2 chxR and CL16 (P2) MAT a ura3 were co-cultured on dilute PDA for 96 hours and recombinant prototrophic progeny were selected on YNB medium. Shown above is a random sampling of progeny. Strains were FACS analyzed and found to be either haploid (H) or diploid (D) as indicated. Sensitivity to cycloheximide (chx) was also tested and strains were scored as either resistant (chxR) or sensitive (chxS). Shading indicates regions of recombination. See Figure 4 for all 94 F1 progeny data.
CHR, chromosome number
Dist, chromosomal position of locus. Distance in number of kb from the beginning of the appropriate chromosomal contig sequence provided at the C. lusitaniae genome database.
putative centromere proximal markers. Chromosome 1, 1106 kb is 40 kb from the putative centromere, Chromosome 6, 303 kb is 20 kb from the putative centromere.
URA3 locus, since all progeny analyzed are prototrophic, all inherited the URA3 locus from parental strain JLR610 (P1).
MAT locus, located 203 kb from the left telomere/ contig end.
% Rec, frequency of recombination between the two indicated loci.
kb/cM, average kb per centiMorgan (1cM= 1% recombination) calculated from the frequency of recombination between the indicated loci.
% spo11, frequency of recombination in spo11 × spo11 crosses. Progeny were typed for every marker on chromosome 6 to determine recombination frequencies, but only one marker each for chromosomes 1 and 5 to establish independent assortment of chromosomes.
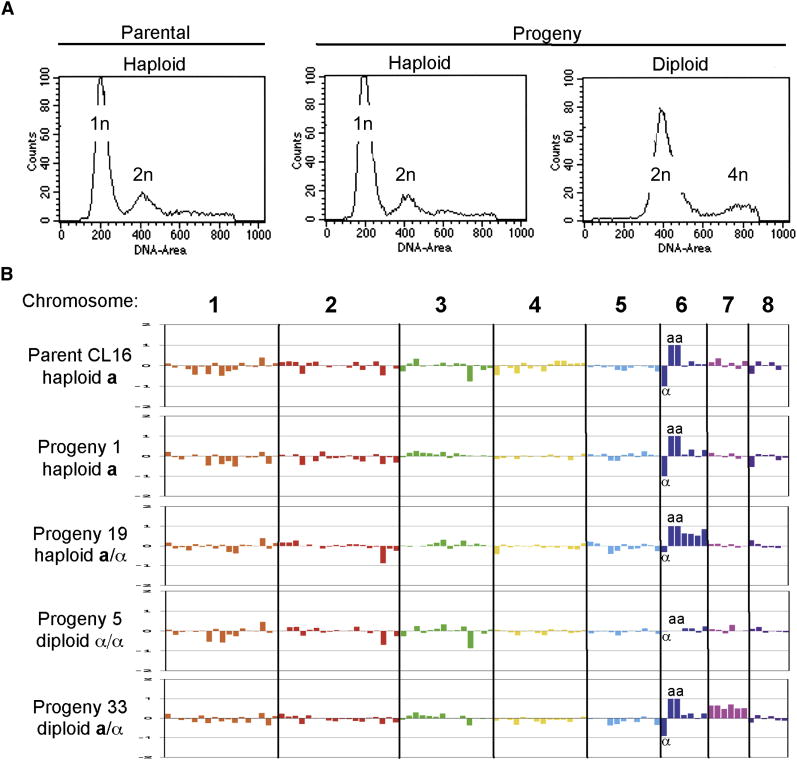
Further analysis provides evidence these progeny are euploid, haploid isolates. First, FACS analysis of representative isolates revealed haploid (or near haploid) DNA content (Fig. 3a). Second, because RFLP mapping provides limited information about a subset of chromosomes, a low-density 70-mer array was generated containing 96 probes spanning all 8 chromosomes for comparative genome hybridization analysis (CGH). By CGH, the parental a and α strains are euploid and 6 of 67 presumptive haploid isolates were also found to be euploid, containing one copy of each of the eight parental chromosomes (Fig. 3b and Supplemental Fig. S6). However, some of the presumed haploid isolates that were not analyzed by CGH, may be aneuploid for chromosomes which were not tested for by RFLP. Thus, during sexual reproduction haploid strains fuse to produce a transient diploid zygote intermediate, and complete genome reduction then occurs producing haploid recombinant spores.
Figure 3. Ploidy and CGH analysis reveal euploid, aneuploid, and diploid progeny.
a, Representative FACS plots of parental and progeny strains. The FACS plot of parental haploid α strain JLR610 is shown. Of the 27 progeny typing as a and α for at least one RFLP loci, 6 strains were haploid by FACS (representative strain, progeny p2 shown), and 21 were diploid by FACS analysis (representative strain, progeny p33 shown). b, CGH analysis plots of log2 ratio of medians for each experimental strain versus parental strain JLR610 for which the array was designed. The 8 chromosomes are plotted in order and separated by thick black lines. For the mating-type specific primers, if only one strain hybridized to a particular probe the log2 ratio was set to -1 (α1) or 1 (a1 or a2). Traditional CGH analysis plots a scanning window average of several genes, however due to the low-density nature of this array each probe is plotted individually. Aneuploid strains were apparent based on an increased log2 ratio for all probes along an entire chromosome. Shown from top to bottom are parental strain CL16, haploid progeny p1, aneuploid progeny p19, diploid progeny p5, and diploid (2N+1) progeny p33. α denotes the α1 probe, and aa donates the a1 and a2 probes.
Spo11 is required for recombination during the C. lusitaniae sexual cycle
Mutation of SPO11 impaired sporulation and abolished recombination, further supporting the hypothesis that meiotic recombination occurs during C. lusitaniae sexual reproduction. SPO11 encodes a meiosis-specific topoisomerase homolog highly conserved throughout eukaryotes that inflicts DNA double strand breaks to provoke meiotic recombination [21, 22]. In bilateral spo11 × spo11 mutant crosses, the production of spores and meiotic progeny was considerably reduced and no recombination was observed in germinated spore products compared with wild-type (a × α) (Table 1) or wild-type × spo11 unilateral crosses (date not shown). Wild-type × wild-type crosses yielded ~4 to 16% spores, whereas spo11 × spo11 strains had an ~15- to 80-fold reduction (0.23 to 0.31%). This analysis was repeated with an independently derived spo11 × spo11 mutant pair with similar results (data not shown).
Three independent lines of evidence support the conclusion that meiosis occurs during C. lusitaniae sexual reproduction: 1) recombination is frequent, on par with meiosis in other organisms, 2) recombination is Spo11-dependent, and 3) 70% of F1 progeny are euploid and haploid (based on RFLP, FACS, CGH) and therefore complete genome duplication and reduction occurs.
C. lusitaniae meiosis is unfaithful resulting in a high frequency of aneuploid progeny
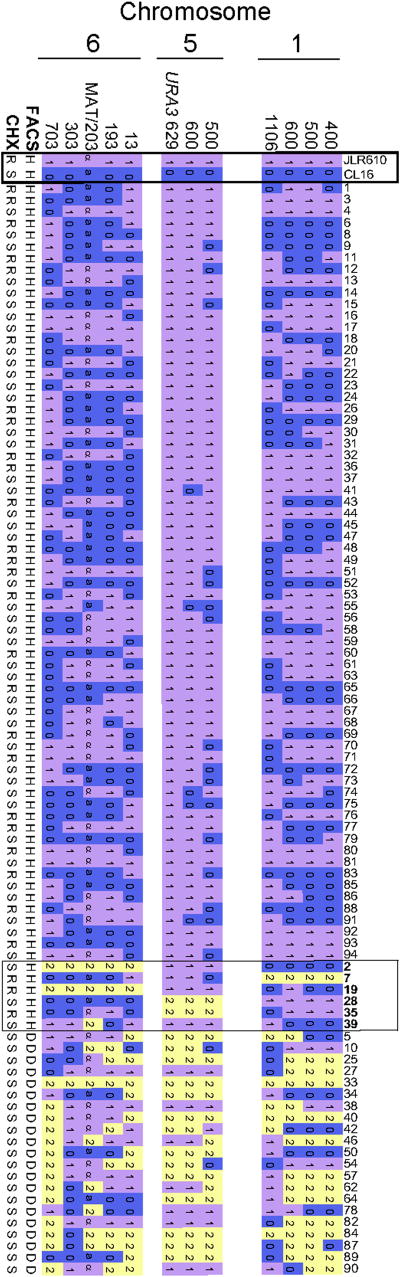
Although ~70% of the F1 progeny are haploid, the remaining progeny (27/94, or ~30%) inherited both parental alleles at one or more loci. By FACS analysis, one-fourth are haploid/aneuploid (6/27), and three-fourths are diploid (21/27) (Fig. 3a). The majority of haploid/aneuploid progeny (except progeny 39) could contain one or a few extra chromosomes (i.e. 1N+1) as all markers typed as heterozygous along one of the three chromosomes analyzed (Fig. 4). Based on RFLP or CGH analysis, 1N+1 aneuploid progeny containing an extra copy of chromosomes 1, 5, or 6 were observed (Table 1, Fig. 4, Fig. 3b). These aneuploid progeny could arise via concerted chromosome loss by a parasexual process from a diploid intermediate, or from precocious sister chromatid segregation or meiotic nondisjunction. Given that the haploid majority of progeny were produced by meiosis, it seems less likely a concomitant parasexual process occurs, but infrequent parasexual reduction cannot be definitively excluded. Recombination frequency in these aneuploid segregants was similar to isolates that had undergone a complete genome reduction, consistent with meiotic reduction. Moreover, the aneuploid progeny observed were all 1N+1, and no examples of higher order aneuploidy (other than diploid) were observed, consistent with concerted meiotic loss of 7 of 8 parental chromosomes via meiosis rather than a parasexual cycle. Finally, analysis of chromosomal configuration in 1N+1 aneuploid isolates revealed heterozygosity at putative centromere-proximal markers on Ch. 1 and 6, which could be consistent with either precocious sister segregation or meiosis I nondisjunction(Fig. 4).
Figure 4. Complete RFLP typing of CL16 × JLR610 F1 progeny.
All strains were typed at each RFLP locus as possessing the allele of the α parent (JLR610) (1) or the allele of the a parent (CL16) (0), or both (2). The parental strains are indicated by a bold box on the left. The first 67 progeny are haploid by FACS analysis, the next 6 enclosed within the box were haploid (H) by FACS and found to be aneuploid, and the last 21 strains were diploid (D) by FACS analysis. The cycloheximide (CHX) resistance of all strains was also tested and strain were scored as either sensitive (S) or resistant (R). Chromosome 1, marker 1106 is ~40 kb from the putative centromere, and chromosome 6, marker 303 is ~20 kb from the putative centromere. The paucity of (0) alleles seen on chromosome 5 is due to the linkage of these markers to the URA3 allele, which was selected for in the initial cross. Thus all progeny were selected to inherit at least one copy of URA3 from the α parent. The configuration of the diploid/ aneuploid strains at the URA3 locus was evaluated by growth of strains on both SD-ura and 5-FOA medium. All strains were capable of growth on SD-ura medium and thus contained at least one copy of the URA3 allele as expected. However, some diploid/ aneuploid strains readily papillated on 5-FOA media (converting to a ura- state) suggesting that these strains contained a copy of the ura3 gene from the a parent and could easily lose the wild-type URA3 allele becoming 5-FOA resistant. Thus, the diploid/ aneuploid strains that readily papillated on 5-FOA were scored as 2 at this locus.
The final progeny class (21/94, 22%) are diploid, based on RFLP, FACS, and CGH analysis (Table 1, Fig. 3, Fig. 4). Mating could produce diploids via several routes. First, cell-cell fusion during mating produces diploid zygotes that might be recovered before meiosis and sporulation occur. Second, diploids could be products of meiosis involving only one reductive division, similar to S. cerevisiae spo12,13 mutants (Supplemental discussion). Third, diploids could be products of F1 spore-spore matings. Seven diploid Spo+ progeny (capable of sporulation in the absence of mating) are a/α MAT heterozygous but homozygous at some other loci and thus could have resulted from either meiosis or subsequent inter-progeny mating of haploid meiotic products. The remaining 14 diploid progeny are Spo- (unable to sporulate in the absence of mating) MAT homozygous (a/a, α/α), and thus unlikely the result of a-α progeny spore matings, suggesting these are direct products of meiosis. All 14 appear to have undergone meiotic recombination based on the observation of homozygous and heterozygous parental marker configurations (Table 1, Fig. 4). Because all 14 are homozygous at the chromosome 6 putative centromere-proximal marker, a parsimonious explanation could be that they result from meiosis II non-disjunction or a failure to undergo meiosis II, but clearly further studies will be required. Both diploid and haploid F1 progeny have been recovered from micromanipulated asci/spores, providing further evidence that some meiotic products are indeed diploid (Supplemental discussion).
Discussion
We conclude that meiosis occurs during sexual reproduction of C. lusitaniae, despite the absence of many key meiotic components. Even with a limited meiotic component repertoire, ~70% euploid, haploid progeny are (faithfully) produced. Previous studies document that haploid parental strains mate to produce diploid zygote intermediates in which DNA replication occurs on mating/sporulation media, based on appearance of 2N/4N ploidy by FACS [12]. Incubation of the a/α diploid progeny strain p33 on PDA media containing hydroxyurea blocked sporulation, providing additional evidence that replication precedes meiosis and sporulation (Supplemental Fig. S5). Our studies provide evidence that Spo11 acts to provoke meiotic DNA recombination. The absence of Dmc1 and its conserved co-factors (Mei5/Sae3) suggests DNA DSB repair is mediated by Rad51-dependent mechanisms, similar to U. maydis, C. elegans, and Drosophila [23-25]. Genome reduction to euploid haploid progeny occurs faithfully in the majority of progeny in the absence of conserved synaptonemal complex proteins, possibly analogous to S. pombe in which LinE elements promote chromosomal pairing and segregation [26, 27]. At the same time, sexual reproduction of C. lusitaniae produces a surprisingly high level of aneuploid (~6%) and diploid (~22%) progeny via a spectrum of meiotic errors, possibly as a consequence of the loss of conserved meiotic components. These aneuploid/diploid isolates can be stably propagated on rich media (YPD), and analysis of F1 progeny (1N, 1N+1, 2N) has not revealed obvious differences in susceptibility to fluconazole or amphotericin B.
Taken together, our findings challenge the notion that genomic inspection alone can provide insights into whether species are capable of undergoing meiosis, and illustrate the mechanistic and evolutionary plasticity of this process central to genetic exchange and sexual reproduction. The level of aneuploidy produced during C. lusitaniae mating is similar to that observed during human oogenesis (estimates range from 7 – 35%), which leads to miscarriage and birth defects associated with trisomy, and in tumor cells leading to metastasis [28]. Aneuploidy is deleterious in these situations, however the effect of aneuploidy in C. lusitaniae are less clear. Aneuploidy could generate diversity that is beneficial, deleterious, or neutral, acting over short or long evolutionary time spans. Aneuploidy in S. cerevisiae triggers common phenotypic/genotypic responses [16], which may be adaptive, or maladaptive, depending on environment.
Our findings have implications on the sexual nature of C. albicans and other microbial pathogens. Frequent aneuploid progeny recovered from C. albicans matings has fueled hypotheses that this process is parasexual and lacks meiosis [1, 2, 5]. However, in light of the findings presented here, the life cycle of C. albicans invites further scrutiny. Given that C. lusitaniae undergoes meiosis yet lacks more than a dozen key meiotic genes retained in C. albicans, and that aneuploidy is frequently generated during sexual reproduction, C. albicans sex might involve an unrecognized form of meiosis. Spo11-dependent recombination does occur during the tetraploid-diploid reduction [2], suggesting the C. albicans parasexual cycle might turn out to be sexual. Finally, there are parallels with the recently discovered genetic exchange akin to sex in the protozoan parasite Giardia [29]. During encystment, two paired diploid nuclei fuse (without cell fusion), key meiotic homologs (Spo11, Dmc1) are expressed, and genetic exchange and genome reduction occurs [29], illustrating the plasticity of sexual reproduction in microbial pathogens, and its potential impact on virulence and population structures beyond the fungi.
Experimental Procedures
Genes contained within and immediately flanking the C. albicans mating type locus MTL were used to search the C. lusitaniae and C. guilliermondii genome databases (Broad Institute) to identify the MAT locus of the sequenced isolate. The MAT allele from a fertile strain of the opposite mating type from the genome database strain was isolated by a degenerate PCR approach, sequenced, and annotated. Gene disruption cassettes were generated by overlap PCR and consisted of either the C. albicans SAT1 gene or the URA3 gene flanked by 800 to 1000 bp of homology surrounding the gene of interest. Strains were transformed by electroporation, and transformants were screened by PCR and Southern hybridization. For mating analysis strains were co-cultured on dilute potato dextrose agar (dilute PDA) at 25°C for the indicated time. Ninety-four progeny from a genetic cross between strain CL16 a ura3 and JLR610 α leu2 chxR were typed using allele specific primers to determine MAT configuration and were further analyzed by RFLP for markers on chromosomes 1, 5, and 6. All progeny were additionally analyzed for cycloheximide resistance, mating/sporulation ability, and FACS to determine DNA ploidy. A low density CGH array, consisting of 96 primers, spanning all 8 chromosomes of C. lusitaniae was used to confirm the euploid nature of the parental strains, and progeny from each of three subgroups (FACS haploid, euploid by RFLP; FACS haploid, aneuploid by RFLP, and FACS diploid) were competitively hybridized against array reference strain JLR610 to determine genome configuration. For detailed methods see Supplemental Methods.
Strains
All strains are described in Supplemental Table S2 and are derived from strain ATCC 38533, an environmental isolate (MAT a), or strain ATCC 42720 (MATα, a clinical isolate (Broad sequence strain). Two or three independent disruption mutants of the a1 (JLR806, JLR808, JLR809), a2 (JLR760, JLR763), and α1 (JLR741, JLR755) transcription factor genes were generated and phenotypically analyzed. Three independent spo11 strains were constructed, 2 in the MATα background (JLR843, JLR834), and one in the MAT a background (JLR805). Strains were confirmed by PCR and Southern blot analysis. Standard practice in the field for analyzing gene disruptions includes either comparing multiple independent disruption strains or complementation of disruptions by reintroduction of a wild-type copy of the gene of interest. Due to the current paucity of selectable markers for genetic analysis, we chose to use the former approach of isolating and characterizing multiple independent mutants to validate disruption phenotypes. For details of strain construction, see Supplemental Methods. All strains were routinely grown on YPD (1% yeast extract, 2% Bacto Peptone, 2% glucose, 2% BactoAgar (Difco)) medium at 30°C unless otherwise indicated.
Annotation and isolation of MAT loci
Genes from the characterized C. albicans MTL locus were used in BLAST searches of the C. lusitaniae and C. guilliermondii genome databases (Broad Institute) and homologs were identified based on best reciprocal BLAST matches. Mating-competent strains of C. lusitaniae (strain CL143) and C. guilliermondii (NRRL Y-2075) were used to isolate the opposite MAT allele. To isolate the opposite MAT allele exact primers were designed to the MAT flanking genes HIP1/MAS2 and RCY1, while degenerate primers were designed for PAP1. A fragment of PAP1 was amplified for both species using degenerate primers. For C. guilliermondii, exact primers to PAP1 were then used in combination with primers to HIP1 and RCY1 to generate overlapping PCR fragments spanning the MAT locus. The C. lusitaniae MAT a allele was further amplified in three overlapping segments. The first fragment was amplified using exact primers to PAP1 and MAS2, the second using an exact primer to PAP1 and a degenerate primer to PIK1. Following sequencing of the second region, exact primers were designed to OBP1 and used in combination with RCY1 specific primers to amplify the right border of the MAT a locus. Multiple independent PCR fragments were sequenced to generate at least 3X coverage across the MAT locus. PAP1, PIK1, and OBP1 were annotated based upon homology and ORF annotation. In addition, a second C. guilliermondii MAT a allele was sequenced from strain NRRL-Y2076 and found to be similar in gene content and structure to the Broad sequence strain. The structures of the α1, a2, and a1 genes were determined by 5’ and 3’ RACE (GeneRacer™; Invitrogen) to determine the intronic structure, and translational start and stop sites for these genes. Primers are listed in Supplemental Table S4.
Matings/ Genetic Crosses
Strains of opposite mating type were co-cultured on dilute Potato Dextrose Agar (1:10 dilution PDA, 14.5 g Bactoagar/1L (Difco)) at room temperature. Overnight cultures were grown at 30°C, washed with PBS, and equivalent cell counts were mixed together and then spotted onto dilute PDA media. All mating progeny were selected on media (detailed below) which supports the growth of either recombinant mating progeny or parental fusion products, but not either parental strain. Unlike S. cerevisiae there is no recognizable stable diploid fusion intermediate that can be isolated from mating reactions, rather once cells of opposite mating type conjugate, they appear to proceed directly into meiosis. For the isolation of individual recombinant progeny, cells were scraped from plates, resuspended in water, serially diluted and replated onto selective media, either Yeast Nitrogen Base (YNB; DIFCO) or Synthetic Dextrose lacking leucine (SD-leu)) with 10 μg/ml cycloheximide (Sigma). To quantitate efficiency of mating, after 72 hours co-incubation on dilute PDA, matings were serially diluted to selective media and efficiency of mating was calculated [31]. Briefly, each mating was plated onto media to differentially select for the parental strains. For instance, in a URA3 leu2 × ura3 LEU2, the matings were plated onto SD-ura and SD-leu to estimate the number of each parental strain in the mating mixture (these numbers will also include some progeny that are prototrophic for leucine or uracil as well). Matings were also plated to media that would only support the growth of recombinant URA3 LEU2 progeny, but not either parental strain (YNB). The number of recombinant progeny were counted and multiplied by 4 since we were selecting for only one-fourth of the progeny based upon independent assortment of chromosomes (the possible progeny genotypes are URA3 leu2, URA3 LEU2, ura3 LEU2, or ura3 leu2). Thus, to calculate the efficiency of mating and progeny formation the following formula was used: (4 × # recombinant progeny)/ (limiting parent + recombinants). Some recombinants will be counted along with the parents in the denominator, however these will only account for 1 to 4% of the total number.
Progeny Mapping
An RFLP map was generated by sequencing multiple 3 kb regions spaced across chromosomes 1, 5, and 6 (Primers are listed in Supplemental Table 3). Target regions were amplified by PCR from strains CL16 and JLR610, and three independent PCR reactions were used to generate at least 3X sequence coverage for each region and restriction maps were generated using MacVector® software. DNA was isolated from single colonies of each progeny strain. To determine the MAT configuration, a common primer that anneals to both MAT alleles was used in conjunction with a MAT a or MATα specific primer. For RFLP typing, each locus was PCR amplified, digested, and identified as deriving from either parent a (0), parent α(1), or both a/α (2). The progeny set was generated by mating strains CL16 × JLR610 on dilute PDA for 96 hours, resuspending the mating mixture in water, and plating various dilutions onto YNB media to select for recombinant prototrophic progeny. 94 independent colonies were selected and single colony purified prior to subsequent analysis. Recombination frequencies were calculated for each pair of loci. An independent mating was performed similarly on Yeast Carbon Base medium (YCB; Difco) and 94 progeny were analyzed and showed similar patterns of recombination, aneuploidy, and diploidy to the cross discussed in this paper (data not shown).
The markers used for selection of progeny were located on independent chromosomes. MAT is located on chromosome 6, URA3 on chromosome 5, LEU2 on chromosome 7, and CYH2 (presumptive gene responsible for conferring cycloheximide resistance) on chromosome 4. Both MAT and chxR/S should segregate independently in these progeny, thus an additional confirmation that the strains selected were progeny and not parental contamination is that 53.7% of the progeny were MAT a (36/67) and 46.3% were MATα (31/67). 50.7% of the progeny were non-parental MAT a chxR or MATα chxS, while 49.3% were parental MAT a chxS or MATα chxR. For spo11 crosses, 78 progeny were isolated from each bilateral and unilateral mutant cross and typed for the MAT locus and all RFLP markers on Chromosome 6, and for one RFLP marker each on chromosomes 1 and 5 to establish independent assortment of chromosomes.
Supplementary Material
Acknowledgments
We thank Geraldine Butler for providing information on meiotic genes and centromeric positions, and for discussions. We acknowledge Sheri Frank, and Mark Palmeri for technical assistance, and Tom Petes, Christina Hull, Geraldine Butler, Pat Pukkila and Christina Cuomo for critical reading of our manuscript. This work was supported by National Institutes of Health grants AI39115 and AI50113.
Footnotes
Supplemental Data Supplemental Data includes further methods, discussion, 4 tables, and 8 figures which can be found online.
Accession Numbers Sequences for the reported MAT loci have been deposited with GenBank under accession numbers: Candida guilliermondii MATα FJ524851, and Candida lusitaniae MAT a FJ524850.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 4.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 5.Johnson A. The biology of mating in Candida albicans. Nat Rev Microbiol. 2003;1:106–116. doi: 10.1038/nrmicro752. [DOI] [PubMed] [Google Scholar]
- 6.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 7.Francois F, Noel T, Pepin R, Brulfert A, Chastin C, Favel A, Villard J. Alternative identification test relying upon sexual reproductive abilities of Candida lusitaniae strains isolated from hospitalized patients. J Clin Microbiol. 2001;39:3906–3914. doi: 10.1128/JCM.39.11.3906-3914.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gargeya IB, Pruitt WR, Simmons RB, Meyer SA, Ahearn DG. Occurrence of Clavispora lusitaniae, the teleomorph of Candida lusitaniae, among clinical isolates. J Clin Microbiol. 1990;28:2224–2227. doi: 10.1128/jcm.28.10.2224-2227.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtzman CP, Fell JW. The yeasts : a taxonomic study. 4. Amsterdam ; New York: Elsevier; 2000. [Google Scholar]
- 10.Noel T, Favel A, Michel-Nguyen A, Goumar A, Fallague K, Chastin C, Leclerc F, Villard J. Differentiation between atypical isolates of Candida lusitaniae and Candida pulcherrima by determination of mating type. J Clin Microbiol. 2005;43:1430–1432. doi: 10.1128/JCM.43.3.1430-1432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reedy JL, Heitman J. Evolution of MAT in the Candida species complex: sex, ploidy, and complete sexual cycles in C. lusitaniae, C. guilliermondii, and C. krusei. In: Heitman J, Kronstad JW, Taylor and JW, Casselton LA, editors. Sex in Fungi: Molecular determination and evolutionary implications. Washington, D.C.: ASM; 2007. pp. 189–200. [Google Scholar]
- 12.Young LY, Lorenz MC, Heitman J. A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae. Genetics. 2000;155:17–29. doi: 10.1093/genetics/155.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candida guilliermondii Sequencing Project. Broad Institute of Harvard and MIT. http://www.broad.mit.edu.
- 14.Candida lusitaniae Sequencing Project. Broad Institute of Harvard and MIT. http://www.broad.mit.edu.
- 15.Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and virulence in eight Candida genomes. Nature. 2009 doi: 10.1038/nature08064. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 17.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzung KW, Williams RM, Scherer S, Federspiel N, Jones T, Hansen N, Bivolarevic V, Huizar L, Komp C, Surzycki R, Tamse R, Davis RW, Agabian N. Genomic evidence for a complete sexual cycle in Candida albicans. Proc Natl Acad Sci U S A. 2001;98:3249–3253. doi: 10.1073/pnas.061628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klapholz S, Esposito RE. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics. 1980;96:589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 22.Klapholz S, Waddell CS, Esposito RE. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson ME, Saville BJ. Bioinformatic identification of Ustilago maydis meiosis genes. Fungal Genet Biol. 2008;45:S47–S53. doi: 10.1016/j.fgb.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 25.Holloman WK, Schirawski J, Holliday R. The homologous recombination system of Ustilago maydis. Fungal Genet Biol. 2008;45:S31–S39. doi: 10.1016/j.fgb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F, McFarlane RJ, Loidl J. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci. 2004;117:3343–3351. doi: 10.1242/jcs.01203. [DOI] [PubMed] [Google Scholar]
- 28.Hunt PA, Hassold TJ. Human female meiosis: what makes a good egg go bad? Trends Genet. 2008;24:86–93. doi: 10.1016/j.tig.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 30.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 31.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.