Abstract
The proteins of the Bcl-2 family are important regulators of apoptosis, or programmed cell death. These proteins regulate this fundamental biological process via the formation of heterodimers involving both pro- and anti-apoptotic family members. Disruption of the balance between anti- and pro-apoptotic Bcl-2 proteins is the cause of numerous pathologies. Bcl-xl, an anti-apoptotic protein of this family, is known to form heterodimers with multiple pro-apoptotic proteins, such as Bad, Bim, Bak, and Bid. To elucidate the molecular basis of this recognition process, we used molecular dynamics simulations coupled with the Molecular Mechanics/Poisson-Boltzmann Surface Area approach to identify the amino acids that make significant energetic contributions to the binding free energy of four complexes formed between Bcl-xl and pro-apoptotic Bcl-2 homology 3 peptides. A fifth protein-peptide complex composed of another anti-apoptotic protein, Bcl-w, in complex with the peptide from Bim was also studied. The results identified amino acids of both the anti-apoptotic proteins as well as the Bcl-2 homology 3 (BH3) domains of the pro-apoptotic proteins that make strong, recurrent interactions in the protein complexes. The calculations show that the two anti-apoptotic proteins, Bcl-xl and Bcl-w, share a similar recognition mechanism. Our results provide insight into the molecular basis for the promiscuous nature of this molecular recognition process by members of the Bcl-2 protein family. These amino acids could be targeted in the design of new mimetics that serve as scaffolds for new antitumoral molecules.
Introduction
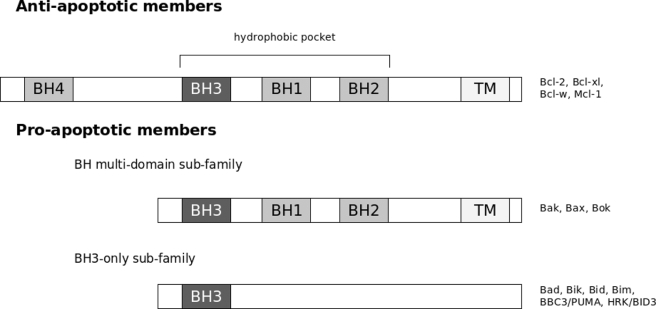
The Bcl-2 protein family plays a pivotal role in the regulation of apoptosis (1, 2). The protein family is made up of two classes, one that contains those proteins that suppress cell death and the other that contains those proteins that promote cell death. In the former class are anti-apoptotic proteins such as Bcl-2 and Bcl-xl, and in the latter class are pro-apoptotic proteins such as Bak and Bax. The Bcl-2 proteins are characterized by regions of specific sequence homology termed Bcl-2 homology (BH)3 domains that number from 1 to 4 and that are critical for function (see Fig. 1).
FIGURE 1.
Schematic representation of the Bcl-2 homology domains of the different Bcl-2 proteins.
Particularly prominent is the BH3 domain, which has been identified as a mediator of cell death and has led to the identification of a family of proteins that contain only this domain (see Table 1). Referred to as BH3-only proteins (BOP), these proteins are believed to be part of a regulatory network serving to integrate cell survival and death signals. The importance of BH3 domains as mediators of cell death was identified in studies aimed at determining the molecular requirements for the interaction of the pro-apoptotic Bcl-2 family member Bak with the cell death suppressor Bcl-xl (3). It was found that only the BH3 domain of Bak was required for its physiological activity and, in particular, for its interaction with Bcl-xl. Subsequent sequence analysis identified homologous domains in other pro-apoptotic proteins such as Bax, Bad, and Bik. Other members of the BOP family include the proteins Bid, Bim, Bik, Bmf, Puma Noxa, and Hrk (4).
TABLE 1.
BH3 sequence identities of the Bcl-2 proteins
Although much remains to be understood of the detailed mechanisms of apoptosis, experimental results suggest that the BH3-only proteins regulate cell death by binding to Bcl-2-type cell death suppressors such as Bcl-2 or Bcl-xl via the BH3 domain resulting in the eventual release of cytochrome c from the mitochondria and subsequent activation of caspases (3, 5). Given that the BH3-only proteins are key modulators of survival and death signals, these proteins are important targets in the development of therapeutics for the treatment of diseases where cell-death regulation breaks down, such as cancer, certain neurodegenerative diseases, and heart disease. One goal of therapeutic design is to develop compounds that activate a BOP-like response and trigger cell death by overcoming inappropriate death suppressor function in these cells. This approach was shown to be valid by the preclinical and clinical studies of antisense strategies toward Bcl-2 anti-apoptotic proteins (6–8). However, antisense molecules present intrinsic problems such as in vivo instability and toxicity. Modified BH3 peptides seem to have more suitable pharmacological potential. In fact, BH3 peptides constrained to an α-helical conformation (9, 10) or mimicked by β-amino acids (11, 12) have been used to induce the apoptosis. BH3 peptides derived from Bid have also demonstrated their ability to activate Bax in vitro (10). Several studies have shown that small molecules that bind to the hydrophobic groove of anti-apoptotic proteins are able to induce or restore apoptosis and have the potential to be used as anticancer drugs. In one study, Oltersdorf et al. (13) found that ABT-737, a thioethylamino-2–4-dimethylphenyl analog, causes tumor regression by triggering apoptosis by binding to the anti-apoptotic protein Bcl-xl. Recently, it has been demonstrated that small molecules antagonizing the anti-apoptotic Bcl-2 proteins are able to overcome the drug resistance that results from the overexpression of anti-apoptotic Bcl-2 proteins (14). The overexpression of anti-apoptotic Bcl-2 proteins is believed to be a general mechanism for the development of chemotherapy resistance.
It is recognized that anti-apoptotic proteins, such as Bcl-xl, have a significant number of binding partners. To better understand the mechanism of regulation, it is important to establish the principles that govern this recognition process. A concept that has emerged over the past number of years is that of the interaction energy hot spot (15, 16). The idea is that only a handful of amino acids at the binding interface make the dominant contribution to the binding affinity. Indeed, mutation studies, mostly alanine scanning studies, have shown that only a few mutations affect the free energy of binding by more than 2 kcal·mol−1 (17). Computational methods have been developed to identify interaction energy hot spots, thus providing detailed insight into the energetics of protein complex formation (18–25). Referred to as the Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) or Molecular Mechanics/Generalized Born Surface Area methods (26), these computational approaches rely on empirical free energy functions (20, 21) and the use implicit representations of the solvent to reduce the computational demands. Complementary to experiments, these physically based models permit the determination of structural and energetic consequences of mutations, as well as yielding insight into the intermolecular interactions that drive binding. Here we applied the MM/PBSA method to investigate the factors that give rise to promiscuous binding by the Bcl-xl anti-apoptotic protein. We used a protocol presented earlier (24) to decompose the free energy of binding into individual amino acid contributions to map out the interactions that have a dominant role in the formation of these complexes, i.e. between anti-apoptotic proteins (Bcl-xl and Bcl-w) and BH3 peptides derived from the pro-apoptotic proteins Bid (27), Bad (28), Bim (30), and Bak (31). We identified consensus interactions that were present in all the complexes studied, suggesting that these interactions are at the basis of the promiscuous recognition process observed in the anti-apoptotic proteins, such as Bcl-xl. Some differences were observed between the different peptides that may have consequences for specificity. Knowledge of these energetic hot spot interactions is of general interest for the development of small molecule mimics of the BH3-only peptide.
MATERIALS AND METHODS
In this study, we use a protocol based on the MM/PBSA method (22, 24, 26, 32) where conformations extracted from molecular dynamics simulations are processed using a simplified description for the solvent to yield an estimate of binding free energy. Individual contributions of each amino acid to the complex formation are estimated, and important energetic amino acid “hot spots” are identified.
Structures
The coordinates of five Bcl-xl/BH3 peptide complexes were obtained from the Protein Data Bank (33). These include the following two structures of Bcl-xl in complex with BH3 peptides from the BOP protein Bad (PDB codes 1G5J (28) and 2BZW) that differ in the length of the BH3 peptides: one structure of Bcl-xl in complex with the BH3 peptide from Bak (PDB code 1BXL) (31), and one structure of the complex Bcl-xl and the BH3 peptide from Bid (PDB code 1PQ1) (30). The structures 2BZW and 1PQ1 are of mouse Bcl-xl in complex with the BH3 peptides from mouse Bad and Bid, respectively, rather than the human analogs. Given the high sequence homology with the human isoforms, these structures were used to develop homology models of the human complex, see below. The fifth structure studied was of the anti-apoptotic protein Bcl-w in interaction with the BH3 peptide from Bim (PDB code 1ZY3) (27). Throughout this study, the different structures, and their respective simulations, will be referred to by their PDB identifiers, see Table 2 for details.
TABLE 2.
Details of the experimental structures
| PDB code | Experimental method | Organism | Bcl-2 protein | BH3 peptide |
|---|---|---|---|---|
| 1G5J | NMR | Human | Bcl-xl | Bad |
| 2BZW | X-ray (2.3 Å) | Mouse | Bcl-xl | Bad |
| 1PQ1 | X-ray (1.65 Å) | Mouse | Bcl-xl | Bim |
| 1BXL | NMR | Human | Bcl-xl | Bak |
| 1ZY3 | NMR | Human | Bcl-w | Bid |
Homology Modeling
The alignment of the Bcl-xl sequence of mouse and human shows an identity rate of 97%. The difference arises from five residues, none of which are located near the BH3 peptide binding groove. This high identity rate led us to construct a model of human Bcl-xl complexed to BH3 peptides Bim and Bad. However, the mouse structures were determined in the absence of the loop between the α-helix 1 and α-helix 2 (i.e. residues 29–81 for 2BZW and residues 28–78 for 1PQ1). Human homology models were built for the 2BZW and 1PQ1 complexes using 1G5J as a template structure and version 9.2 of MODELLER program (34). Fifty models were generated and refined. The lowest energy model proposed by the MODELLER energy function was used as the starting structure for the MD simulations of these two complexes.
Determination of Protonation State
The pKa values of titratable groups were determined for both isolated proteins and complexes using continuum electrostatics as described by Schaefer et al. (35). Using the University of Houston Brownian Dynamics program (36), the titration curves were obtained using a series of c-shell scripts provided by M. Schaefer.4 The protonation states of selected residues were determined at a physiological pH of 7.4. The dielectric constant for the solvent was 80. Four values of the dielectric constant for the protein were tested (4, 8, 12, and 20), and all gave similar results. It was found that all amino acids were in their standard protonation state, and all His were neutral.
Molecular Dynamics Simulations
Molecular dynamic simulations were done using the CHARMM program (37) employing the all-atom parameter set of CHARMM22 (38) with two-dimensional dihedral cross-term energy correction map corrections (39). Hydrogen atoms were added to the crystal structure using the HBUILD module (40) after imposing the protonation states determined by the previously described pKa calculations. The terminal residues of the proteins were built in their charged state (NH3+ and COO−), whereas the BH3 peptide termini were neutralized using the acetyl group and N-methylamide for the N and C termini, respectively. The protein-peptide complexes were solvated in pre-equilibrated box of TIP3P water (41, 42). The size of the water box was chosen to have a minimal distance of 10 Å between the atoms of the complex and the extremities of the water box. Water molecules closer than 2.8 Å to any of the protein or peptide heavy atoms were deleted.
Molecular dynamics simulations were performed at 300 K with periodic boundary conditions. Bond lengths involving bonds between heavy atoms and hydrogen atoms were constrained using the SHAKE algorithm (43). The nonbonded van der Waals interactions were truncated at a cutoff distance of 12.5 Å using a vshift function (44). Electrostatic interactions were truncated at 14.0 Å using an fswitch function (44). The minimization phase was divided into two steps. During the first step, the protein was fixed, and the water was energy-minimized by 100 steps of steepest descent algorithm followed by 400 steps using the adopted basis Newton Raphson algorithm. During the second step, the water was fixed, and the protein was minimized using both the same algorithms and same number of steps as above. The constraints were then removed, and the entire system was minimized with 500 and 3000 steps of steepest descent and adopted basis Newton Raphson, respectively. The system was gradually heated over 15 ps to reach 300 K. The system was equilibrated in two 20-ps stages. In the early equilibration, velocities were assigned every 50 steps according to a Gaussian distribution function, and in the late equilibration, the velocities were scaled by a single factor only when the average temperature was lying outside the 300 ± 10 K window. A time step of 2 fs was used, and the simulation time was 2 ns for each system. The purpose of the simulations was to generate multiple structures around the experimental structure to improve the statistical sampling of the MM/PBSA analysis.
Free Energy Decomposition of Interactions between Bcl-xl(w) and the BH3 Peptides
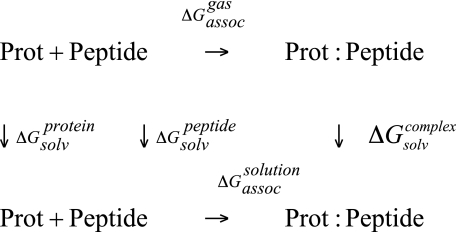
To obtain a semi-quantitative estimate of the contributions of all amino acids to the binding free energy for the formation of the Bcl-xl(w)-BH3 peptide complex, a molecular free energy decomposition scheme based on the MM/PBSA analysis was performed, following the approach presented by Lafont et al. (24). From this analysis, an estimation of the free energy of binding for molecular complexes can be obtained. Briefly, in the MM/PBSA approach, the free energy is estimated using a standard thermodynamic cycle of the form shown in Scheme 1, where the binding free energy is calculated according to Equation 1,
where ΔEMMgas is the difference in the gas phase energy; ΔSMM is the change in entropy upon complex formation, and ΔGsolvation is the change in solvation free energy. The gas phase energy differences contain terms for the intermolecular electrostatic and van der Waals energies, as well as the equivalent internal energy terms. These terms are based on the CHARMM force field in the present approach. The solvation free energy is divided into two contributions, an electrostatic and a nonpolar contribution. This latter term is approximated by an empirical relationship based on solvent-accessible surface area, and the electrostatic contribution is calculated here using the Poisson-Boltzmann equation. We verified, as a function of the relevant parameters, that the Poisson-Boltzmann equation, yielded convergent results. As discussed below, the change in entropy upon binding is neglected in the current model. A study by Zoete and Michielin (45) showed that inclusion of entropy can improve agreement with experimental ΔG values but does not necessarily change the global energy profile.
SCHEME 1.
We analyzed the binding free energies in terms of individual energetic contributions. Decomposition of the binding free energy to individual amino acid contributions leads to the identification of amino acids that play a dominant role in binding and can contribute to reliable predictions of the role of particular amino acids in stabilizing complexes.
Several approximations are introduced in the MM/PBSA method. The first was the neglect of conformational change upon complex formation, which is dictated by the absence of experimental structures for the unbound protein and peptides. To account for the unbound species in the calculations, their respective structures were obtained from the complex generated during the molecular dynamics simulations. With this approximation, there are no changes to the internal energy terms. The second approximation is the neglect of changes in configuration entropy because of binding. Although these simplifications preclude calculations of absolute values of the binding free energies, they have been shown in previous work to be satisfactory in the context of identifying interaction energy hot spots in protein-protein and protein-ligand complexes. Similar simplifications have been employed in other studies (19, 24, 46). Via this approach, the total binding free energy can be decomposed into individual energetic contributions per residue, yielding a more detailed energetic map of the binding interface.
For the MM/PBSA analysis, the structures are extracted from the molecular dynamics simulations by the clustering method described below. The electrostatic component was calculated using the University of Houston Brownian Dynamics program (36). The atomic charges and radii used for the protein were those of the CHARMM 22 force field (38).
The total free energy and its individual components for each individual system were averaged and weighted based on the cluster populations, i.e. a higher weight was given to conformations extracted from more populated clusters (i.e. the weight of a conformation is proportional to the ratio of the number of conformations with that energy to the total number of conformations extracted from the MD trajectory). Equally, the standard deviation of each amino acid contribution was calculated taking into account the statistical weight of the conformation (47).
Clustering of Conformations
As described in Lafont et al. (24), the different energy terms used in the MM/PBSA method fluctuate significantly with the small structural changes linked to thermal fluctuations. It is therefore necessary to use an ensemble of structures rather than one experimental structure in free energy decompositions based on the MM/PBSA scheme. In this work, we used a protocol that identified structures that would show large variations in the MM/PBSA free energy. We computed the Coulomb interaction energy in vacuo between the protein and the peptide for all conformations saved from the MD trajectory (coordinates were collected every 0.1 ps from the MD) using the CHARMM program. A dielectric constant of 1 and a nonbonded cutoff of 12.5 Å were used with a shift truncation function for electrostatics. The conformations of each trajectory were clustered in 10 groups based on their electrostatic interaction energy. The conformation with vacuum interaction energy closest to the cluster average value was extracted and processed using the MM/PBSA procedure. In all, 10 conformations were extracted from molecular dynamics trajectories in this nonlinear fashion. This procedure reduces the computational cost of performing MM/PBSA calculations while allowing an estimate of the effect of conformational variation on the decomposition analysis. Although the energy used for selecting the conformations is a vacuum electrostatic term, which differs from the solvent term computed by continuum electrostatics, the two terms show a significant statistical anti-correlation, so that conformations that show large variation in vacuum energy also show significant variation in solution energy. Tests using twice the number of structures showed that the free energies that we calculate here are indeed convergent when 10 structures are used (data not shown).
Amino Acid Efficiency
Inspired by the definition of ligand efficiency, which is usually defined as the binding affinity per heavy atoms (48), we defined the amino acid efficiency (24) as the free energy contribution of each amino acid divided by its number of non-hydrogen atoms (i.e. 4 for Gly, 5 for Ala, 14 for Trp, etc.). The amino acid efficiency is a metric for assessing whether the contribution of a particular amino acid is derived from an optimal fit with the protein target or simply as a result of making many contacts.
RESULTS
The interactions between the Bcl-2 proteins are believed to modulate the onset of apoptosis. Given the important biological consequences resulting from the interactions between these proteins, a detailed knowledge of the individual amino acid contribution to the binding provides unique and significant insight that has both fundamental as well as applicative implications. Using an MM/PBSA-based approach that has been described in detail elsewhere (24), we present the energy interaction maps between the anti-apoptotic and pro-apoptotic proteins of the Bcl-2 family. Critical hot spot interactions that give rise to the versatile recognition capability were identified across several different Bcl-2-peptide complexes. In the following sections, we first present the analysis of the molecular dynamics simulations followed by the results of post-simulation analysis using the MM/PBSA approach.
MD Simulations Are Stable
Despite the possibility to perform the MM/PBSA analysis on the individual experimental structures, previous work has shown that more relevant results can be obtained using an ensemble of conformations generated, for example, by a molecular dynamics simulation (24). Here, molecular dynamics simulations of five protein-peptide complexes were done as described under “Materials and Methods.” The simulations were run for up to 2 ns to be able to generate a sampling of conformation in the vicinity of the equilibrium experimental structures. The backbone root mean square difference (RMSD) time series was calculated during the production phase using the respective initial nonminimized experimental structure as the reference structure. For each frame in the trajectory, the complexes were reoriented over the entire backbone. The 1G5J, 1BXL, 1PQ1, and 2BZW complexes show a stable RMSD during the 2.0 ns of the trajectory. After 0.3 ns of simulation time, the RMSD of the 1ZY3 complex (Bcl-w/Bid) complex is also stable. The time necessary to reach a stable RMSD is due to the dynamic behavior of the N-terminal end which, during the early stages of the MD simulation, adopts more folded conformations than those in the initial structures. Overall, the simulations show stable RMSD profiles reaching a plateau values between 1.3 and 2.7 Å.
The root mean square fluctuations of the backbone atoms averaged by residue obtained for each complex are compared with the experimental fluctuations derived from B-factors or calculated from the ensemble of NMR structures (supplemental Fig. S1). The experimental and theoretical fluctuations are in good agreement along the entire protein. Although the loops between the α-helixes show significant flexibility, the fluctuations of the residues within the α-helixes are lower in amplitude. The major differences involve the fluctuations of the N-terminal part and the fluctuations of the loop between α1 and α2. The experimental fluctuations of the N-terminal part are larger than the theoretical fluctuations when the structure was resolved by NMR spectroscopy. Indeed, there are probably few nuclear Overhauser effect data concerning the N-terminal part, and several models are then compatible with the NMR constraints. The long loop between α1 and α2 presents flexibility differences between experimental and calculated fluctuations in 1BXL and 1G5J structures.
Another measure of the stability of the structures during the simulations is the degree to which secondary structural elements present in the experimental structures are maintained. Using the STRIDE program (49) and snapshots from the simulations, the secondary structure was assessed along each simulation. The results show that the secondary structure remains close to the experimental structure with the continuous presence of the α-helices (the time series are shown in supplemental Fig. S2). The structural differences are mainly observed in the loops between the α-helices. The loops are either unstructured or in β-turn conformations in the experimental structures. Additional β-turn or α-helix formation occurs in some loops during the simulations, in particular, in the loop between α1 and α2. This could well be a transient event. Further exploration would require longer simulations, but this question is not within the scope of this work. Based on RMSD. time series, root mean square fluctuations, and on secondary structure stability observed during the trajectories, we conclude that the MD simulations are sufficiently stable for the purpose of extracting structures that exhibit small conformational fluctuations around the experimental structures for the MM/PBSA analysis of the complexes.
Free Energy Decomposition of the Protein Complexes Shows Consistent Hot Spot Interactions
We present here the MM/PBSA results obtained for the complexes formed by anti-apoptotic proteins and peptides derived from the BH3 domains of pro-apoptotic proteins. These results identify amino acids at the interface that make significant contributions to the binding free energy associated with complex formation. In all, simulations of five protein-peptide complexes were run. Four complexes of Bcl-xl with BH3 peptides and one of Bcl-w/Bim were analyzed. The peptides correspond to the BH3 domain from the pro-apoptotic proteins Bad, Bak, Bim, and Bid. For the purpose of this work, we define a hot spot amino acid as one that makes a ±1 kcal·mol−1 contribution to the binding free energy. We first present the amino acids identified as important for the anti-apoptotic proteins, Bcl-xl or Bcl-w, then for the pro-apoptotic peptides.
Free Energy Decomposition of the Contribution of Bcl-xl to the Binding Free Energy
The decomposition of the binding free energy for the complexes composed of the anti-apoptotic protein, Bcl-xl, and the BH3 peptide from pro-apoptotic proteins identified amino acids that contribute more than 1 kcal·mol−1 to the binding energy. The first 4 columns of Table 3 contain the by-residue energies for Bcl-xl; the last column contains the results for Bcl-w (see below). The by-residue results for all amino acids in the protein and the peptides are shown in supplemental Fig. S3, a–d, and the results averaged by residue are shown in supplemental Fig. S4. The energy decomposition for the significant contributions are given in supplemental Tables S1–S5.
TABLE 3.
Amino acids of the anti-apoptotic proteins that contribute significantly to the ΔG of binding with BH3 peptides
| Residue (Bcl-xl) | Domain | Free energy componentab |
||||
|---|---|---|---|---|---|---|
| Bad (1G5J)c | Bad (2BZW)c | Bim (1PQ1)c | Bak (1BXL)c | Bid (1ZY3)d | ||
| Glu92 | BH3 | – | −1.8 | – | – | |
| Glu96 | BH3 | – | 1.7 | −1.4 | – | −1.7 (Glu52) |
| Phe97 | BH3 | −3.4 | −3.4 | −3.5 | −2.9 | −2.3 (Phe53) |
| Arg100 | BH3 | −1.3 | 1.8 | 2.2 | −4.6 | −7.3 (Arg56) |
| Tyr101 | −2.7 | −4.1 | −4.0 | −2.8 | −2.9 (Phe57) | |
| Ala104 | – | −2.4 | – | – | – | |
| Phe105 | – | −1.1 | – | −1.9 | – | |
| Leu108 | – | – | – | – | −1.4 (Leu64) | |
| Gln111 | −2.3 | – | – | – | – | |
| Leu112 | −1.8 | −1.7 | −1.9 | – | −1.5 (Leu68) | |
| His113 | – | −1.0 | – | – | – | |
| Gln125 | – | −2.8 | −1.3 | −2.1 | – | |
| Val126 | −2.9 | −3.0 | −2.9 | −1.7 | −2.0 (Val82) | |
| Glu129 | BH1 | −1.3 | −9.2 | – | – | −5.0 (Glu85) |
| Leu130 | BH1 | −3.3 | −4.6 | −3.5 | −1.7 | −1.5 (Leu86) |
| Asn136 | BH1 | −1.2 | −1.6 | – | – | – |
| Trp137 | BH1 | – | – | – | −2.0 | – |
| Gly138 | BH1 | – | – | – | −1.6 | – |
| Arg139 | BH1 | – | −2.5 | −6.6 | −7.7 | −(Arg95) |
| Ala142 | BH1 | – | – | −1.1 | −1.1 | – |
| Phe146 | BH1 | −1.9 | −1.3 | −1.1 | – | – |
| Glu193 | BH2 | – | – | – | – | −1.7 (Ala149) |
| Leu194 | BH2 | – | −1.6 | – | −1.6 | −2.1 (Leu150) |
| Tyr195 | BH2 | −1.3 | −1.3 | −2.5 | −3.0 | – |
| Ser203 | – | – | – | −1.0 | – | |
| Lys205 | – | – | – | – | −2.7 (Arg161) | |
| Gly206 | – | – | – | −1.6 (Leu162) | ||
a Energies are in kcal·mol−1. Dashes indicate that the |ΔG| for the amino acid was less than 1 kcal·mol−1.
b Hot spot amino acids are denoted by boldface
c Bcl-xl.
d Bcl-w; the corresponding amino acids of Bcl-w are shown in parentheses.
For the two complexes of Bcl-xl/Bad (PDB codes 1G5J and 2BZW), the results are essentially the same for the core interactions. In fact, most of the residues identified as being important contributors to the binding free energy in the complex with the shorter Bad peptide were also identified in the complex with the longer Bad peptide. The exceptions are Gln111, Glu96, and Arg100. For the latter two residues, in the structure 2BZW, the charged side chains of Glu96 and Arg100 are located in the BH3 domain of Bcl-xl where they form an intramolecular salt bridge. As such, they are unable to interact favorably with the Bad peptide. Similarly, variation of Gln111 conformation between the two structures results in a different interaction profile for this amino acid. In the structure of Bcl-xl with Bim (PDB code 1PQ1), the BH3 peptide from Bim has 33 residues, making it the longest peptide in this study. From the decomposition analysis, 12 residues from Bcl-xl were identified as making important contributions to the free energy of binding; these are given in Table 3. Among them, only Arg100 makes an unfavorable contribution due primarily to its large desolvation cost, which is only partially compensated by the interaction with the phenolic group of Tyr22 from the Bim peptide.
Finally, in the complex of Bcl-xl with the Bak peptide (PDB code 1BXL), 14 residues from Bcl-xl show up as hot spots following the free energy decomposition analysis (see Table 3). The BH3 peptide from Bak is the shortest BH3 peptide in this study having only 16 amino acids.
The van der Waals components of the interaction energies are summed by structural domains (see Table 4). This shows that the BH1 and BH3 domains form the majority of the van der Waals contacts in the complexes with the different BH3 peptides.
TABLE 4.
van der Waals energy component of the binding free energy for the different BH domains
| van der Waals component |
|||||
|---|---|---|---|---|---|
| 1G5J | 2BZW | 1PQ1 | 1BXL | 1ZY3 | |
| kcal·mol−1 | |||||
| BH4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| BH3 | −7.6 | −5.5 | −8.6 | −6.3 | −5.7 |
| BH1 | −17.4 | −16.8 | −16.5 | −14.8 | −4.6 |
| BH2 | −3.3 | −5.2 | −7.0 | −4.8 | −1.7 |
Study of the Complex Bcl-w-BH3 Peptide from Bid (PDB Code 1ZY3)
Bcl-w is another anti-apoptotic protein of the Bcl-2 family, which plays an essential role in spermatogenesis (50). Its up-regulation is also associated to the colorectal cancer (51). Bcl-w is functionally similar to Bcl-xl and Bcl-2, but Bcl-w is located exclusively on the mitochondria. This complex is the only one available of an anti-apoptotic protein of the Bcl-2 family other than Bcl-xl at the time of this work. Bcl-w and the BH3 peptide from Bid contain 178 and 20 residues, respectively. From the free energy decomposition, 14 residues making favorable contributions were identified (Table 3; supplemental Fig. S3e). The equivalent residues in the Bcl-xl protein are indicated in Table 3. From the summed van der Waals components (Table 4), we find that for Bcl-w, the BH1 helix makes fewer contacts with the BH3 peptide of the partner protein than in Bcl-xl.
Contributions of Amino Acids from BH3 Peptide
Given that the lengths of the different BH3 peptides bonded to Bcl-xl or Bcl-w are different, only the energetic contributions of the central residues of each BH3 peptides are reported in Table 5. The amino acids are noted alongside the free energy values. The BH3 peptides are localized in a hydrophobic groove at the surface of Bcl-xl or Bcl-w. Most of the residues from the BH3 peptide form favorable interactions with the amino acids from Bcl-xl or Bcl-w (see Table 5), which maintains the stability of the complexes.
TABLE 5.
Total energy and amino acid efficiencies for the central residues of the BH3 peptides
Energies and absolute values of amino acid efficiencies are given in kcal·mol−1.
| Residue no. | BH3 peptides |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1G5J (Bad 25)a |
2BZW (Bad 27)b |
1PQ1 (Bim 33)b |
1BXL (Bak 16)b |
1ZY3 (Bid 20)b |
|||||||||||
| AAb | ΔGAA | Fc | AA | ΔGAA | F | AA | ΔGAA | F | AA | ΔGAA | F | AA | ΔGAA | F | |
| 8 | Ala | −2.3 | 0.5 | Ala | −2.6 | 0.5 | Glu | 1.6 | 0.2 | Ile | −4.0 | 0.5 | |||
| 9 | Gln | −0.4 | 0.0 | Gln | −2.5 | 0.3 | Ile | −2.3 | 0.3 | Gly | −1.0 | 0.3 | Lys | −3.5 | 0.4 |
| 10 | Arg | 1.8 | 0.2 | Arg | −1.0 | 0.1 | Trp | −0.7 | 0.1 | Gln | −0.5 | 0.1 | Asn | 0.3 | 0.0 |
| 11 | Tyr | −1.6 | 0.1 | Tyr | −3.3 | 0.3 | Ile | −3.2 | 0.4 | Val | −1.1 | 0.2 | Ile | −3.5 | 0.4 |
| 12 | Gly | −1.8 | 0.5 | Gly | −2.0 | 0.5 | Ala | −2.7 | 0.5 | Gly | 0.0 | 0.0 | Ala | −1.5 | 0.3 |
| 13 | Arg | −1.3 | 0.1 | Arg | −1.8 | 0.2 | Gln | −0.7 | 0.1 | Arg | 0.8 | 0.1 | Arg | −0.6 | 0.1 |
| 14 | Glu | 0.1 | 0.0 | Glu | 0.1 | 0.0 | Glu | 0.4 | 0.0 | Gln | −2.0 | 0.2 | His | 0.1 | 0.0 |
| 15 | Leu | −5.0 | 0.6 | Leu | −4.7 | 0.6 | Leu | −4.3 | 0.5 | Leu | −5.8 | 0.7 | Leu | −3.4 | 0.4 |
| 16 | Arg | −0.1 | 0.0 | Arg | −3.2 | 0.3 | Arg | −0.6 | 0.1 | Ala | −2.7 | 0.5 | Ala | −2.0 | 0.4 |
| 17 | Arg | −0.7 | 0.1 | Arg | −0.7 | 0.1 | Arg | −1.0 | 0.0 | Ile | −0.8 | 0.1 | Gln | −0.3 | 0.0 |
| 18 | Met | −2.8 | 0.4 | Met | −2.6 | 0.3 | Ile | −2.9 | 0.4 | Ile | −5.0 | 0.6 | Val | −1.8 | 0.3 |
| 19 | Ser | −1.1 | 0.1 | Ser | −0.7 | 0.1 | Gly | −1.9 | 0.5 | Gly | −2.0 | 0.5 | Gly | 0.4 | 0.1 |
| 20 | Asp | −1.1 | 0.1 | Asp | −5.0 | 0.6 | Asp | −4.8 | 0.5 | Asp | −4.1 | 0.5 | Asp | −6.0 | 0.8 |
| 21 | Glu | −4.8 | 0.5 | Glu | −1.4 | 0.2 | Glu | −1.9 | 0.2 | Asp | −2.0 | 0.3 | Ser | −0.7 | 0.1 |
| 22 | Phe | −5.5 | 0.5 | Phe | −5.5 | 0.5 | Phe | −6.2 | 0.6 | Ile | −8.2 | 1.0 | Met | −2.6 | 0.3 |
| 23 | Val | −2.0 | 0.3 | Val | −1.5 | 0.2 | Asn | −3.4 | 0.5 | Asn | −5.4 | 0.7 | Asp | 1.2 | 0.2 |
| 24 | Asp | 0.5 | 0.1 | Asp | 0.7 | 0.1 | Ala | 0.0 | 0.0 | Arg | −3.4 | 0.3 | Arg | −2.7 | 0.2 |
| 25 | Ser | 0.3 | 0.1 | Ser | 0.0 | 0.0 | Tyr | −3.9 | 0.7 | Ser | −4.7 | 0.8 | |||
| 26 | Phe | −4.7 | 0.4 | Phe | −2.8 | 0.3 | Tyr | −6.3 | 0.5 | Ile | −5.4 | 0.7 | |||
a BH3 peptide and its amino acid length are shown.
b AA means amino acid.
c This is the absolute value of the amino acid efficiency.
To facilitate the following discussion, all the residues of the BH3 peptides have been renumbered with respect to the alignment shown in Fig. 2, where counting begins from the first amino acid of the longest peptide (Bad from PDB code 2BZW). Throughout the remainder of this study, we will indicate the position of a particular amino acid of the BH3 peptides by (+)X, where X corresponds to its position with respect to amino acid 1 of the alignment. For example, the first amino acid of the Bim peptide would then be referred to as (+)4 (Fig. 2). Only (+)Leu15 and (+)Asp20 are conserved among all these peptides, therefore these will be specifically noted. According to our selection criterions for hot spot amino acid, the majority of the BH3 peptide residues make significant contributions to the binding free energy. Under “Discussion,” the amino acid hot spots of BH3 peptide will be discussed in the context of their amino acid efficiency, which was defined under “Materials and Methods.”
FIGURE 2.
BH3 peptide numbering. Also indicated are the names of the proteins from which the peptides are derived along with the number of residues present in the peptides. The alignment was obtained using ClustalW 1.83.
DISCUSSION
Among the proteins of the Bcl-2 family, there is a significant conservation of the BH domains (52, 53). From a structural analysis, it has been observed that the BH1–BH3 domains form a hydrophobic groove by which interactions with other proteins are mediated. Unlike the other domains, the BH4 domain is present only in anti-apoptotic Bcl-2 family members, such as Bcl-2 and Bcl-xl, where it is indispensable for function; deletion of this domain results in the abrogation of anti-apoptotic function (54). The BH4 domain, unlike the other three domains (BH1, BH2, and BH3), does not participate in the formation of the protein central core. From the free energy decomposition, no amino acids from this domain were determined to make any significant contributions to the binding free energy between the protein and the pro-apoptotic peptides. This is consistent with the observation that deletion of the BH4 domain from Bcl-2 and Bcl-xl renders the proteins apoptotically inactive but still able to associate with pro-apoptotic proteins such as Bax (54). It has been suggested that the role of the BH4 domain is to sequester other proteins, for example CED-4 (54). In the study by Sorenson (55), the BH4 domain is shown to be necessary for binding paxillin, an essential protein for development, linking cell adhesion processes to apoptosis.
The BH1–BH3 domains form the hydrophobic groove that mediates protein-protein interactions. Experimental studies have shown that mutations in these domains lead to the loss of the anti-apoptotic activity of Bcl-2 proteins, indicating that the BH1–BH3 domains are crucial for the heterodimerization with other Bcl-2 family members (56, 57).
The BH1 domain is the longer domain with 20 residues, and it is located between the α4 C-terminal end and the N-terminal of α5. Structurally, this domain contributes significantly to the formation of the hydrophobic groove. This domain makes significant energetic contributions to the interaction with BH3 peptides. In particular, the BH1 domain of Bcl-xl makes significant van der Waals contact with the BH3 peptides of the partner proteins (see Table 4) and also contains recurrent interaction energy hot spots (cf. Table 3, see detailed discussion below). On the other hand, the van der Waals interactions arising from the BH1 domain of Bcl-w are of lesser magnitude in these calculations, and this domain contains a fewer number of hot spots (see Tables 3 and 4 and detailed discussion below).
The 15 residues of the BH3 domain are localized to α2 where the N-terminal part of this helix makes contact with the ligand. In most of the complexes, this domain is the second most important domain with respect to interactions with BH3 peptides. An exception is the Bcl-w/Bim complex, where the contributions of the BH1 domain are less important than in the other complexes and are roughly on the same order of magnitude as the BH3 domain interactions.
The BH2 domain sequence is composed of 15 residues and is formed by α6 and α7 helices. According to the free energy decomposition, this domain makes a more variable contribution to the binding energy.
Analysis of the Hot Spots Present in the Different Complexes of Bcl-xl
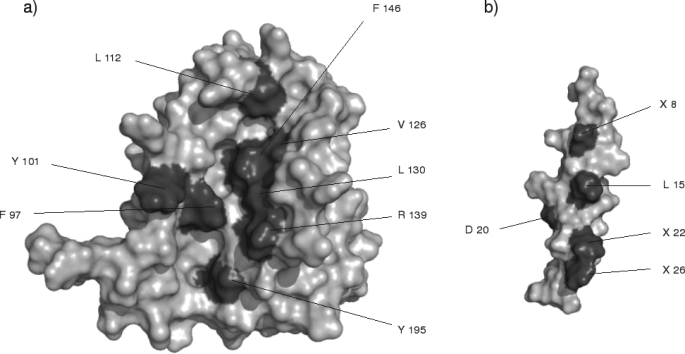
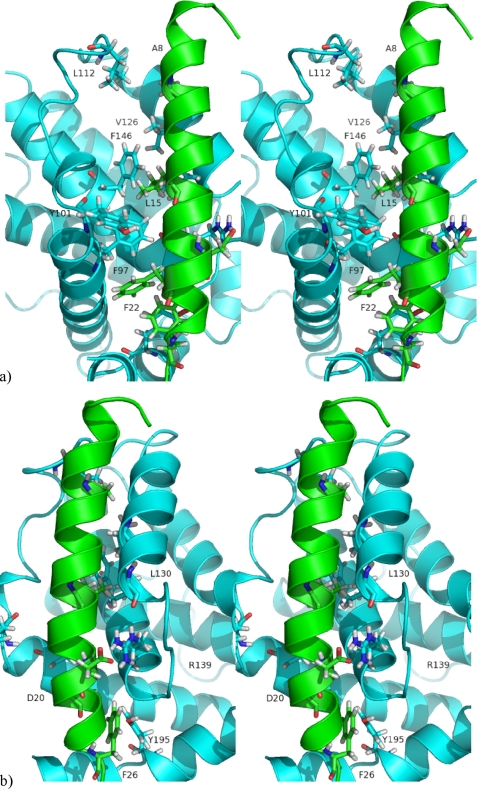
From the free energy decomposition carried out on the molecular dynamics simulations of the different Bcl-xl-BH3 peptide complexes, multiple amino acids were identified that recurrently contribute significantly to the binding free energy associated with complex formation (see Table 3). All of amino acids, referred to as energetic hot spots, are located in either the BH1 or the BH3 domains. These hot spot amino acids are illustrated in Figs. 3 and 4.
FIGURE 3.
a, surface representation of a Bcl-2 anti-apoptotic protein; b, BH3 peptide derived from Bcl-2 pro-apoptotic protein. The BH3 peptide has been rotated 180° from the anti-apoptotic protein. The dark gray colored amino acids are those amino acids that make the most significant contribution to the binding free energy (X being a residue with several possible variants in the sequence alignment).
FIGURE 4.
a, cross-eyed stereo representation of the experimental structure of Bcl-xl with the Bak BH3 peptide (PDB code 1BXL) zooming on the groove interactions. Indicated as sticks are the hot spot interactions arising from Bcl-xl (blue) and the BH3 peptides (green). b, as in a, but the protein complex was rotated clockwise along an axis running from top to bottom to better visualize interactions hidden in a.
Located in the BH3 domain, Phe97 is positioned in α2. This amino acid makes a significant hydrophobic contribution in all four Bcl-xl complexes. It makes a perpendicular T-stacking interaction with the (+)Phe22 in the Bim and the two Bad peptides (Fig. 4a). Interestingly, (+)Phe22 from these two pro-apoptotic BH3 peptides is the sequence equivalent to the Phe97 in the BH3 domains of anti-apoptotic Bcl-2 proteins. In other words, Phe97 interacts strongly with its sequence counterpart in these two peptides. Although (+)Phe22 is not conserved in all the pro-apoptotic BH3 peptide sequences, other hydrophobic amino acids are found in place of Phe, for example (+)Ile22 from the Bak peptide is able to make a comparably favorable van der Waals interaction with Phe97 from Bcl-xl. In addition, Phe97 makes significant van der Waals interactions with (+)Leu15, which is one of most conserved residues among the BH3 domain of the Bcl-2 proteins.
Another conserved interaction found in the different protein-peptide complexes implicates Tyr101 of Bcl-xl. This residue is found in the loop between α2 and α3. This amino acid is conserved in the five known sequences of Bcl-xl, in the seven known sequences of Bcl-2, and in apoptosis regulator R11, an anti-apoptotic protein from Xenopus (58). Interestingly, in all four Bcl-w sequences, a Phe substitutes for this Tyr. In all the complexes studied here, Tyr101 makes a conserved van der Waals interaction with either Met or Ile in position 18 of the BH3 peptides.
Leu112 is in α3 of the Bcl-xl protein (Fig. 4b). This amino acid binds the N-terminal region of the BH3 peptide, where it is in interaction with the residues (+)7, 8, and 11, which form a hydrophobic pocket around Leu112. Despite a favorable contribution (total ΔG, −0.4 kcal·mol−1), this residue makes a smaller contribution in the complex Bcl-xl-Bak with respect to the other complexes. This is probably for two reasons. One, in this particular structure, the peptide is too short to be sufficiently close to Leu112, and second, its N terminus is more disordered than the other peptides, and it does not form a helical structure. This again is likely to be related to its short length.
The Val126 is situated in α4, near the BH1 domain. It makes favorable interactions with the residues (+)8, (+)13, and (+)Leu15 of the BH3 peptides. (+)Leu15 is conserved across the BH3 peptides found in 1G5J, 2BZW, 1PQ1, and 1ZY3 complexes. The N-terminal part of the BH3 peptide from 1BXL complex is shorter than the other BH3 peptides, thus the lack of residue (+)8.
Leu130 belongs to the BH1 domain in α4. It makes strong van der Waals interactions with the residues (+)12, (+)Leu15, and (+)16 in the five complexes. These residues form a hydrophobic pocket into which the side chain of Leu130 is inserted. When residue (+)16 of the BH3 peptide is an Arg, as in the case of 1G5J, 2BZW, and 1PQ1, the van der Waals interactions arise from the interaction of Leu130 with the side chain of this amphipathic amino acid.
Arg139 is localized in α5 of the BH1 domain (Fig. 4b), and it makes significant contributions to the binding free energy in the complexes 2BZW, 1PQ1, and 1BXL. The electrostatic contributions are highly favorable in the complexes 1PQ1 and 1BXL, because of the presence of a salt bridge with the carboxylic group of the residue (+)Asp20 of the BH3 peptides. In the 2BZW complex, initially there is no salt bridge between Arg139 and (+)Asp20. However, during the molecular dynamics simulation, these residues move to form a salt bridge, although the mean electrostatic contribution remains around zero because of the close presence of (+)Arg16 from BH3 peptide. In the 1G5J complex, Arg139 makes an unfavorable contribution to the binding free energy as its electrostatic component is not counter-balanced by the summation of its nonpolar solvation and van der Waals contributions. The unfavorable electrostatic contribution is due the close proximity of the (+)Arg16 from the Bad peptide. The structure available in the PDB deposition 1G5J is, in fact, an energy-minimized average structure coming from NMR experiments. It would be interesting to look at the variation of this geometry around Arg139 in the individual structures to see if this geometry is an artifact of the average structure. Unfortunately, the individual structures are not available in the Protein Data Bank.
Besides this possible artifactual result, Arg139 makes a significant contribution to the binding in the other three complexes involving Bcl-xl and can therefore be labeled an energetic hot spot. This conclusion is further supported by the fact that Arg139 forms a salt bridge with residue (+)Asp20 and is a highly conserved residue among the BH1 domains of Bcl-2 protein as determined by a sequence alignment done using ClustalW 1.83 (59, 60).
Phe146 is found in α5 of the BH1 domain of Bcl-xl (Fig. 4a). In the structures 1G5J, 2BZW, and 1PQ1 complexes, Phe146 makes significant van der Waals interaction with residue (+)11 and (+)Leu15 of the BH3 peptide. In other complexes, such as the 1BXL complex, the ΔG value for Phe146 was slightly less favorable than in the other complexes with its value of −0.8 kcal·mol−1. However, some principal interactions observed in this complex are conserved with respect to the other Bcl-xl complexes in that it makes van der Waals interactions with (+)Leu15 of the Bak peptide. However, as (+)Val11 is located in the unfolded N-terminal tail of the peptide, this particular interaction with Phe146 is lost.
Tyr195 is positioned at α7 in the BH2 domain (Fig. 4b). This amino acid makes significant van der Waals interactions with the aromatic residues (+)Phe26 and (+)Tyr26 in the Bad and Bim peptides, respectively.
Other van der Waals interactions are made with the residue (+)23 of the BH3 peptide. The BH3 peptide is shorter in the 1BXL complex than in the other complexes, and residue (+)26 is not present. However, in this structure, van der Waals interactions do occur between Tyr195 and the three last amino acids.
Study of the Complex Bcl-w-BH3 Peptide from Bid (PDB Code 1ZY3)
In addition to the study of the complexes between the Bcl-xl/BH3 peptides, we also carried out an analysis of the complex between the anti-apoptotic protein Bcl-w and the BH3 peptide from Bid. The results show that, in contrast to Bcl-xl, the BH1 domain of Bcl-w is not the most critical for binding. In the Bcl-w-Bid peptide complex, the Bid peptide is positioned in a more parallel manner to the groove than in the complexes with Bcl-xl. The calculations show that the residues constituting the BH1 domain make fewer contacts with the BH3 peptide residues and, as a result, contribute less to the interaction. The BH2 domain contributes significantly to binding interactions with the BH3 peptide (entry 1ZY3 in Table 4). As seen in the Bcl-xl complexes, the BH4 domain contributes essentially nothing to the energetic anchoring of the Bid peptide. Interestingly, however, many of the hot spots found in the Bcl-w-Bid complex are analogous to those found in the Bcl-xl-BH3 peptide complexes, in particular residues such as Phe53, Phe57, Leu68, Val82, and Leu86. Other conserved interactions include Arg56 of Bcl-w, which corresponds to the Arg100 of Bcl-xl. Arg56 forms a salt bridge with (+)Asp20 of BH3 peptide during the entire molecular dynamics simulation. On one hand, the contribution of Arg95 to the free energy of association is not sufficiently high to classify it, in the strict sense used in this work, as a hot spot. This result could be explained by an unfavorable electrostatic contribution, due to surrounding nonpolar side chains of the residues Asn92 and Gly94 from Bcl-w and (+)Ala16 from the Bid peptide. However, its presence is necessary for the favorable contribution of (+)Asp20 to the binding free energy. This, coupled with its similarity to Arg100 of Bcl-xl, dictates that it should be considered as an important contributing amino acid in the binding interface. The observation of common hot spots in Bcl-xl and Bcl-w confirms the importance of these residues in the interaction between anti-apoptotic proteins and BH3 peptide.
Efficient Amino Acids on the Surface of the BH3 Peptide
The BH3 peptide binds to the hydrophobic groove of the anti-apoptotic proteins Bcl-xl or Bcl-w. To identify amino acids of the BH3 peptides that make significant contributions, we employ the concept of amino acid efficiency, which was described under “Materials and Methods.” The amino acid efficiency has been calculated for each central residue ranging from (+)8 to (+)26 of the BH3 peptides (Table 5). We found that the amino acids before and after in sequence made very small contributions to the binding energy.
To better distinguish the amino acids of the BH3 peptide that make the most important energetic contributions, we used the amino acid efficiencies. This criterion identifies the amino acids that have the most favorable binding energy per heavy atom. This concept has proved very useful in drug design, where efficient ligands are sought as they have an optimal binding affinity with respect to their molecular weight (48). Analysis of experimental binding data for large sets of ligands (61–63) indicates that the maximum affinity per heavy atom for organic compounds is −1.5 kcal·mol−1. It is generally considered that ligand efficiency values above 0.5 (in absolute value) correspond to optimized interactions, whereas values above 1 are indicative of extremely good interactions. In the present context, it is of interest to identify the amino acids that make efficient interactions, as they would be particularly suitable for small molecule mimicry.
To simplify our discussion, we will employ the residue numbering of the Bcl-xl protein. The positions of the important amino acids at the surface of BH3 peptide are shown in Figs. 3 and 4. From a comparison of the different peptides, it is immediately apparent that the most conserved amino acids of the BH3 peptides, (+)Leu15 and (+)Asp20, are consistently identified as efficient for all the systems.
The free energy contribution of (+)Leu15 is among the most favorable for binding due to van der Waals contacts with the anti-apoptotic protein, and its side chain is inserted into hydrophobic pocket constituted by Phe97, Val126, Leu130, and Phe146 (see Fig. 3). (+)Leu15 is highly conserved among all the BH3 domains of the Bcl-2 proteins (both anti- and pro-apoptotic), being substituted by Met only in the Bcl-w protein (see Table 1); (+)Leu15 is strictly conserved in all the BOP and pro-apoptotic BH3 domains. The correspondence between the high conservation and the high efficiency for this hydrophobic amino acid stresses the usefulness of the amino acid efficiency concept, and it suggests that the interactions that evolve to a high efficiency are conserved because they reflect an optimal fit between the ligand and the binding pocket (specially in the case of nonpolar amino acids).
Likewise, (+)Asp20 is a hot spot for all complexes, except for the complex Bcl-xl with the short Bad peptide (PDB code 1G5J). Indeed, in this particular complex, the (+)Asp20 position is slightly different from the other complexes, which subsequently prohibits the formation of a salt bridge with Arg139 from the anti-apoptotic protein. As we discussed above, this is likely a structural artifact because of the averaging of the NMR structures. With this consideration, then (+)Asp20 can be considered as a common hot spot of the BH3 peptide. In this complex, the acidic (+)Glu21 is also identified as efficient. During the molecular dynamics simulations, (+)Glu21 forms an H-bond with Tyr101. As for (+)Leu15, the high efficiency (+)Asp20 is linked to the high conservation, suggesting an optimized interaction network that is not tolerant to mutations.
The (+)Ala8 residue in Bad peptide was determined to be an interaction hot spot, as well as an efficient amino acid. Its side chain interacts with side chains from Leu112, Ser122, Gln125, and Val126, which form a hydrophobic pocket. The equivalent residue, (+)Ile8, in Bid peptide shows an equivalent interaction and also make favorable contributions to the binding energy. Mutagenesis studies of the Bad peptide performed by Petros et al. (28) showed that when (+)Ala8 was mutated to Gly, a 4.0-fold decrease of affinity with respect to the wild-type Bad peptide was observed. This mutation leads to the decrease of van der Waals interactions with the amino acids constituting the hydrophobic pocket at the surface the Bad peptide. This is reflected in its relatively high efficiency value. Interestingly, on the Bim peptide, (+)Glu8 is found in this position, and in the present calculations it makes unfavorable energetic contributions to the binding affinity with Bcl-xl, and it has a lower efficiency value. This is likely due to the effects of desolvation, and it suggests that this amino acid may be important in the differential values of affinity noted in binding assays of these peptides with different anti-apoptotic proteins (64). The residue (+)12 is not considered a hot spot because, in the 1BXL complex, it plays no role in the complex stability (ΔG = 0.0 kcal·mol−1). In this complex, (+)Gly12 is localized at the α-helix extremity, whereas its carbonyl group forms an H-bond with (+)Ala16; the potential hydrogen bond from its amino group is unsatisfied. In the other complexes, where the peptide is longer, this amino group forms an intramolecular H-bond. This suggests that (+)12 should remain as an amino acid of interest, especially in the context of a ligand design project. The residue (+)22 on the other hand is a conserved hot spot. It makes strong interactions in all systems, but in Bcl-w with the Bid peptide. It interacts primarily with Phe97, Tyr101, and Tyr195. In the case of the BH3 peptide from Bak, (+)Ile22 is slightly displaced in comparison with the other complexes. Therefore, interaction with Tyr101 is not possible. However, (+)Ile22 forms an H-bond between its backbone carbonyl and the hydroxyl group of the Tyr195 side chain. The lack of helical structure at the N-terminal end of this peptide might explain this unusual interaction, which is not made in the longer BH3 peptide, the (+)Ile22 backbone carbonyl being implicated in H-bond with the i + 4 amino acid. Thus, these observations could be considered as an artifact because of the short length of the Bak peptide.
Residue (+)26, which is Phe, Tyr, and Ile, respectively, in Bad, Bim, and Bid peptides, makes mainly van der Waals contacts with Tyr195, a common hot spot detected in anti-apoptotic protein. These N-terminal interactions appear somewhat stronger in the Bim and Bid peptides; however, it is unclear whether this would be the case in the presence of the full protein. It is important to note that although residue (+)26 is lacking in the Bak peptide, it is an aromatic residue (Tyr) in the sequence and would be present in the full protein. In a previous experimental study (28), (+)Phe26 from Bad peptide has been mutated to an Ala. The affinity experiments have shown that the affinity of the mutated Bad peptide was 3.5-fold lower than the wild-type Bad peptide, suggesting that the individual contribution of (+)Phe26 in the interaction is important.
The anti-apoptotic protein Bcl-xl has been shown to be able to bind diverse BH3 domains derived from different BOP (BH3-only proteins), which are believed to play a role in regulating apoptosis in cells. Our results demonstrate that the binding of this diverse set of domains is done by essentially the same set of interactions via a set of interfacial hot spots.
Several notable differences arise between the BH3 peptides, and these may contribute to their differential binding to the anti-apoptotic proteins (64). Apart from the discussion earlier of (+)Ala8 versus (+)Glu8 in the Bad and Bim peptides, respectively (see above), other differences are evident at the level of amino acid efficiencies. In particular, competitive binding assays show that the IC50 value for Bid binding to Bcl-xl is an order of magnitude larger than those for Bad and Bim (64). In the current analysis of BH3 peptide amino acid efficiencies, several notable differences between Bad and Bim on one hand and Bid on the other are evident. Most notable is the amino acid at (+)23, which is a Val in the Bad and Bim peptide and contributes favorably to binding. In Bid, the corresponding amino acid is an Asp, and its contribution is unfavorable. This is probably due in large part to the cost of desolvating the charged group and placing it near the amphipathic side chain of an Arg. A more detailed study of differential binding of BH3 peptides that includes studies of other anti-apoptotic proteins is underway and will be presented elsewhere.
Comparison with Mutation Studies
Mutagenesis studies have identified some amino acids that are important for Bcl-xl heterodimerization with pro-apoptotic Bcl-2 protein. The comparison between the mutation results and our results could contribute to a better understanding of the observed consequences of the mutations on the biological activity of Bcl-xl. Mutations that lead to the loss of Bcl-xl anti-apoptotic activity have been identified; these include the mutations of 135VNW137 to AIL, 138GRI140 to ELN (57) and Gly138 to Ala (56).
The present analysis of ΔG values obtained for the VNW sequence shows that although Val135 makes a small contribution to the interaction energy, the two latter have mean ΔG values of −0.8 and −0.9 kcal·mol−1, respectively. This lack of significant contribution by Val135 may be explained by the observation that the side chain of Val135 points away from of the BH3 peptide in the experimental structure and maintains this orientation during the molecular dynamics simulations. On the other hand, the side chain of Asn136 forms H-bond with a carboxylate group of the highly conserved residue (+)Asp20 of the BH3 peptide (Fig. 4). These H-bonds are maintained during the simulations. The mutation of Asn136 to Ile eliminates the formation of the H-bond and leads to the loss of a stabilizing interaction. The Trp137 side chain makes strong van der Waals interactions with the residue (+)23 from the BH3 peptide. The replacement of this residue by one with a smaller side chain, such as Leu, most likely leads to the reduction of van der Waals interactions. Although Asn136 and Trp137 have not been classified as conserved hot spots by the definition and energy cutoffs used here, these residues are clearly important for the binding activity of Bcl-xl. In two of the complexes, (Bcl-xl/Bad, PDB codes 1G5J and 2BZW), Asn136 is a canonical hot spot, whereas in the Bcl-xl/Bak (PDB code 1BXL) and Bcl-xl/Bim (PDB code 1PQ1) complexes, Trp137 appears as a hot spot. It could also be that the loss of Bcl-xl anti-apoptotic activity is because of cumulative effects arising from the mutation of these three residues.
The mutation of 138GRI140 to ELN results in the loss of Bcl-xl anti-apoptotic activity. Gly138, which is highly conserved among the Bcl-2 protein family, inserts in a small pocket formed by residues (+)19, (+)22, (+)23, and (+)26 from the BH3 peptide. The computational results yield an average ΔG of 0.8 kcal·mol−1 for the contribution of this amino acid to the binding free energy. The size of the pocket into which it inserts would therefore suggest that Glu is too bulky and thus disrupts the interaction between Bcl-xl and the pro-apoptotic proteins. The replacement of the Arg139, which is proposed to be a hot spot for the interaction, causes the loss of the salt bridge with the carboxylic group of the residue (+)Asp20, thus destabilizing the interaction. From the calculations, Ile140 appears to have little influence on the interaction as its mean ΔG is around 0.0 kcal·mol−1. Its lack of importance is confirmed by structural analysis of the complexes; this residue is localized at the opposite side of the α-helix 5 and is not in a position to interact with the BH3 domain of the pro-apoptotic protein.
The mutation of the Gly138 to Ala in Bcl-xl leads to inactivation of its anti-apoptotic properties (56). This study confirms the importance of having a small side chain at this position. This residue cannot be considered as a hot spot for the interaction in the strict sense of the definition used, because it is the mutated residue that most likely makes an unfavorable contribution to the binding energy. This arises from the fact that the Ala (in the mutant) is bulkier that the native Gly. The fact that the mutation of Gly alone is sufficient to lead to the loss of Bcl-xl anti-apoptotic activity does not mean that Arg139 in the mutation G138E/R139L/I140N has no influence on the loss of the Bcl-xl binding activity. The effect of Arg139, identified as a hot spot in the present calculations, can be assessed by mutation of it alone.
In the study by Cheng et al. (57), two double mutations, each implicating Phe146, reduced the Bcl-xl anti-apoptotic activity by about 80%. These mutagenesis experiments show the importance of the Phe146 for the Bcl-xl binding activity. This particular amino acid was identified as an interaction hot spot in the Bcl-2 complexes.
Conclusions
By comparing the complexes formed between anti-apoptotic proteins Bcl-xl and Bcl-w, and BH3 peptides from the pro-apoptotic proteins Bad, Bim, Bak, and Bid, we identified consensus amino acids that make important energetic contributions to their respective free energies of association. The interactions between these proteins are critical for the regulation of apoptosis. When compared with available experimental data, the residues identified here as energetic hot spots are indeed crucial for the interaction between Bcl-xl and pro-apoptotic Bcl-2 proteins.
Some variation arose in the comparison of the two complexes containing the Bad peptide. One complex (PDB code 1G5J) contained a shorter peptide than the other (PDB code 2BZW). The structure of the former was determined by NMR spectroscopy, and the energy-minimized average structure was made available in the Protein Data Bank. The second complex contained a longer Bad peptide, and its structure was determined by x-ray crystallography. Between the two structures, some structural differences were observed. Most of variations in the results for these two structures were due to structural differences between residues at the terminal ends of the short peptide. This underlines the necessity of using multiple structures from difference sources, whether from molecular dynamics simulations, from experimental sources, or from both to obtain robust results.
We expect these results to be robust across different force fields. Studies by the Guvench and MacKerell (65) and Rueda et al. (29) have shown that the major forces fields used in protein dynamics yield essentially the same behavior when tested on a large variety of different proteins.
Deregulation of apoptosis characterized by the high expression of Bcl-2 anti-apoptotic proteins has been implicated in numerous cancers. The identification of molecules able to mimic the pro-apoptotic BH3 domain and to restore apoptosis in cancer cells is an appealing approach to the development of new therapeutic compounds. There is, for example, ABT-737, which is currently one of the few small molecule BH3 mimetics that have a high affinity for Bcl-2 anti-apoptotic proteins (Bcl-xl, Bcl-2, and Bcl-w) with a dissociation constant below 1 nm (13, 53). However, to obtain other efficient BH3 mimetics, an increased understanding of the binding interface between Bcl-2 proteins is required. Our results identified interfacial residues (Phe97, Tyr101, Leu112, Val126, Leu130, Arg139, Phe146, and Tyr195), which, if targeted in a design process, could lead to the development of new molecules that disrupt the Bcl-2 anti-apoptotic-pro-apoptotic protein complex. Furthermore, the BH3 residues (+)8, (+)Leu15, (+)Asp20, (+)22, and (+)26, which have been identified as being important for complex formation, can be the cornerstone of new BH3 mimics.
An equally important question concerns the elucidation of the factors that give rise to the differential binding properties of BH3 domain binding by the anti-apoptotic proteins. To address this question, we are currently working on the study of Mcl-1/BH3 binding. Mcl-1 has been shown to have binding tendencies against BH3 domains that are markedly different from Bcl-xl (64). The comparison should provide interesting insight and will be presented elsewhere.
Supplementary Material
Acknowledgments
We thank the Centre Informatique National de l'Enseignement Supérieur, the Institut du Développement et des Ressources en Informatique Scientifique, and the Centre d'Etude du Calcul Parallèle de Strasbourg of the University Louis Pasteur for generous allocations of computer time. We thank the reviewers for their helpful comments and Cedric Grauffel for technical assistance.
This work was supported by the Université de Strasbourg, CNRS, INSERM, and the Agence Nationale de la Recherche.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S4.
4 M. Schaefer, personal communication.
- BH
- Bcl-2 homology
- α
- α-helix
- CHARMM
- Chemistry at Harvard Molecular Mechanics
- MM/PBSA
- Molecular Mechanics/Poisson-Boltzmann Surface Area
- PDB
- Protein Data Bank
- RMSD
- root mean square difference
- BOP
- BH3-only protein.
REFERENCES
- 1.Willis S., Day C. L., Hinds M. G., Huang D. C. ( 2003) J. Cell Sci. 116, 4053– 4056 [DOI] [PubMed] [Google Scholar]
- 2.van Delft M. F., Huang D. C. ( 2006) Cell Res. 16, 203– 213 [DOI] [PubMed] [Google Scholar]
- 3.Reed J. C. ( 2000) Am. J. Pathol. 157, 1415– 1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang D. C., Strasser A. ( 2000) Cell 103, 839– 842 [DOI] [PubMed] [Google Scholar]
- 5.Kirkin V., Joos S., Zornig M. ( 2004) Biochim. Biophys. Acta 1644, 229– 249 [DOI] [PubMed] [Google Scholar]
- 6.Tanabe K., Kim R., Inoue H., Emi M., Uchida Y., Toge T. ( 2003) Int. J. Oncol. 22, 875– 881 [PubMed] [Google Scholar]
- 7.Kim R., Emi M., Tanabe K., Toge T. ( 2004) Cancer 101, 2177– 2186 [DOI] [PubMed] [Google Scholar]
- 8.Kim R., Tanabe K., Emi M., Uchida Y., Toge T. ( 2005) Cancer 103, 2199– 2207 [DOI] [PubMed] [Google Scholar]
- 9.Walensky L. D., Kung A. L., Escher I., Malia T. J., Barbuto S., Wright R. D., Wagner G., Verdine G. L., Korsmeyer S. J. ( 2004) Science 305, 1466– 1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. ( 2006) Mol. Cell 24, 199– 210 [DOI] [PubMed] [Google Scholar]
- 11.Sadowsky J. D., Schmitt M. A., Lee H. S., Umezawa N., Wang S., Tomita Y., Gellman S. H. ( 2005) J. Am. Chem. Soc. 127, 11966– 11968 [DOI] [PubMed] [Google Scholar]
- 12.Sadowsky J. D., Fairlie W. D., Hadley E. B., Lee H. S., Umezawa N., Nikolovska-Coleska Z., Wang S., Huang D. C., Tomita Y., Gellman S. H. ( 2007) J. Am. Chem. Soc. 129, 139– 154 [DOI] [PubMed] [Google Scholar]
- 13.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. ( 2005) Nature 435, 677– 681 [DOI] [PubMed] [Google Scholar]
- 14.Doshi J. M., Tian D., Xing C. ( 2006) J. Med. Chem. 49, 7731– 7739 [DOI] [PubMed] [Google Scholar]
- 15.Wells J. A. ( 1991) Methods Enzymol. 202, 390– 411 [DOI] [PubMed] [Google Scholar]
- 16.Clackson T., Wells J. A. ( 1995) Science 267, 383– 386 [DOI] [PubMed] [Google Scholar]
- 17.Bogan A. A., Thorn K. S. ( 1998) J. Mol. Biol. 280, 1– 9 [DOI] [PubMed] [Google Scholar]
- 18.Honig B., Nicholls A. ( 1995) Science 268, 1144– 1149 [DOI] [PubMed] [Google Scholar]
- 19.Hendsch Z. S., Tidor B. ( 1999) Protein Sci. 8, 1381– 1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortemme T., Baker D. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 14116– 14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerois R., Nielsen J. E., Serrano L. ( 2002) J. Mol. Biol. 320, 369– 387 [DOI] [PubMed] [Google Scholar]
- 22.Gohlke H., Kiel C., Case D. A. ( 2003) J. Mol. Biol. 30, 891– 913 [DOI] [PubMed] [Google Scholar]
- 23.Zeeh J. C., Zeghouf M., Grauffel C., Guibert B., Martin E., Dejaegere A., Cherfils J. ( 2006) J. Biol. Chem. 281, 11805– 11814 [DOI] [PubMed] [Google Scholar]
- 24.Lafont V., Schaefer M., Stote R. H., Altschuh D., Dejaegere A. ( 2007) Proteins 67, 418– 434 [DOI] [PubMed] [Google Scholar]
- 25.Browning C., Martin E., Loch C., Wurtz J. M., Moras D., Stote R. H., Dejaegere A. P., Billas I. M. ( 2007) J. Biol. Chem. 282, 32924– 32934 [DOI] [PubMed] [Google Scholar]
- 26.Kollman P. A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D. A., Cheatham R. T., III ( 2000) Acc. Chem. Res. 33, 889– 897 [DOI] [PubMed] [Google Scholar]
- 27.Denisov A. Y., Chen G., Sprules T., Moldoveanu T., Beauparlant P., Gehring K. ( 2006) Biochemistry 45, 2250– 2256 [DOI] [PubMed] [Google Scholar]
- 28.Petros A. M., Nettesheim D. G., Wang Y., Olejniczak E. T., Meadows R. P., Mack J., Swift K., Matayoshi E. D., Zhang H., Thompson C. B., Fesik S. W. ( 2000) Protein Sci. 9, 2528– 2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rueda M., Ferrer-Costa C., Meyer T., Perez A., Camps J., Hospital A., Gelpi J. L., Orozco M. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 796– 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. ( 2003) Immunity 19, 341– 352 [DOI] [PubMed] [Google Scholar]
- 31.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. ( 1997) Science 275, 983– 986 [DOI] [PubMed] [Google Scholar]
- 32.Vorobjev Y. N., Almagro J. C., Hermans J. ( 1998) Proteins 32, 399– 413 [PubMed] [Google Scholar]
- 33.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. ( 2000) Nucleic Acids Res. 28, 235– 242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sali A., Blundell T. L. ( 1993) J. Mol. Biol. 234, 779– 815 [DOI] [PubMed] [Google Scholar]
- 35.Schaefer M., van Vlijmen H. W., Karplus M. ( 1998) Adv. Protein Chem. 51, 1– 57 [DOI] [PubMed] [Google Scholar]
- 36.Davis M. E., Madura J. D., Luty B. A., McCammon J. A. ( 1991) Comput. Phys. Commun. 62, 187– 197 [Google Scholar]
- 37.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. ( 1983) J. Comp. Chem. 4, 187– 217 [Google Scholar]
- 38.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. ( 1998) J. Phys. Chem. 102, 3586– 3616 [DOI] [PubMed] [Google Scholar]
- 39.Mackerell A. D., Jr., Feig M., Brooks C. L., III ( 2004) J. Comput. Chem. 25, 1400– 1415 [DOI] [PubMed] [Google Scholar]
- 40.Brunger A. T., Karplus M. ( 1988) Proteins 4, 148– 156 [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. ( 1983) J. Chem. Phys. 79, 926– 935 [Google Scholar]
- 42.Neria E., Fischer S., Karplus M. ( 1996) J. Chem. Phys. 105, 1902– 1921 [Google Scholar]
- 43.Ryckaert J. P., Ciccotti G., Berendsen H. J. C. ( 1997) J. Comput. Phys. 23, 327– 341 [Google Scholar]
- 44.Steinbach P. J., Brooks B. R. ( 1994) J. Comp. Chem. 15, 667– 683 [Google Scholar]
- 45.Zoete V., Michielin O. ( 2007) Proteins 67, 1026– 1047 [DOI] [PubMed] [Google Scholar]
- 46.Gouda H., Kuntz I. D., Case D. A., Kollman P. A. ( 2003) Biopolymers 68, 16– 34 [DOI] [PubMed] [Google Scholar]
- 47.Spiegel M. R. ( 1975) in Probability and Statistics, Schaum's Outline Series, McGraw-Hill Inc., New York [Google Scholar]
- 48.Hopkins A. L., Groom C. R., Alex A. ( 2004) Drug Discov. Today 9, 430– 431 [DOI] [PubMed] [Google Scholar]
- 49.Frishman D., Argos P. ( 1995) Proteins 23, 566– 579 [DOI] [PubMed] [Google Scholar]
- 50.Wilson-Annan J., O'Reilly L. A., Crawford S. A., Hausmann G., Beaumont J. G., Parma L. P., Chen L., Lackmann M., Lithgow T., Hinds M. G., Day C. L., Adams J. M., Huang D. C. ( 2003) J. Cell Biol. 162, 877– 887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson J. W., Nostro M. C., Balzi M., Faraoni P., Cianchi F., Becciolini A., Potten C. S. ( 2000) Br. J. Cancer 82, 178– 185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato T., Hanada M., Bodrug S., Irie S., Iwama N., Boise L. H., Thompson C. B., Golemis E., Fong L., Wang H. G., Reed J. ( 1994) Proc. Natl. Acad. Sci. U. S. A. 91, 9238– 9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L., Ming L., Yu J. ( 2007) Drug Resist. Updates 10, 207– 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D. C., Adams J. M., Cory S. ( 1998) EMBO J. 17, 1029– 1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorenson C. M. ( 2004) J. Biol. Chem. 279, 11368– 11374 [DOI] [PubMed] [Google Scholar]
- 56.Sedlak T. W., Oltvai Z. N., Yang E., Wang K., Boise L. H., Thompson C. B., Korsmeyer S. J. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 7834– 7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng E. H., Levine B., Boise L. H., Thompson C. B., Hardwick J. M. ( 1996) Nature 379, 554– 556 [DOI] [PubMed] [Google Scholar]
- 58.Cruz-Reyes J., Tata J. R. ( 1995) Gene (Amst.) 158, 171– 179 [DOI] [PubMed] [Google Scholar]
- 59.Thompson J. D., Higgins D. G., Gibson T. J. ( 1994) Nucleic Acids Res. 22, 4673– 4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. ( 2007) Bioinformatics (Oxf.) 23, 2947– 2948 [DOI] [PubMed] [Google Scholar]
- 61.Kuntz I. D., Chen K., Sharp K. A., Kollman P. A. ( 1999) Proc. Natl. Acad. Sci. U. S. A. 96, 9997– 10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reynolds C. H., Bembenek S. D., Tounge B. A. ( 2007) Bioorg. Med. Chem. Lett. 17, 4258– 4261 [DOI] [PubMed] [Google Scholar]
- 63.Reynolds C. H., Tounge B. A., Bembenek S. D. ( 2008) J. Med. Chem. 51, 2432– 2438 [DOI] [PubMed] [Google Scholar]
- 64.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. ( 2005) Mol. Cell 17, 393– 403 [DOI] [PubMed] [Google Scholar]
- 65.Guvench O., MacKerell A. D., Jr. ( 2008) Methods Mol. Biol. 443, 63– 88 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.