SUMMARY
N-Methyl-D-aspartate receptors (NMDARs) play important functions in neural development. NR2B is the predominant NR2 subunit of NMDAR in the developing brain. Here we use MADM (Mosaic Analysis with Double Markers) to knock out NR2B in isolated single cells and analyze its cell-autonomous function in dendrite development. NR2B mutant dentate gyrus granule cells (dGCs) and barrel cortex layer 4 spiny stellate cells (bSCs) have similar dendritic growth rates, total length and branch number as control cells. However, mutant dGCs maintain supernumerary primary dendrites resulting from a pruning defect. Furthermore, while control bSCs restrict dendritic growth to a single barrel, mutant bSCs maintain dendritic growth in multiple barrels. Thus, NR2B functions cell-autonomously to regulate dendrite patterning to ensure that sensory information is properly represented in the cortex. Our study also indicates that molecular mechanisms that regulate activity-dependent dendrite patterning can be separated from those that control general dendrite growth and branching.
Keywords: barrel cortex, spiny stellate cells, dentate gyrus, granule cells, dendrite growth, dendrite patterning, activity dependent, MADM, NMDA receptor, NR2B, structural plasticity, cell-autonomous function
INTRODUCTION
The dendrites of CNS neurons play a critical role in integrating synaptic inputs from a multitude of presynaptic partners and, subsequently, in determining the extent to which a neuron transmits this information to its postsynaptic partners. Characteristic dendritic arborization patterns allow neurons to perform signal processing and computation appropriate for their functions. For example, in the adult somatosensory cortex of rodents, most layer 4 stellate neurons orient their dendrites towards a single barrel center to maximize contacts with thalamocortical afferents representing a single whisker (Woolsey and Van der Loos, 1970; Woolsey et al., 1975). The development of dendritic trees characteristic to specific neuronal types is believed to result from the interplay between intrinsic genetic programs, extracellular signals and electrical activity (reviewed in Scott and Luo, 2001; Wong and Ghosh, 2002; Jan and Jan, 2003; Parrish et al., 2007). Despite our increasing understanding of dendrite development, it is unclear if mechanisms that sculpt specific dendrite patterns are an integral part of those that control dendrite growth and branching, or if independent mechanisms can regulate these two aspects of dendrite development.
N-methyl-D-aspartate-type glutamate receptors (NMDARs) play a central role in activity-dependent regulation of dendrite development (reviewed in Constantine-Paton et al., 1990; Cline, 2001; Wong and Ghosh, 2002). NMDARs function mainly as heterotetramers of two obligate NR1 subunits and a combination of two NR2 subunits (A-D) (Kutsuwada et al., 1992; Monyer et al., 1992). Each NMDAR subunit combination confers distinct functional properties, including the regulation of unitary conductance, binding affinity, and gating and desensitization kinetics. For example, compared to NR2A-containing receptors, NR2B-containing NMDARs have a 3- to 4-fold slower decay time course of NMDAR-mediated excitatory postsynaptic currents resulting in a larger Ca2+ influx (reviewed in Cull-Candy and Leszkiewicz, 2004). In the mammalian brain, the NR1 mRNA is found ubiquitously, whereas the NR2 subunits are differentially expressed, both temporally and spatially. At embryonic stages, the NR2B subunit is expressed in the entire brain, while the NR2D subunit is expressed selectively in the diencephalon, mesencephalon, and the spinal cord. From the time of birth to adult, expression of NR2B becomes restricted to the forebrain and NR2D expression peaks 1-week after birth but is then strongly reduced. During this time, the expression levels increase for NR2A in the forebrain and for NR2C in the cerebellum (Watanabe et al., 1992; Monyer et al., 1994). The expression pattern of NR2B suggests that it plays a more important role during development than other NR2 subunits. Indeed, NR2B knockout mice die shortly after birth (Kutsuwada et al., 1996) similar to NR1 knockout mice (Forrest et al., 1994; Li et al., 1994), but knockout mice for NR2A, 2C, or 2D are fully viable (Ikeda et al., 1995; Sakimura et al., 1995; Ebralidze et al., 1996; Kadotani et al., 1996).
Pharmacological agents that block NMDARs have been used to study their function in dendrite development. In the Xenopus retinotectal system, NMDAR antagonists inhibit dendritic arborization of tectal neurons during development (Rajan and Cline, 1998) or in response to visual stimulation (Sin et al., 2002). NMDAR function in dendrite development has also been examined in knockout mice. In the cortex-specific NR1 knockout, individual layer 4 stellate cells lose oriented arborization and grow exuberant dendrites and spines (Datwani et al., 2002). NMDARs are also necessary for dendritic spine formation induced by sensory activity and long-term potentiation (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999). Cortex-specific NR1 knockout results in reduced spine densities (Ultanir et al., 2007). Although these studies have revealed important functions for NMDARs in multiple aspects of dendrite development, it is unclear to what extent the observed defects are caused by the cell-autonomous perturbation of NMDAR function. These experiments cannot exclude secondary consequences of perturbing the NMDAR in other neurons in the circuit. In the Xenopus retinotectal system, NMDAR blockade also affects the arborization of the retinal ganglion cell axon termini (Cline and Constantine-Paton, 1990; Ruthazer et al., 2003), which may indirectly perturb tectal cell dendrite development. In cortex-specific NR1 knockout mice, although thalamocortical axons are genetically unperturbed, their terminal arborization patterns are grossly altered in response to NR1 knockout in cortical cells (Lee et al., 2005) and barrels do not form properly (Datwani et al., 2002). It is therefore difficult to determine if the unoriented dendrites of layer 4 stellate neurons reflect the cell-autonomous requirement for NMDAR, or if they are a secondary consequence of the general pattern formation defects in the barrel cortex. Recently, genetic perturbations of NR2A and NR2B subunits have been reported using overexpression and morpholino-mediated knockdown in single Xenopus tectal cells. Compared to overexpression, knockdown of NR2B has minor consequences on dendrite development (Ewald et al., 2008).
In this study, we use the MADM system (Mosaic Analysis with Double Markers) to knock out NR2B in isolated single neurons to assess the cell-autonomous function of NR2B in dendrite development. MADM permits simultaneous gene inactivation and distinct labeling of homozygous mutant cells and their wild-type siblings in the same animal through Cre/LoxP-mediated interchromosomal mitotic recombination events (Figure S1A). Moreover, infrequent recombination generates isolated single knockout cells allowing us to unambiguously assess cell-autonomous function of genes (Zong et al., 2005; Muzumdar et al., 2007). We find that in two types of neurons analyzed, dentate gyrus granule cells and barrel cortex layer 4 spiny stellate cells, NR2B is dispensable for general dendrite growth and branching, but is required for dendrite patterning critical for information processing. Our study also indicates that molecular mechanisms that regulate activity-dependent dendrite patterning are separable from those that control general dendrite growth and branching.
RESULTS
Validation of MADM knockout of NR2B
To investigate the role of NMDARs in dendrite development, we employed the MADM system to generate mice in which isolated single cells lack the NR2B subunit (Figure S1). Phenotypic analyses were performed on two types of MADM mice. In NR2B-GR/RG;Cre (MADM-Green-KO) mice, green (GFP+ only) cells are homozygous mutant for NR2B, red (Dsred2-myc+ only) cells are homozygous wild type, and yellow (GFP+ and Dsred2-Myc+) cells and all unlabeled cells are heterozygous (Figure 1A, top; Figure S1). In NR2B-RG/GR;Cre (MADM-Red-KO) mice, red cells are homozygous mutant for NR2B, green cells are wild type, while yellow and unlabeled cells are heterozygous (Figure 1A, middle). In several experiments, we found that the phenotypes segregate with NR2B genotypes rather than with the color of fluorescent proteins expressed. GR/RG;Cre (MADM-WT) mice (Figure 1A, bottom) were used as an additional control.
Figure 1. Validation of MADM-Mediated NR2B Knockout In Labeled Cells.
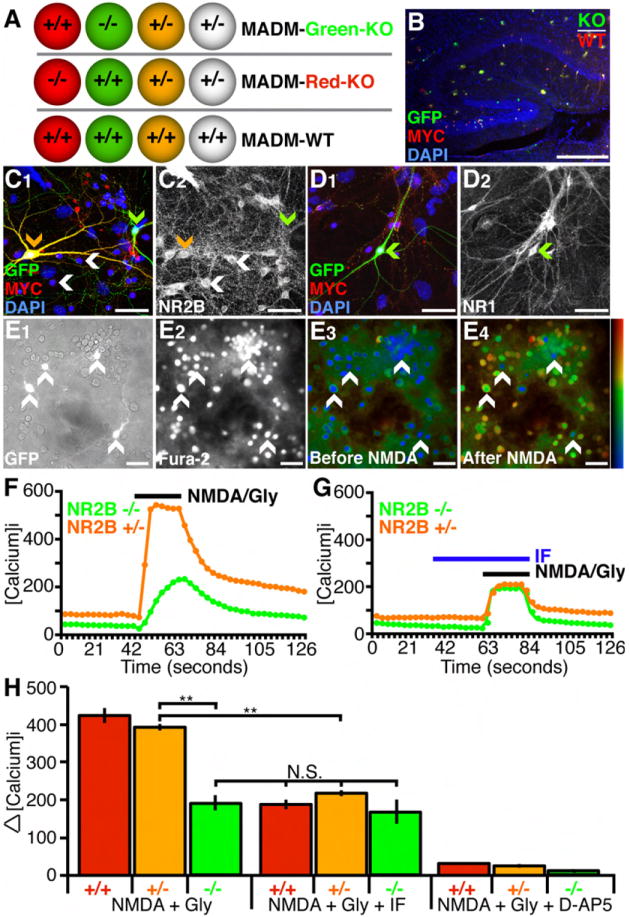
(A) Schematic of three MADM mice used in this study. In the first two, wild type (NR2B+/+), heterozygous (NR2B+/-), and homozygous knockout (NR2B-/-) cells are labeled with different fluorescent markers as indicated. In the third, MADM-WT, all cells are wild type.
(B) Sparse MADM labeling in the hippocampus using nestin-Cre (see Figure S1 for details). In this case (NR2B-GR/RG;nestin-Cre), red cells are NR2B+/+, yellow cells are NR2B+/-, green cells are NR2B-/-. Scale bar, 200 μm.
(C and D) Hippocampal cells were immunostained for GFP, Myc (C1 and D1), NR2B (C2), NR1 (D2), and counterstained with nuclear marker DAPI. NR2B protein is present in labeled NR2B+/- neurons (C2, orange arrowhead) and unlabeled NR2B+/- neurons (C2, white arrowhead), but not detectable in NR2B-/- neurons (C2, green arrowhead). NR1 protein is present in all neurons (D2, green arrowhead). Scale bar, 50 μm.
(E) Calcium imaging of hippocampal cells from MADM-Green-KO mice. GFP fluorescent cells (E1) represent NR2B-/- and NR2B+/- neurons (white arrowheads in all panels). Fura-2 was loaded into cells for [Ca2+]i measurement (E2). Marked elevation of [Ca2+]i is evident when comparing cells before (E3) and after (E4) application of 100 μM NMDA and 10 μM Glycine. Scale bar, 50 μm.
(F and G) Representative [Ca2+]i traces of single NR2B+/- (orange) and NR2B-/- (green) hippocampal cells. The presence of 3 μM IF reduces NMDA/Gly response in NR2B+/-cells to the level of NR2B-/- cells (G).
(H) Quantification of [Ca2+]i for different stimulation conditions and genotypes. NR2B+/- and NR2B+/+ neurons have significantly larger responses compared to NR2B-/- neurons. Addition of 3 μM IF significantly reduces the average response in NR2B+/- and NR2B+/+ neurons, to the level indistinguishable from NR2B-/- neurons without the drug, but does not significantly change the responses of NR2B-/- cells. Addition of 50 μM D-AP5 reduces the response in all cell types to near baseline levels. N for each condition (number of cells for each genotype and condition analyzed) are as follows: NMDA + Gly (100 NR2B+/+; 378 NR2B+/-; 26 NR2B-/- cells), NMDA + Gly + IF (40 NR2B+/+; 214 NR2B+/-; 12 NR2B-/- cells), and NMDA + Gly + D-AP5 (39 NR2B+/+; 79 NR2B+/-; 8 NR2B-/- cells). **p<0.01, ANOVA/Tukey HSD multiple comparison test; N.S., not significant. Results shown in this and all subsequent figures are mean ± SEM.
To validate the loss of NR2B in NR2B-/- cells as predicted by the MADM scheme, we used nestin-Cre (Petersen et al., 2002) to generate MADM labeled cells in all regions of the brain including the hippocampus (Figure 1B). We cultured neurons from dissociated hippocampi of postnatal day (P)0 NR2B-GR/RG;nestin-Cre mice. Triple-immunostaining using antibodies against NR2B, GFP and Myc revealed that all neurons display strong NR2B-immunoreactivity except for green NR2B-/- neurons (Figures 1C1 and 1C2; n>50 cells for each genotype). NR1 expression in NR2B-/- neurons was indistinguishable from other neurons (Figures 1D1-1D2; n>30 cells for each genotype).
To ensure that cells identified as NR2B-/- are indeed altered in their NMDA sensitivity, we measured the [Ca2+]i response to NMDA in cultured hippocampal neurons, with TTX present throughout to block spiking (Figure 1E). We found that application of 100 μM NMDA along with 10 μM of the co-agonist Glycine results in an elevation of [Ca2+]i in all GFP+ cells, as assessed with Fura-2, a ratiometric fluorescent dye that binds to free intracellular calcium (Figures 1E2-1E4, arrows; Grynkiewicz et al., 1985). The rise of [Ca2+]i required NMDAR function, as it was blocked by application of an NMDAR antagonist D-AP5 in cells of all genotypes (Figure 1H).
We compared [Ca2+]i response in NR2B-/- cells with controls. Since we cannot detect Dsred2-Myc in live cells, GFP+ cells are a mix of green and yellow cells (Figure S1A; Zong et al., 2005). Thus, following calcium imaging, we stained for Myc to distinguish GFP+ only cells from GFP+/Dsred2-Myc+ cells. Figure 1F shows a sample trace of [Ca2+]i for one green and one yellow cell in response to NMDA application; all cells are quantified in Figure 1H (left). NR2B-/- cells exhibited a reduction in NMDA-mediated [Ca2+]i response, consistent with the genetic loss of NR2B resulting in decreased NMDAR function. If the decreased [Ca2+]i response was indeed due to the loss of NR2B-containing NMDARs, we reasoned that ifenprodil (IF), an NR2B-specific antagonist (Williams, 1993; Tovar and Westbrook, 1999), should have no effect on NR2B-/- neurons. Indeed, application of 3 μM IF had no significant effect on the [Ca2+]i response of NR2B-/- cells (Figure 1G, 1H). By contrast, IF reduced the [Ca2+]i response of NR2B+/- cells to the level found in NR2B-/- cells (Figure 1G, 1H).
The immunocytochemical and pharmacological data confirmed our MADM strategy for generating NR2B knockout in singly labeled cells. Previous findings with whole animal (Kutsuwada et al., 1996) and conditional (von Engelhardt et al., 2008) NR2B knockout mice showed that expression of other NR2 subunits are unaffected. Our data are consistent with this lack of compensation as the [Ca2+]i response to NMDA in NR2B-/- neurons (presumably mediated by other NR2 subunits) was indistinguishable from that in NR2B+/- neurons acutely treated with IF. Of note, NR2B+/- and NR2B+/+ cells were indistinguishable in their response to NMDA application, suggesting that NR2B is not dosage sensitive. Consistently, all of our in vivo assays showed no significant difference between NR2B+/+ and NR2B+/- cells (see below). Hence, we sometimes used NR2B+/- cells as controls for NR2B-/- cells because GFP labels processes better than Dsred-Myc in adult neurons (Zong et al., 2005).
Quantitative analysis of dendrite morphology in NR2B-/- dentate gyrus granule cells
We performed MADM analysis using nestin-Cre to examine NR2B-/- cells throughout the brain. Compared to control (NR2B+/+ and NR2B+/-), NR2B-/- neurons had no gross defects in survival, migration and dendritic morphogenesis (data not shown). To examine potential quantitative differences, we focused our study on dentate gyrus granule cells (dGCs), as we could generate sufficient number of labeled cells in isolation to trace the entire dendrites and quantitatively compare morphometrics. We observed dGCs with single and multiple primary apical dendrites in both control and mutant cells (Figures 2A-2D). To analyze a more homogenous cell population (see Claiborne et al., 1990), we first focused our analysis on dGCs with a single primary dendrite, whose cell bodies are positioned in the top third of the granule cell layer within the suprapyramidal blade.
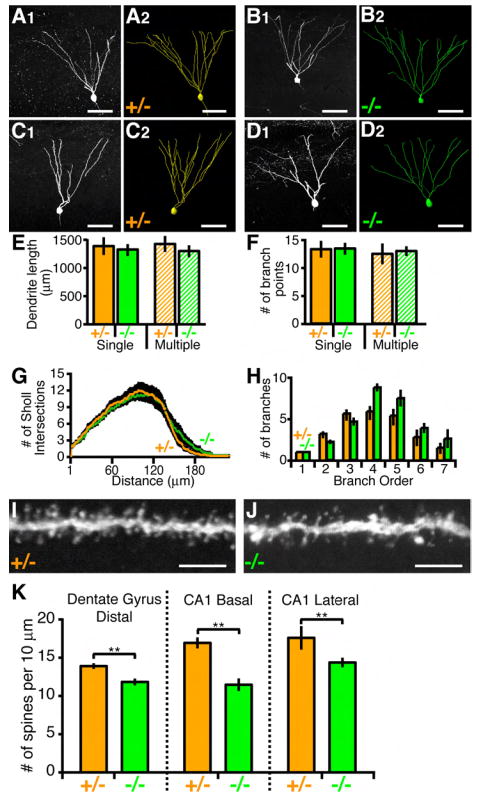
Figure 2. Dendrite Morphology of Hippocampal Neurons.
(A and B) Representative images of NR2B+/- and NR2B-/- P21 dGCs. (A1 and B1) Confocal images from single 60-μm sagittal sections. (A2 and B2) Corresponding 3D reconstructions spanning several 60-μm sections. Scale bar, 50 μm.
(C and D) Same as (A and B) except for dGCs with two primary dendrites are shown.
(E-H) Quantification of dendritic parameters of P21 dGCs. Only dGCs with one primary dendrite were selected for analysis except the hatched bars of (E) and (F). (E and F) ANOVA reveals no significant differences between NR2B+/- (n=14 for single; n=7 for multiple) versus NR2B-/- (n=14 for single; n=7 for multiple) cells. (G and H). ANOVA with repeated measures reveals no significant difference between NR2B+/- versus NR2B-/- for Sholl analysis and branch order.
(I and J) Representative confocal images of spines in the distal portion of NR2B+/- (I) and NR2B-/- (J) P21 dGCs. Scale bar, 5 μm.
(K) Spine counts from 10 μm segments. For each genotype, n > 20 cells. **p<0.01, t-test.
Consistent with our qualitative examination from single cell tracings of dGCs (Figures 2A and 2B), the total dendrite length and branch point number are indistinguishable between NR2B-/- and NR2B+/- cells (Figures 2E and 2F, solid bars). We performed 3D Sholl analysis (1953), which counts the number of intersections of dendrites with successively larger concentric spheres centered at the cell body (Figure 2G), as well as branching order analysis (Figure 2H). In neither case was there a statistically significant difference between NR2B-/- and NR2B+/- cells.
We also quantified spine density in the distal dendritic segments of dGCs. We found a slight but significant reduction in spine density in NR2B-/- cells compared to NR2B+/- cells (Figures 2I and 2J; quantified in Figure 2K). The reduction in spine density was also found in other hippocampal neurons, such as the basal and lateral dendritic segments of CA1 pyramidal neurons (Figure 2K). Spine density reduction was previously reported in cortical neurons from animals in which NR1 was knocked out in the entire cortex (Ultanir et al., 2007). Our MADM analysis indicated that the function of NR2B in establishing a normal density of dendritic spines is cell autonomous.
NR2B regulates primary apical dendrite number
Compared with the relatively mild phenotypes described in the previous section, we found a more striking phenotype in NR2B-/- dGCs in affecting the number of primary apical dendrites. More than 90% of control dGCs had a single primary dendrite (Figures 3A and 3E). By contrast, ~25% of NR2B-/- dGCs had two or more primary dendrites (Figures 3B and 3E). This phenotype was also present in CA1 and CA3 pyramidal neurons with a similar 2.5-4 fold increase in neurons extending multiple primary apical dendrites (Figures 3C and 3D; quantified in Figure 3E). We did not however find an equivalent phenotype in NR2B-/- cortical pyramidal cells (data not shown).
Figure 3. Increased Primary Dendrite Number In NR2B-/- Hippocampal Neurons.
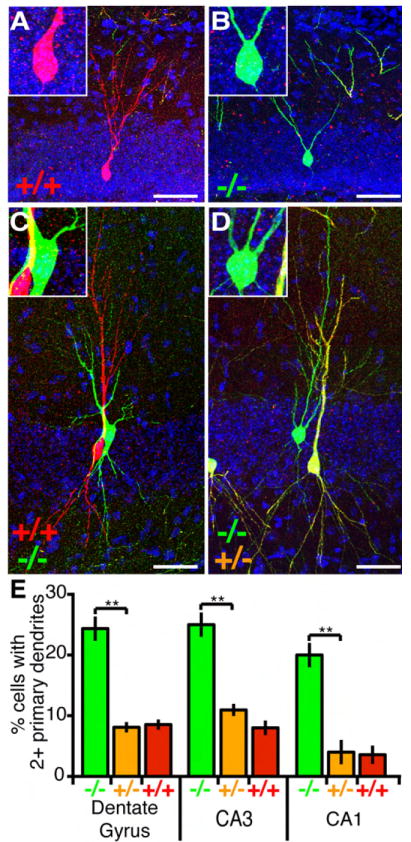
(A-D) Representative confocal images of hippocampal cells at P21: an NR2B+/+ dGC with a single primary dendrite (A), an NR2B-/- dGC with two primary dendrites (B), NR2B+/+ (red) and NR2B-/- (green) CA3 pyramidal cells with one and two primary apical dendrites (C), and NR2B+/- (yellow) and NR2B-/- (green) CA1 pyramidal cells with one and three primary apical dendrites (D). Insets are magnifications highlighting primary dendrites exiting the cell body. Scale bar, 50 μm.
(E) Quantification of primary apical dendrite number in dGCs and in CA3/CA1 pyramidal neurons. NR2B-/- cells have a significantly higher fraction of cells with multiple primary dendrites compared to NR2B+/+ or NR2B+/- cells. **p<0.01 ANOVA/Tukey HSD multiple comparison test. Dentate gyrus: 192 NR2B-/-, 1143 NR2B+/-, 132 NR2B+/+ cells from 6 brains; CA3: 105 NR2B-/-, 824 NR2B+/-, 112 NR2B+/+ cells from 9 brains; CA1: 103 NR2B-/-, 798 NR2B+/-, 99 NR2B+/+ cells from 9 brains.
NR2B regulates pruning of supernumerary primary dendrites of dGCs
The marked difference in the number of primary dendrites in hippocampal neuronal types prompted us to examine the underlying mechanism. The overall increase in primary dendrite number in NR2B-/- neurons could reflect a role of NR2B in limiting the generation of additional primary dendrites after one has already formed, or in pruning supernumerary primary dendrites down to a single one. To distinguish these possibilities, we tracked the developmental history of dendrite growth following neuronal birth. We focused our analysis on dGCs because these neurons are born continuously over a long period of time starting from embryonic development into adulthood (Zhao et al., 2008). In addition, newly born neurons are added from the basal side of the granule cell layer, thus the cell body position can be used to estimate the age of the neurons (Altman and Bayer, 1990).
We have confirmed a previous report (Green and Juraska, 1985) that dGCs having multiple primary dendrites tend to locate in the upper granule cell layers, suggesting that earliest-born neurons contribute more to dGCs with multiple primary dendrites. To test this experimentally, we injected the thymidine analog CldU, which labels dividing cells in the S phase (Vega and Peterson, 2005), into MADM mice at different developmental time points. Three weeks later, we examined the morphology of MADM-labeled neurons co-labeled with CldU (Figure 4A). Neurons strongly labeled with CldU are likely born shortly after the pulse injection, as further division dilutes CldU levels. Indeed, dGCs strongly labeled with CldU from early injections tend to locate in the apical granule cell layers, whereas those from later injections are progressively more basal (Figures 4B1-4B4), although this rule is not strict (arrowheads in Figures 4B1-4B4).
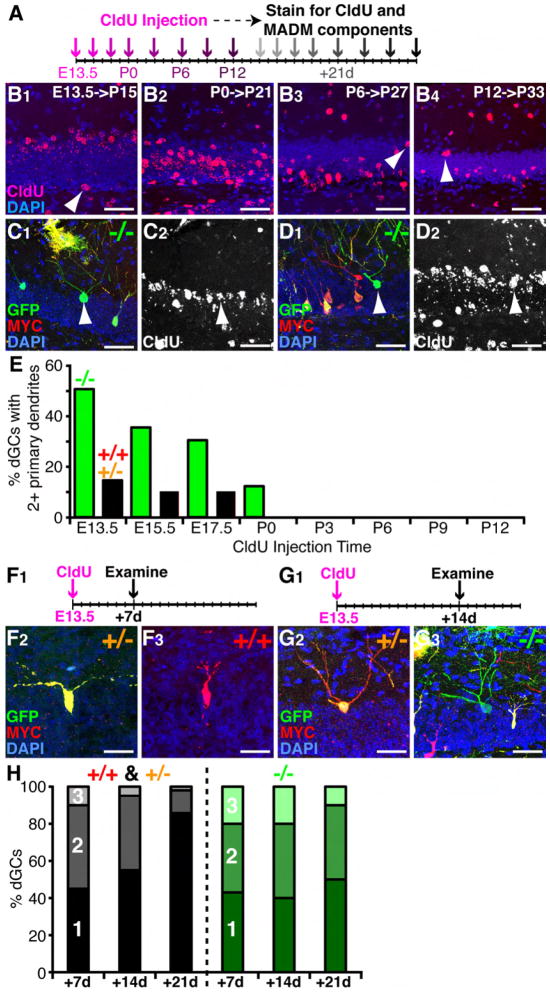
Figure 4. Embryonic Born NR2B-/- dGCs Fail to Prune Excessive Primary Dendrites.
(A) Schematic of birthdating MADM-labeled neurons. CldU was injected at 8 developmental times (purple arrows), and brains were analyzed 21 days post injection (gray arrows).
(B) CldU staining shows the locations of dGCs born at 4 different times and examined 21 days after as indicated. Some CldU labeled cells deviate from the general trend (white arrowheads). Scale bar, 50 μm.
(C and D) Representative confocal images of NR2B-/- dGCs 21 days after birth at E13.5 showing the MADM labeling (C1 and D1) or corresponding CldU staining (C2 and D2). NR2B-/- dGCs with two primary dendrites (C1, arrowhead) or a single primary dendrite (D1, arrowhead) in the upper granule cell layers are co-labeled with strong levels of CldU (C2 and D2, arrowheads). Scale bar, 50 μm.
(E) Percentage of dGCs with 2 or more primary dendrites born at different developmental times. n>20 for each genotype per injection time point.
(F) Representative confocal images of dGCs 7-days after birth at E13.5 (F1). An NR2B+/- dGC with two primary dendrites (F2) and an NR2B+/+ dGC with a single primary dendrite (F3) are co-labeled with strong CldU. Scale bar, 50 μm.
(G) Representative confocal images of dGCs 14-days after birth at E13.5 (G1). An NR2B+/- dGC (G2) and an NR2B-/- dGC each with three primary dendrites (G3) are co-labeled with strong CldU. Scale bar, 50 μm.
(H) Fractions of dGCs with 1, 2, 3+ primary dendrites at 7-, 14-, and 21-days after birth.
By correlating birth timing with the morphology of MADM labeled NR2B-/- and control dGCs (Figures 4C and 4D), we found that dGCs born at P3 or later always had a single primary dendrite, regardless of the genotypes (Figure 4E). However, the earlier the neurons were born, the more likely they were to have multiple primary dendrites. NR2B affected the primary dendrite number only in dGCs from the early-born cohort. 50% of NR2B-/- neurons born shortly after E13.5 had multiple primary dendrites compared to only 12% in NR2B+/- or NR2B+/+ cells (Figure 4E).
We next tracked the developmental time course of the multiple primary dendrite phenotype using the early born cohort of dGCs. We injected CldU at E13.5 and analyzed the morphology of MADM and CldU labeled neurons 1 and 2 weeks later (Figures 4F and 4G). We found that both control and NR2B-/- dGCs have multiple primary dendrites at early stages in dendrite development (Figure 4F-G). One week after birth, more than half of neurons had multiple primary dendrites in both control and NR2B-/- cells (Figure 4H). Two weeks after birth, the fraction of cells with multiple primary dendrites stayed roughly the same among both control and NR2B-/- dGCs; however, the number of cells with 3 primary dendrites was reduced in control cells, but not in NR2B-/- dGCs (Figure 4H). By this time, primary dendrites in both control and NR2B-/- dGCs had begun to branch further (compare Figures 4F and 4G), although usually only one of the two primary dendrites had >5 branch points. Three weeks after birth, only ~15% of control cells had two primary dendrites, and the ones with 3 primary dendrites are very rare. By then, these cells had reached dendrite arborization comparable to adult dGCs and they exhibit mature spine morphology. By contrast, 3-week old NR2B-/- dGCs showed no reduction in the number of primary dendrites compared to 1- and 2-week old dGCs. The proportion of 3-week old dGCs with multiple primary dendrites was similar to the levels seen among NR2B-/- dGCs in 1-year old mice (data not shown). Taken together, these data indicate that early-born dGCs initially extend multiple primary dendrites and subsequently prune supernumerary primary dendrites between 2-3 weeks after birth. NR2B function is required for the elimination of excessive primary dendrites.
Interestingly, NR2B-/- and NR2B+/- dGCs with multiple primary dendrites still possessed the same total dendritic length and branch number as NR2B-/- and control dGCs with a single primary dendrite (Figures 2E-2F). This finding implies that mechanisms regulating the pattern of dendrite branching in dGCs are separable from those controlling the total dendritic length and branch number, and that NR2B is necessary only for the former. To further test the effects of NR2B-mediated alterations of dendrite patterning, we next turned to layer 4 spiny stellate cells of the barrel cortex, where the spatial distribution of dendritic branches has a clear role in shaping somatotopic maps.
Cell-autonomous function of NR2B in dendrite targeting of layer 4 spiny stellate cells to a single barrel
The stereotyped somatosensory pathway from whisker to cortex allows us to explore the link between the NMDAR function and refinement of synaptic circuits. The primary somatosensory cortex contains an exquisite somatotopic map where each individual whisker relays information via thalamocortical axons to layer 4 of the primary sensory cortex to form a discrete anatomical unit, the “barrel,” allowing precise delineation of functional organization, development, and plasticity. Cell bodies of layer 4 spiny stellate cells (bSCs), the major postsynaptic partners of thalamocortical axons, are located predominantly around the axon-rich barrel centers. Each bSC located in the barrel wall sends dendrites into a single barrel and, thus, represents sensory information primarily from a single whisker. These patterns are subject to perturbations of the sensory periphery by nerve or whisker lesions during a critical period, suggesting that early neural activity is essential for the pattern formation of the barrel cortex (reviewed in Inan and Crair, 2007).
NMDARs play important roles in the formation of synaptic patterns in the barrel cortex. When NR1 is conditionally knocked out from all cortical neurons, the morphology of the barrels is severely disrupted. Stellate cells are no longer clustered around barrel wall and instead are evenly distributed in layer 4 with unoriented dendrites (Datwani et al., 2002). In these experiments, it is difficult to determine whether dendritic patterning defects are caused by cell-autonomous requirement for NMDARs or if they are a secondary consequence of a general disruption of barrel cortex patterning. MADM offers an opportunity to distinguish between these possibilities by generating sparse NR2B-/- bSCs in an otherwise normal barrel cortex.
We used nestin-CreER line 1 (Imayoshi et al., 2006) to generate isolated MADM-labeled bSCs such that a single barrel has only one labeled cell. For unknown reasons, Cre activity in this line is independent of tamoxifen induction allowing for sparser recombination events compared with nestin-Cre (Imayoshi et al., 2006; our unpublished observation) and thereby making it ideal for tracing the dendrites of single bSCs whose cell bodies are located in the barrel wall. Consistent with previous results (Woolsey et al., 1975; Hickmott and Merzenich, 1999), we found that NR2B+/- and NR2B+/+ bSCs elaborate their dendrites into a single barrel as outlined by DAPI staining (Figures 5A1-5A3; Figure S2). By contrast, NR2B-/- cells sent dendrites to multiple adjacent barrels (Figures 5B1-5B3). For quantification (Figure 5C-5G), we focused on bSCs whose cell bodies were located in the barrel wall of the large barrels that correspond to whiskers. We found that >90% of total dendrite length and 100% of branch points of NR2B+/- bSCs were confined within a single barrel. By contrast, nearly half of total dendrite length and branch point number of NR2B-/- bSCs were located outside the “primary barrel,” defined as the barrel that contains the greatest proportion of dendrite length and branching. The dendrites of NR2B-/- bSCs also extended into the septal region (the region between barrels; Figure 5B3 and 5C) and frequently into more than two barrels (Figure 5B3). NR2B-/- bSCs from older animals (P40) had similar dendritic mistargeting defects (data not shown). These results demonstrate that NR2B is cell-autonomously required in bSCs for oriented dendritic arborization within a single barrel.
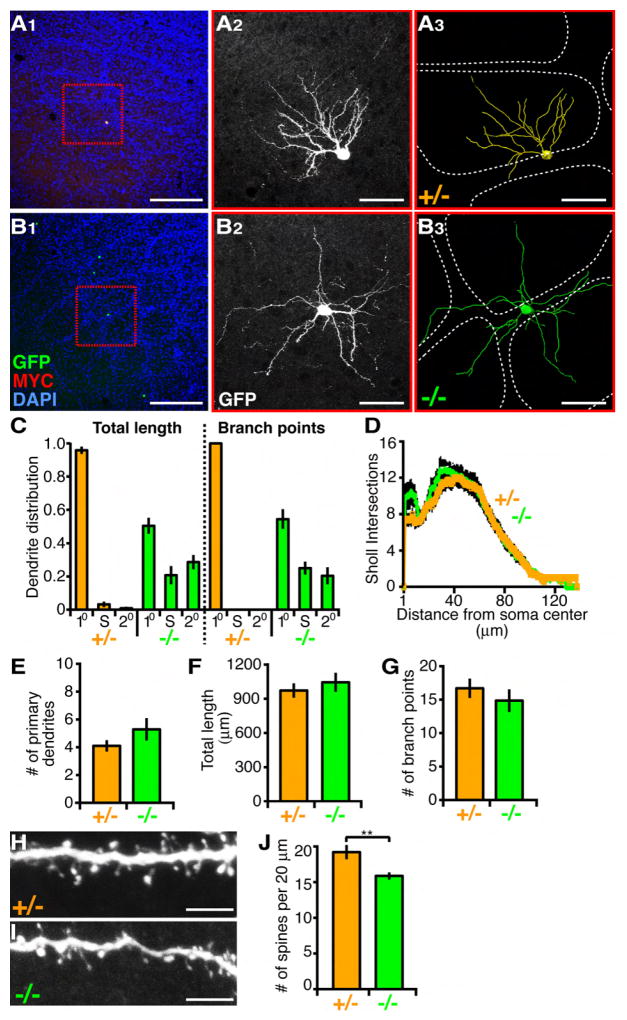
Figure 5. NR2B-/- Layer 4 Spiny Stellate Cells of the Barrel Cortex Mistarget Their Dendrites.
(A and B) Dendrite targeting of P21 bSCs in the barrel cortex. (A1 and B1) Representative low magnification images show barrels outlined by surrounding dense nuclei staining (DAPI in blue), and sparse MADM labeled NR2B+/- (A1) and NR2B-/- (B1) bSCs. (A2 and B2) High magnification confocal images of NR2B+/- (A2) and NR2B-/- (B2) bSCs (same cell as in red box in A1 and B1). (A3 and B3) 3D reconstructions of bSCs in (A2) and (B2) superimposed over barrel walls representing the edge of barrels (dashed white lines). Scale bar, 200 μm for (A1) and (B1); 50 μm for the rest.
(C) Quantification of dendrite distribution of P21 bSCs with respect to barrels. Percentage of dendritic length (left) and branch point number (right) in primary barrel (10), septae (S), and secondary (20) barrels are presented for NR2B-/- (n=12) and NR2B+/- (n=14) cells.
(D-G) Quantification of P21 bSC dendritic parameters. ANOVA with repeated measures reveals no significant difference between NR2B+/- versus NR2B-/- for Sholl analysis. Number of primary dendrites (E), total dendritic length (F) and number of branch points (G) are not significantly different between NR2B-/- (n=12) and NR2B+/- (n=14) cells. p>0.05, t-test.
(H and I) Representative confocal images of spines in the distal portion of NR2B+/- (H) and NR2B-/- (I) P21 bSCs. Scale bar, 5 μm.
(J) Spine counts from 20 μm segments. For each genotype, n > 20 cells. **p<0.01, t-test.
Despite a dramatic dendritic patterning defect, dendritic morphometric parameters were comparable between NR2B-/- cells and NR2B+/- cells. These included branch distribution using 3D Sholl analysis (Figure 5D), number of primary dendrites (Figure 5E), total dendrite length (Figure 5F) and number of branch points (Figure 5G). One exception was a reduction in the spine density in secondary segments of NR2B-/- cells compared to control cells (Figures 5H-5J). Thus, in the case of bSCs, dendritic growth and branching are dissociated from patterning—oriented growth within a single barrel that enables each cell to represent information largely from a single whisker. Together with our data on dGCs, we reinforce the notion that general dendrite growth can be uncoupled from dendrite patterning.
Dendrite development of bSCs
To determine the developmental mechanisms by which NR2B regulates bSC dendrite patterning, we examined the time course of dendrite development in control and NR2B-/- cells (Figure 6). We traced dendritic trees of individual cells every three days starting from P3 (Figures 6A-6H, 6M and 6N; Figure S2) to quantify their morphometric parameters. Between P3 and P15, NR2B+/- cells continually grew their dendritic trees, adding new branch points and augmenting total length (Figures 6I and 6J). Both parameters reach steady state at P15, when they are indistinguishable from young adult (P21; Figure 5). NR2B-/- cells exhibited almost identical growth curves during the same period of time (Figures 6I and 6J), indicating that NR2B does not affect the rate of dendrite growth and branching.
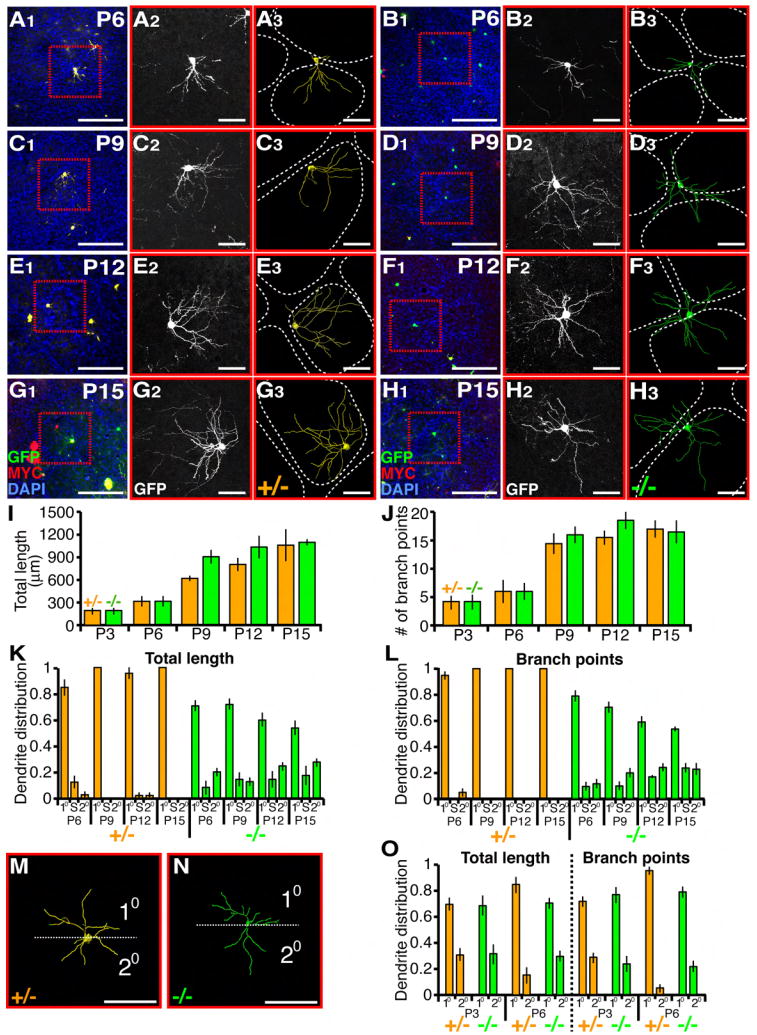
Figure 6. Dendrite Development of Layer 4 Spiny Stellate Cells.
(A-H) Representative images and tracing of sparse MADM labeled NR2B+/- and NR2B-/- bSCs at postnatal dates as indicated. Low magnification images showing labeled cells with DAPI staining outlining the barrel walls (left), confocal images of MADM-labeled cells (middle), corresponding 3D reconstructions (right). Scale bars, 100 μm for (A1-H1); 50 μm for the rest.
(I and J) Quantification of dendritic parameters of P3-P15 bSCs. For each genotype and time point, n>7 cells. ANOVA reveals no significant difference between NR2B+/- versus NR2B-/- cells.
(K and L) Quantification of dendrite distribution of P6-P15 bSCs with respect to barrels. For each genotype and time point, n>7 cells.
(M and N) Dendrite distribution of P3 bSCs. Representative 3D reconstructions are oriented so that maximal dendritic branches reside in one half (10) and the rest reside in the opposite half (20).
(O) Quantification of dendritic length and branch point distribution of P3 and P6 bSCs according to scheme in (M and N). For each genotype and time point, n>7 cells.
We next quantified the distribution of dendrite length and the number of branch points with respect to barrels (Figures 6K and 6L). As early as P6, control cells had already directed their dendrites towards the primary barrel (Figure 6A, Figure S2). At this time, a small minority of dendrites were directed away from the primary barrel; by P9, those are completely pruned and all dendrites were fully confined within a single barrel (Figure 6C, Figure S2). By contrast, NR2B-/- cells at P6 exhibited promiscuous dendritic targeting outside of the primary barrel (Figure 6B). From P6 to P15, the dendritic distribution outside of the primary barrel became increasingly pronounced with the lengthening and branching of dendrites in secondary barrels and in septal regions (Figures 6B, 6D, 6F, and 6H). By P15, NR2B-/- cells had reached a similar degree of mistargeted dendrites as in P21 (Figure 5; Figures 6K and 6L).
Since the difference between NR2B+/- and NR2B-/- cells is already pronounced at P6, we analyzed the dendritic morphology at P3. At this time point, bSCs have already begun dendrite arborization but barrel formation was not complete and boundaries were undetectable. To analyze spatial distribution of dendritic branches, we rotated a plane centered at the cell body to find a position at which dendrites were maximally asymmetric with respect to the plane (Figures 6M and 6N). We found that NR2B+/- and NR2B-/- bSCs had a similar degree of dendrite asymmetry at P3, but deviated from each other by P6 with regard to the asymmetry of both dendrite length and branch points (Figure 6O).
These experiments demonstrate that NR2B is required in bSCs for oriented growth during the time when thalamocortical axons consolidate at the barrel, displacing stellate cells to the barrel walls (between P3 and P6). After the pattern formation of the barrel is initially established (P6), NR2B-/- cells continue to grow and branch outside the primary barrel whereas control cells restrict their growth and branching within the primary barrel. These findings have important implications for the mechanisms of NMDAR action in shaping the pattern of dendritic trees (see Discussion).
DISCUSSION
Previous studies have implicated important functions of NMDARs in many aspects of dendrite development. We report here the use of a genetic mosaic method to analyze the effect of loss of NR2B in isolated single neurons in an otherwise normal brain. In the two model neurons examined in detail (Figures 7A and 7B), we find that NR2B is not cell-autonomously required for dendrite growth and branching, but is essential for dendrite patterning. We discuss these findings and possible underlying mechanisms below.
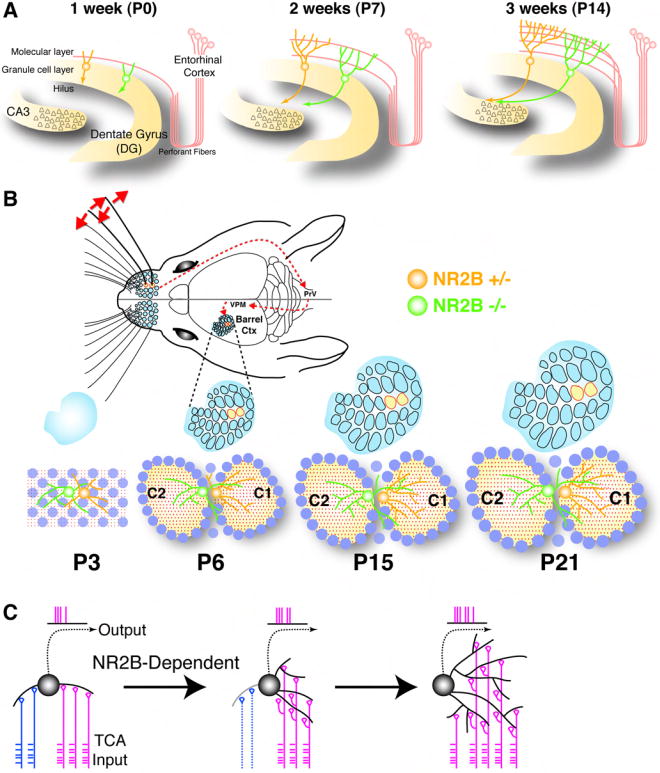
Figure 7. Summary and Model for NR2B Function in Dendrite Patterning.
(A) Schematic summary of dendrite development of a control (yellow) and an NR2B-/- (green) dGC born at E13.5. Pruning of excessive primary dendrite 1-3 weeks after birth (P0-P14) in normal dGCs correlates with the arrival of entorhinal cortical input. This pruning does not occur in NR2B-/- dGCs. Schematic modified after (Zhao et al., 2008).
(B) Schematic summary of dendrite development of a control (yellow) and an NR2B-/- (green) bSC in the developing barrel cortex. At P3, bSCs (blue circles) have not organized into barrels; thalamocortical axons (red dots) are yet to sort out their innervation patterns; normal and NR2B-/- cells exhibit similar degree of asymmetry. At P6, when barrel pattern becomes first apparent, normal cells already direct vast majority of dendrites towards the primary barrel whereas NR2B-/- cells fail to do so. In the next 9 days, normal cells continue to grow their dendritic trees, but limited to the primary barrels. NR2B-/- cells also grow their dendritic trees at the same rate, but in the primary barrel as well as in secondary barrel(s) and septae between barrels. Schematic of the whisker-barrel system modified after (Petersen, 2007).
(C) Model for NR2B-dependent dendrite patterning of a bSC. Dendritic branches are initially contacted by thalamocortical afferents (TCAs) representing different whiskers (blue and red). An initial bias causes the bSC to fire according to the red inputs and, in so doing, strengthens synapses and the stability of the dendrites contacted by red inputs while weakens those contacted by the blue inputs via NR2B-dependent Hebbian mechanisms.
NR2B Is Essential for Dendrite Patterning Relevant to Their Function
NR2B is cell-autonomously required for dendrites to acquire patterns appropriate for their physiological function. bSCs provide a clear example: they direct their dendrites towards a single barrel in order to maximize the input from thalamocortical axons representing a single whisker. Loss of NR2B disrupts this pattern, causing individual bSCs to reduce total length of dendrite branches in the primary barrel, and to contact and likely receive input from thalamocortical axons representing multiple neighboring whiskers (Figure 7B). Since summation of weak but synchronous postsynaptic potentials at the synapses between thalamocortical axons and stellate cell dendrites is necessary to drive the firing of stellate cells (Bruno and Sakmann, 2006), the reduction of total dendritic arborization in the primary barrel would reduce the effectiveness of a stellate cell to represent the appropriate whisker. Moreover, spreading the dendrites into multiple barrels and into the septa would degrade the receptive field of stellate cells, compromising spatial discrimination of whisker sensation.
The functional consequence of the multiple primary dendrites in NR2B-/- dGCs is less clear. One can however speculate about the biophysical and computational properties of dendrites, based on analogy with bSCs. dGC cell bodies are the preferred targets of major inhibitory interneurons (Freund and Buzsaki, 1996) and may present impediments to interactions between inputs onto dendritic trees derived from different primary dendrites. Branches that originate from the same primary dendrite could, given only moderate attenuation of EPSPs (Schmidt-Hieber et al., 2007), summate their synaptic input more effectively to drive the firing of dGCs than branches originating from different primary dendrites. Multiple primary dendrites in a single dGC could thus effectively behave as different compartments, each integrating input independently. The fact that a small fraction of dGCs has multiple primary dendrites in normal animals suggests that this type of dendrite pattern may be useful for some aspect of hippocampal information processing. Loss of NR2B expands this population of dGCs and could thus alter information transfer from the entorhinal cortex to the hippocampus.
Developmental Mechanisms of NR2B Action in Dendrite Patterning
Comparison of the developmental time course of NR2B-/- and NR2B+/- dGCs and bSCs (Figures 7A and 7B) reveals a common feature: NR2B-/- cells cannot prune dendritic branches that might detect minor input not coincident with the input detected by the major dendritic trees. For both cell types, NR2B-mediated pruning of dendrites coincides with the development of afferents and synaptogenesis. In bSCs, NR2B is required for pruning of mistargeted dendrites between P3 and P9. During this period, thalamocortical afferents targeting cortical layer 4 develop into the characteristic barrel pattern: they segregate into dense terminal clusters by selective arborization of collateral branches in the primary barrel, while simultaneously pruning the branches extending to secondary barrels (Erzurumlu and Jhaveri, 1990; Senft and Woolsey, 1991; Schlaggar and O’Leary, 1994). Similarly, dGC dendrite pruning correlates with the arrival of ipsilateral associational and commissural entorhinal afferents in the first postnatal week (Fricke and Cowan, 1977).
We propose that NR2B-mediated dendrite patterning follows Hebb’s rule and its extension. “When an axon of cell A is near enough to excite a cell B and repeatedly and persistently takes part in firing it, some growth or metabolic change takes place in one or both cells such that A’s efficiency, as one of the cells firing B, is increased” (Hebb, 1949). Conversely, “when the presynaptic axon of cell A repeatedly and persistently fails to excite the postsynaptic cell B while cell B is firing under the influence of other presynaptic axons, metabolic change takes place in one or both cells such that A’s efficiency, as one of the cells firing B, is decreased” (Stent, 1973). Although Hebb’s rule has mostly been applied to adjusting synaptic strength, in the developing brain, dendritic growth and branch stabilization are tightly associated with, and perhaps a consequence of, synapse formation on the dendrites (Vaughn, 1989; Niell et al., 2004). Thus, Hebb’s rule can be readily extended to dendritic growth: strengthening of synapses leads to stabilization and extension of dendrites; weakening of synapses leads to destabilization and elimination of dendritic branches (reviewed in Cline and Haas, 2008).
Calcium influx through the NMDAR channel relies on simultaneous release of glutamate from presynaptic neurons and depolarization of postsynaptic neurons to relieve the magnesium block (Nowak et al., 1984). NMDAR is therefore an ideal candidate to execute Hebb’s rule as a molecular coincidence detector of correlated pre- and postsynaptic activity. In addition, the long decay time of NR2B-containing NMDARs allows greater calcium influx and a wider window of coincidence detection. Thus, we envision (Figure 7C) that in normal bSCs, dendritic branches that extend to the primary barrel would receive correlated input from thalamocortical axons representing the same whisker, driving them to fire and, consequently, strengthen their synapses and stabilize their dendritic growth. At the same time, dendrites extending to neighboring barrels or septa would receive input uncorrelated with the primary whisker input. Initial selection of the primary barrel for a particular bSC is probably the result of stochastic or genetically controlled biases at the outset. However, once one of the barrels starts providing sufficiently more input than others, the other inputs coming from non-primary barrels are not as effective in driving the bSCs to fire. The synapses in non-primary barrels are weakened over time according to the extension of Hebb’s rule, and as a consequence, the dendrites corresponding to “weaker” input barrels are destabilized and pruned. In NR2B-/- cells, the ability to detect coincidence is severely compromised, thus the “reward” program to strengthen growth in the primary barrel, and perhaps more importantly, the “punishment” program to prune dendrites in the non-primary barrel, cannot be properly executed.
NMDARs could regulate dendrite growth and pruning through strengthening and weakening of synapses using mechanisms akin to those intensely studied in long-term synaptic potentiation and depression (reviewed in Malenka and Bear, 2004). In addition, calcium entry through NMDARs can affect dendrite development through local action on the cytoskeleton and through transcriptional programs (reviewed in Konur and Ghosh, 2005; Zheng and Poo, 2007). The calcium-induced transcriptional programs require the discrimination of dendrites that are to be strengthened or weakened, but this can in principle be achieved by the tagging mechanism proposed in protein synthesis-dependent long-term synaptic plasticity (Frey and Morris, 1997; Martin et al., 1997).
Although we cannot rule out the possibility that the dendrite patterning phenotypes are caused by a reduction of total NMDAR activity, rather than a specific loss of NR2B, we favor the latter possibility. In the barrel cortex, NR2A, the other major NR2 subunit, is not prominently expressed until P7 (Liu et al., 2004), after the patterning defect is already evident (Figure 6). Moreover, NR2B couples to distinct intracellular scaffolding molecules (reviewed in van Zundert et al., 2004) and signal transduction pathways as compared to other NR2 subunits. For instance, NR2B but not NR2A or NR1 form a complex with autophosphorylated CaMKII (Strack and Colbran, 1998) that sustains calcium/calmodulin independent CaMKII kinase activity (Bayer et al., 2001). Interestingly, CaMKII phosphorylates NeuroD at Ser336, a site conserved in NeuroD2 (Gaudilliere et al., 2004), and NeuroD2 has been shown to be essential for barrel cortex pattern formation (Ince-Dunn et al., 2006).
NR2B Is Not Essential for General Dendrite Growth and Branching
Despite profound dendrite patterning defects, the dendrites of NR2B-/- dGCs and bSCs grow to their normal length and acquire normal number of branch points. Although it is possible that NR2B may regulate dynamics of branch elongation and retraction at a fast time-scale, developmental studies at intervals of every few days reveal a similar rate of dendrite elongation and branch addition. The difference between our findings and previous studies implicating NMDAR function in promoting (e.g., Rajan and Cline, 1998; Sin et al., 2002) or limiting (e.g., Luthi et al., 2001; Datwani et al., 2002) dendrite growth can be accounted for by the following two factors. First, non-autonomous effects of NMDAR perturbation may affect dendrite growth. For example, cortex-specific knockout of NR1 also affects thalamocortical axon terminal arborization (Lee et al., 2005), which can in turn affect stellate cell dendrite growth. Second, other NR2 subunits may contribute to dendrite growth. For example, even though NR2B is highly expressed in layer 4 barrel cortex, with a 2-fold increase in expression from P2 to P7, NMDAR consisting of NR2A subunits are expressed from P7 onward (Liu et al., 2004), and these NR2A-containing NMDARs could regulate general dendrite growth and branching. Consistent with this interpretation, a recent study reported that knockdown of NR2A, but not NR2B, in isolated Xenopus tectal neurons causes dendrite growth defects (Ewald et al., 2008). Future genetic mosaic studies on genes encoding the obligate NR1 subunit should help address these possibilities.
As discussed in the previous section, regulation of dendrite patterning by NMDARs must eventually be realized through local growth or elimination of dendritic branches according to their activity patterns. Thus, the fact that NR2B mutant cells have profound patterning defects without affecting the total dendritic length and branching points imply the existence of an NR2B-independent homeostatic program that regulates the size of dendritic trees appropriate for particular cell types.
Mosaic Analysis in Mammalian Neural Development
Our study illustrates the utility of the MADM system to reveal cell-autonomous functions of important genes in mammalian neural development. Although Cre-mediated excision of floxed alleles can be used for tissue-specific knockout of genes to bypass lethality and determine tissue autonomy, it is difficult to create and label sparse knockout cells to address cell autonomy in a rigorous manner. MADM, as well as the recently developed SLICK method (Young et al., 2008), permit the generation of isolated single mutant cells, while at the same time uniquely labeling these cells so their morphology and physiological properties can be analyzed in an otherwise normal tissue. The limitation of the current MADM system is that candidate genes must be located distal to ROSA26 on chromosome 6, where the original MADM cassettes were inserted (Zong et al., 2005). We are in the process of creating MADM cassettes on other mouse chromosomes so that more genes can be subjected to similar mosaic analysis.
EXPERIMENTAL PROCEDURES
Tissue preparation and 5-Chloro-2-deoxyuridine analysis
All animal procedures were based on animal care guidelines and were approved by the Stanford University Administrative Panels on Laboratory Care. Tissue preparation and 5-chloro-2-deoxyuridine (CldU; Sigma: Cat. C6891) staining and analysis was performed according to Espinosa and Luo (2008).
Dendrite reconstructions and analysis
Serial 60-μm sagittal sections of hippocampus and tangential sections of barrel cortex were immunostained and imaged through a 40x (1.3 NA) oil objective by 1 μm optical sectioning using a Zeiss 510 confocal microscopy (Carl Zeiss Inc., Oberkochen, Germany). For hippocampus, isolated P21 dGCs (<4 cells per 60-μm section) were selected that met the following criteria: 1) cell body in the upper 1/3 of the granule cell layer, 2) cell located in the suprapyramidal blade, 3) dendritic tree could be followed to the top of the molecular layer, and 4) mature spines are present. Hippocampal dGCs would traverse approximately four 60-μm thick sections; dendrites within each section were traced in 3D using the Imaris 6.2 software (Bitplane), and then the whole dendritic tree was reconstructed by manually aligning and stitching the sections together. For barrel cortex, isolated bSCs (<5 cells per 60-μm section in the barrel field) located at the edge of the barrel boundary were randomly selected for quantification. Using DAPI as a counterstain and GFP autofluorescence, the barrel boundary was identifiable at low magnification (10x objective). bSCs traverse no more than two sections. Each confocal image was traced with Imaris and sequentially aligned and stitched together to obtain a complete 3D reconstruction. Following 3D reconstructions, number of primary dendrites, dendrite length, branch point number, 3D-Sholl analysis, and branch order complexity were quantified using Imaris. Additionally, 3D reconstructions were superimposed over low magnification DAPI and GFP images. Using Imaris, branch segments are separated into the primary barrel, septae (cell dense region in between barrel centers negative for GFP autofluorescence), or secondary barrel(s) compartments. Dendritic segments within each compartment were quantified for total length, total branch point number, and number of primary dendrites.
Supplementary Material
Acknowledgments
We thank G. Westbrook for NR2B knockout mice, R. Kageyama for nestin-CreER mice, and W. Zhong for nestin-Cre mice. We thank many colleagues for helpful advice and discussion, in particular B. Barres, S. McConnell, M. Schnitzer, and members of the Luo laboratory. We thank K. Svoboda, B. Tasic, K. Miyamichi, S. Hippenmeyer, Y. Chou, and J. Leong for comments on the manuscript. J. Zhong and C. Manalac for technical assistance. J.S.E. is a predoctoral Ruth L. Kirschstein National Research Service Award fellow. L.L. is a Howard Hughes Medical Institute (HHMI) investigator. This work was supported by National Institutes of Health (NIH) grants GM58234 (R.W.T) and NS050835 (L.L.).
Footnotes
SUPPLEMENTAL DATA
The Supplemental Data include Supplemental Results, Experimental Procedures and two Figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. The Journal of comparative neurology. 1990;301:365–381. doi: 10.1002/cne.903010304. [DOI] [PubMed] [Google Scholar]
- Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- Bruno RM, Sakmann B. Cortex is driven by weak but synchronously active thalamocortical synapses. Science (New York, N.Y) 2006;312:1622–1627. doi: 10.1126/science.1124593. [DOI] [PubMed] [Google Scholar]
- Claiborne BJ, Amaral DG, Cowan WM. Quantitative, three-dimensional analysis of granule cell dendrites in the rat dentate gyrus. J Comp Neurol. 1990;302:206–219. doi: 10.1002/cne.903020203. [DOI] [PubMed] [Google Scholar]
- Cline H, Haas K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. The Journal of physiology. 2008;586:1509–1517. doi: 10.1113/jphysiol.2007.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Current opinion in neurobiology. 2001;11:118–126. doi: 10.1016/s0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Cline HT, Constantine-Paton M. NMDA receptor agonist and antagonists alter retinal ganglion cell arbor structure in the developing frog retinotectal projection. J Neurosci. 1990;10:1197–1216. doi: 10.1523/JNEUROSCI.10-04-01197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine-Paton M, Cline HT, Debski E. Patterned activity, synaptic convergence, and the NMDA receptor in developing visual pathways. Annual review of neuroscience. 1990;13:129–154. doi: 10.1146/annurev.ne.13.030190.001021. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- Datwani A, Iwasato T, Itohara S, Erzurumlu RS. NMDA receptor-dependent pattern transfer from afferents to postsynaptic cells and dendritic differentiation in the barrel cortex. Molecular and cellular neurosciences. 2002;21:477–492. doi: 10.1006/mcne.2002.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebralidze AK, Rossi DJ, Tonegawa S, Slater NT. Modification of NMDA receptor channels and synaptic transmission by targeted disruption of the NR2C gene. J Neurosci. 1996;16:5014–5025. doi: 10.1523/JNEUROSCI.16-16-05014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Erzurumlu RS, Jhaveri S. Thalamic axons confer a blueprint of the sensory periphery onto the developing rat somatosensory cortex. Brain Res Dev Brain Res. 1990;56:229–234. doi: 10.1016/0165-3806(90)90087-f. [DOI] [PubMed] [Google Scholar]
- Espinosa JS, Luo L. Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J Neurosci. 2008;28:2301–2312. doi: 10.1523/JNEUROSCI.5157-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald RC, Van Keuren-Jensen KR, Aizenman CD, Cline HT. Roles of NR2A and NR2B in the development of dendritic arbor morphology in vivo. J Neurosci. 2008;28:850–861. doi: 10.1523/JNEUROSCI.5078-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13:325–338. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- Fricke R, Cowan WM. An autoradiographic study of the development of the entorhinal and commissural afferents to the dentate gyrus of the rat. The Journal of comparative neurology. 1977;173:231–250. doi: 10.1002/cne.901730203. [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Green EJ, Juraska JM. The dendritic morphology of hippocampal dentate granule cells varies with their position in the granule cell layer: a quantitative Golgi study. Experimental brain research. Experimentelle Hirnforschung. 1985;59:582–586. doi: 10.1007/BF00261350. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. The Journal of biological chemistry. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hebb DO. The Organization of Behavior: A Neuropsychological Theory. New York: John Wiley; 1949. [Google Scholar]
- Hickmott PW, Merzenich MM. Dendritic bias of neurons in rat somatosensory cortex associated with a functional boundary. The Journal of comparative neurology. 1999;409:385–399. [PubMed] [Google Scholar]
- Ikeda K, Araki K, Takayama C, Inoue Y, Yagi T, Aizawa S, Mishina M. Reduced spontaneous activity of mice defective in the epsilon 4 subunit of the NMDA receptor channel. Brain Res Mol Brain Res. 1995;33:61–71. doi: 10.1016/0169-328x(95)00107-4. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Ohtsuka T, Metzger D, Chambon P, Kageyama R. Temporal regulation of Cre recombinase activity in neural stem cells. Genesis. 2006;44:233–238. doi: 10.1002/dvg.20212. [DOI] [PubMed] [Google Scholar]
- Inan M, Crair MC. Development of cortical maps: perspectives from the barrel cortex. Neuroscientist. 2007;13:49–61. doi: 10.1177/1073858406296257. [DOI] [PubMed] [Google Scholar]
- Ince-Dunn G, Hall BJ, Hu SC, Ripley B, Huganir RL, Olson JM, Tapscott SJ, Ghosh A. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron. 2006;49:683–695. doi: 10.1016/j.neuron.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Kadotani H, Hirano T, Masugi M, Nakamura K, Nakao K, Katsuki M, Nakanishi S. Motor discoordination results from combined gene disruption of the NMDA receptor NR2A and NR2C subunits, but not from single disruption of the NR2A or NR2C subunit. J Neurosci. 1996;16:7859–7867. doi: 10.1523/JNEUROSCI.16-24-07859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M, et al. Molecular diversity of the NMDA receptor channel. Nature. 1992;358:36–41. doi: 10.1038/358036a0. [DOI] [PubMed] [Google Scholar]
- Kutsuwada T, Sakimura K, Manabe T, Takayama C, Katakura N, Kushiya E, Natsume R, Watanabe M, Inoue Y, Yagi T, et al. Impairment of suckling response, trigeminal neuronal pattern formation, and hippocampal LTD in NMDA receptor epsilon 2 subunit mutant mice. Neuron. 1996;16:333–344. doi: 10.1016/s0896-6273(00)80051-3. [DOI] [PubMed] [Google Scholar]
- Lee LJ, Iwasato T, Itohara S, Erzurumlu RS. Exuberant thalamocortical axon arborization in cortex-specific NMDAR1 knockout mice. The Journal of comparative neurology. 2005;485:280–292. doi: 10.1002/cne.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell. 1994;76:427–437. doi: 10.1016/0092-8674(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Liu XB, Murray KD, Jones EG. Switching of NMDA receptor 2A and 2B subunits at thalamic and cortical synapses during early postnatal development. J Neurosci. 2004;24:8885–8895. doi: 10.1523/JNEUROSCI.2476-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi A, Schwyzer L, Mateos JM, Gahwiler BH, McKinney RA. NMDA receptor activation limits the number of synaptic connections during hippocampal development. Nature neuroscience. 2001;4:1102–1107. doi: 10.1038/nn744. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science (New York, N.Y) 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science (New York, N.Y) 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Luo L, Zong H. Modeling sporadic loss of heterozygosity in mice by using mosaic analysis with double markers (MADM) Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4495–4500. doi: 10.1073/pnas.0606491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ. In vivo imaging of synapse formation on a growing dendritic arbor. Nature neuroscience. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annual review of neuroscience. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Petersen CC. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Rajan I, Cline HT. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci. 1998;18:7836–7846. doi: 10.1523/JNEUROSCI.18-19-07836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthazer ES, Akerman CJ, Cline HT. Control of axon branch dynamics by correlated activity in vivo. Science (New York, N.Y) 2003;301:66–70. doi: 10.1126/science.1082545. [DOI] [PubMed] [Google Scholar]
- Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373:151–155. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- Schlaggar BL, O’Leary DD. Early development of the somatotopic map and barrel patterning in rat somatosensory cortex. The Journal of comparative neurology. 1994;346:80–96. doi: 10.1002/cne.903460106. [DOI] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Jonas P, Bischofberger J. Subthreshold dendritic signal processing and coincidence detection in dentate gyrus granule cells. J Neurosci. 2007;27:8430–8441. doi: 10.1523/JNEUROSCI.1787-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Luo L. How do dendrites take their shape? Nature neuroscience. 2001;4:359–365. doi: 10.1038/86006. [DOI] [PubMed] [Google Scholar]
- Senft SL, Woolsey TA. Growth of thalamic afferents into mouse barrel cortex. Cereb Cortex. 1991;1:308–335. doi: 10.1093/cercor/1.4.308. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. Journal of anatomy. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Stent GS. A physiological mechanism for Hebb’s postulate of learning. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:997–1001. doi: 10.1073/pnas.70.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Colbran RJ. Autophosphorylation-dependent targeting of calcium/ calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl-D-aspartate receptor. The Journal of biological chemistry. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. J Neurosci. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultanir SK, Kim JE, Hall BJ, Deerinck T, Ellisman M, Ghosh A. Regulation of spine morphology and spine density by NMDA receptor signaling in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19553–19558. doi: 10.1073/pnas.0704031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert B, Yoshii A, Constantine-Paton M. Receptor compartmentalization and trafficking at glutamate synapses: a developmental proposal. Trends in neurosciences. 2004;27:428–437. doi: 10.1016/j.tins.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Vaughn JE. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse (New York, N.Y) 1989;3:255–285. doi: 10.1002/syn.890030312. [DOI] [PubMed] [Google Scholar]
- Vega CJ, Peterson DA. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nature methods. 2005;2:167–169. doi: 10.1038/nmeth741. [DOI] [PubMed] [Google Scholar]
- von Engelhardt J, Doganci B, Jensen V, Hvalby O, Gongrich C, Taylor A, Barkus C, Sanderson DJ, Rawlins JN, Seeburg PH, et al. Contribution of Hippocampal and Extra-Hippocampal NR2B-Containing NMDA Receptors to Performance on Spatial Learning Tasks. Neuron. 2008;60:846–860. doi: 10.1016/j.neuron.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Williams K. Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Molecular pharmacology. 1993;44:851–859. [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nature reviews. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Woolsey TA, Dierker ML, Wann DF. Mouse SmI cortex: qualitative and quantitative classification of golgi-impregnated barrel neurons. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:2165–2169. doi: 10.1073/pnas.72.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey TA, Van der Loos H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain research. 1970;17:205–242. doi: 10.1016/0006-8993(70)90079-x. [DOI] [PubMed] [Google Scholar]
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nature neuroscience. 2008;11:721–728. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zheng JQ, Poo MM. Calcium signaling in neuronal motility. Annual review of cell and developmental biology. 2007;23:375–404. doi: 10.1146/annurev.cellbio.23.090506.123221. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.