Abstract
Vertebrate retinal progenitor cells (RPCs) are pluripotent, but pass through competence states that progressively restrict their developmental potential (Cepko et al., 1996; Livesey and Cepko, 2001; Cayouette et al., 2006). In the rodent eye, seven retinal cell classes differentiate in overlapping waves, with RGCs, cone photoreceptors, horizontals, and amacrines forming predominantly before birth, and rod photoreceptors, bipolars, and Müller glia differentiating postnatally. Both intrinsic and extrinsic factors regulate each retinal cell type (for review, see Livesey and Cepko, 2001). Here, we conditionally deleted the transcription factor Rbpj, a critical integrator of multiple Notch signals (Jarriault et al., 1995; Honjo, 1996; Kato et al., 1997; Han et al., 2002), during prenatal mouse retinal neurogenesis. Removal of Rbpj caused reduced proliferation, premature neuronal differentiation, apoptosis, and profound mispatterning. To determine the cell autonomous requirements for Rbpj during RGC and cone formation, we marked Cre-generated retinal lineages with GFP expression, which showed that Rbpj autonomously promotes RPC mitotic activity, and suppresses RGC and cone fates. In addition, the progressive loss of Rbpj−/− RPCs resulted in a diminished progenitor pool available for rod photoreceptor formation. This circumstance, along with the overproduction of Rbpj−/− cones, revealed that photoreceptor development is under homeostatic regulation. Finally, to understand how the Notch pathway regulates the simultaneous formation of multiple cell types, we compared the RGC and cone phenotypes of Rbpj to Notch1 (Jadhav et al., 2006b; Yaron et al., 2006), Notch3, and Hes1 mutants. We found particular combinations of Notch pathway genes regulate the development of each retinal cell type.
Introduction
Notch is a major metazoan signaling pathway and key regulator of retinal neurogenesis (for review, see Perron and Harris, 2000; Baker, 2001; Livesey and Cepko, 2001; Lai, 2002). A Notch signal is transmitted between two cells through ligand-receptor binding, which triggers release of the NOTCH intracellular protein domain (ICD) within the receiving cell (for review, see Fortini, 2009; Kopan and Ilagan, 2009). Subsequently, the NOTCH ICD is transported into the nucleus, where it complexes with RBPJ and MAML and activates target genes such as Hes1 or Hes5. The Notch pathway transmits multiple types of signals. The most well studied of these, lateral inhibition, occurs when an equivalent group of cells initially express ligand and receptor uniformly, until one cell stochastically begins to express more ligand, making it the signaling cell. The classic, canonical pathway: Delta ⇒ Notch ⇒ Rbpj(CSL) ⇒ Hes/E(spl), is widely used during neuronal and glial development (for review, see Baker, 2000; Fortini, 2009; Kopan and Ilagan, 2009).
In the rodent retina, multiple Notch pathway genes are expressed (Weinmaster et al., 1991, 1992; Austin et al., 1995; Ahmad et al., 1997; Bao and Cepko, 1997; Rowan et al., 2004; Nelson et al., 2006). In the frog and chick eye, Delta-Notch signaling controls the temporal development of multiple cell classes. However, in the mouse retina, the early lethality of Notch pathway mutations has hampered a deep examination of these genes (Austin et al., 1995; Dorsky et al., 1995; Ahmad et al., 1997; Dorsky et al., 1997; Henrique et al., 1997; Furukawa et al., 2000; Schneider et al., 2001; Silva et al., 2003). Recently, Cre-lox deletion of Notch1 demonstrated a critical role for this receptor in repressing ectopic cone photoreceptor development (Jadhav et al., 2006b; Yaron et al., 2006). In Notch1−/− eyes, RPCs prematurely exited the cell cycle and differentiated as cone photoreceptors as early as E13.5, upregulating three factors: Otx2, Crx and Thrβ2/Thrb (Jadhav et al., 2006b; Yaron et al., 2006). The extra cones arose at the expense of rod fates, although a loss of retinal ganglion cells (RGCs) was also described (Yaron et al., 2006).
While these Notch1 studies advanced our knowledge of retina neurogenesis, additional questions remain. First, are mammalian cones regulated by a canonical Notch signal (DELTALIKE1 ⇒ NOTCH1 ⇒ RBPJ ⇒ HES1) and if not, which ligand, receptor and downstream effector are involved? Why were cone fates selectively derepressed in Notch1 mutant embryos? Finally, why did RGCs decrease in Notch1 conditional mutants, which differed from other retinal studies demonstrating that Delta-Notch signaling blocks RGC genesis (Austin et al., 1995; Dorsky et al., 1995, 1997; Ahmad et al., 1997; Henrique et al., 1997; Schneider et al., 2001; Silva et al., 2003)? Here, we assessed several roles for the pathway integrator Rbpj during mouse RGC and photoreceptor development, as well as the prenatal RGC and photoreceptor phenotypes of Notch3 and Hes1 mutants.
Materials and Methods
Mice
The Rbpjtm1Hon conditional allele (termed RbpjCKO) was generated by Han et al., maintained on a 129/SvJ background, and genotyped as described (Han et al., 2002). Notch3Gt(PST033)Byg gene trap mutant mice (termed Notch3LacZ) were generated by Mitchell and colleagues, and maintained as a homozygous viable stock in a 129/BL6 mixed background (Leighton et al., 2001; Mitchell et al., 2001; Pan et al., 2004; Demehri et al., 2008). Hes1tm1Fqu mutant mice (termed Hes1−/−) were generated by Ishibashi and colleagues, maintained on an ICR background and genotyped as previously described (Ishibashi et al., 1995; Cau et al., 2000). α-Cre transgenic mice were generated by Marquardt et al., maintained on a CD-1 background and PCR genotyped as described (Marquardt et al., 2001). Chx10-Cre BAC transgenic mice in a CD-1 background were obtained from Jackson Laboratories and genotyped as in (Rowan and Cepko, 2004). Z/EG lineage tracing mice also in a CD-1 background were acquired from Jackson Laboratories and genotyped for GFP per (Novak et al., 2000). Images of adult heads were captured on a Leica dissecting microscope with an Optronics digital camera and software.
Retinal phenotype analyses
Embryonic and postnatal tissues were fixed in 4% paraformaldehyde/PBS for 40–60 min at 4°C, processed through a sucrose/PBS series, cryoembedded and sectioned. Primary antibodies used were anti-βgal (Tom Glaser, University of Michigan, Ann Arbor, MI; 1:1000); anti-BrdU (Abd Serotec; 1:500); anti-cleaved PARP (Cell Signaling Technology; 1:500); anti-CRX (Cheryl Craft, University of California, Los Angeles, Los Angeles, CA; 1:1000), anti-S OPSIN (Cheryl Craft; 1:1000), anti-M/L OPSIN (Cheryl Craft; 1:1000) (Zhu and Craft, 2000; Zhu et al., 2003); anti-RHODOPSIN (Millipore Bioscience Research Reagents; 1:1000); anti-POU4F2 (Santa Cruz Biotechnology; 1:50); anti-SOX9 (Millipore Bioscience Research Reagents; 1:200); anti-RXRγ (Santa Cruz Biotechnology; 1:200); anti-THRB/TRβ2 (Douglas Forrest, National Institutes of Health, Bethesda, MD; 1:2500) (Ng et al., 2009); anti-NR2E3 (Anand Swaroop, National Eye Institute, Bethesda, MD; 1:500) (Cheng et al., 2004); anti-TUBB3 (Covance; 1:1000); anti-CCND1 (Santa Cruz Biotechnology; 1:500), anti-GFP (Invitrogen; 1:1000 or Abcam; 1:1000); anti-HES1 (1:1000) (Lee et al., 2005); anti-ISL1 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA; 1:20); anti-PROX1 (Covance; 1:1000); anti-CALRETININ (Millipore Bioscience Research Reagents; 1:200); anti-GLUTAMINE SYNTHETASE (Millipore Bioscience Research Reagents; 1:1000); anti-CALBINDIN (Millipore Bioscience Research Reagents; 1:1000); anti-CHX10 (Exalpha Biologicals; 1:1000), DAPI stain (Sigma Chemical; 1:1000). Secondary antibodies used were directly conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen/Invitrogen) or biotinylated (Jackson Immunologicals) and sequentially labeled with streptavidin Alexa 488 or 594 (Invitrogen).
In situ hybridization on cryosections was performed as described (Brown et al., 1998) using Math5/Atoh7 (Brown et al., 1998), Hes5 (a gift from Kenny Campbell, Cincinnati Children's Research Foundation, Cincinnati, OH), Nrl and Nr2e3 (gifts from Alan Mears, Ottawa Hospital Research Institute, Ottawa, ON, Canada) cDNA plasmids as templates for digoxygenin-labeled antisense riboprobes. For S-phase analyses, BrdU (Sigma Chemical) was injected intraperitoneally as described in the study by Mastick and Andrews (2001) and animals were killed 1.5 h later for tissue processing that included 2N hydrochloric acid treatment of sections before antibody staining. To visualize LacZ activity in Notch3LacZ embryos, X-gal staining of cryosections followed (Brown et al., 2001). Standard histology on paraffin embedded adult eyes was also performed. All microscopic imaging was performed on a Zeiss fluorescent microscope with Zeiss camera and Apotome deconvolution device. Images were processed using Axiovision (v5.0) and Adobe Photoshop software (v7.0) and electronically adjusted for brightness, contrast and pseudocoloring.
Cell counting
Labeled tissue sections were imaged and cells quantified using Axiovision (v5.0) software. Three or more animals were analyzed per genotype and age, with ≥2 sections from each control or mutant littermate animal. Retinal sections were judged to be of equivalent depth in the eye by anatomical landmarks in the head and other eye tissues, with only the nasal side of the retina imaged for consistency of mutant phenotypes. The cell autonomy of HES1+, BrdU-labeled, cPARP+, POU4F2+, RXRγ+, CRX+, THRB2+ or NR2E3+ cells at E16.5 or P3 was determined within a 200× field of each section containing the distal retina. The percentage of marker+/GFP+; marker+/DAPI or GFP+/DAPI cells ± SEM was determined, with GFP reporting either IRES-GFP or Z/EG expression. The percentages of HES1+/DAPI; POU4F2+/DAPI and CRX+/DAPI nuclei were determined in 200× fields within in the central retinas of E16.5 wild-type and Notch3LacZ/LacZ embryos. The percentage of CRX+/DAPI nuclei was also determined in wild-type and Hes1−/− retinal sections. A two-tailed Student's t test and Welch post hoc test were used to determine p values (Instat Software, v3.0).
Results
Conditional deletion of Rbpj causes microphthalmia and retinal mispatterning
To define the roles of Notch signaling in the mammalian retina more fully, we conditionally deleted the common pathway component, Rbpj, using a conditionally mutant allele (RbpjCKO) (Han et al., 2002) and the α-Cre retinal driver, which initiates Cre and IRES-GFP expression in distal optic cup at E10.5 (Fig. 1A,B) (Marquardt et al., 2001). In our experiments, control animals were littermates of the genotype RbpjCKO/CKO (lacking the Cre transgene), or α-Cre;RbpjCKO/+;Z/EG, which had no histologic or molecular marker abnormalities (E13.5 to P21; n ≥ 6 animals per age). Adult α-Cre;RbpjCKO/CKO eyes were microphthalmic (Fig. 1C–F; supplemental Fig. 1, available at www.jneurosci.org as supplemental material), with severely mispatterned distal retinas that contained large rosettes (Fig. 1F, boxed area; H,J, arrows). These areas of mispatterning were also apparent in Rbpj retinal mutants at E16.5 (Fig. 2B,J), and were more severe than those of Notch1 retinal mutants (Jadhav et al., 2006b; Yaron et al., 2006).
Figure 1.
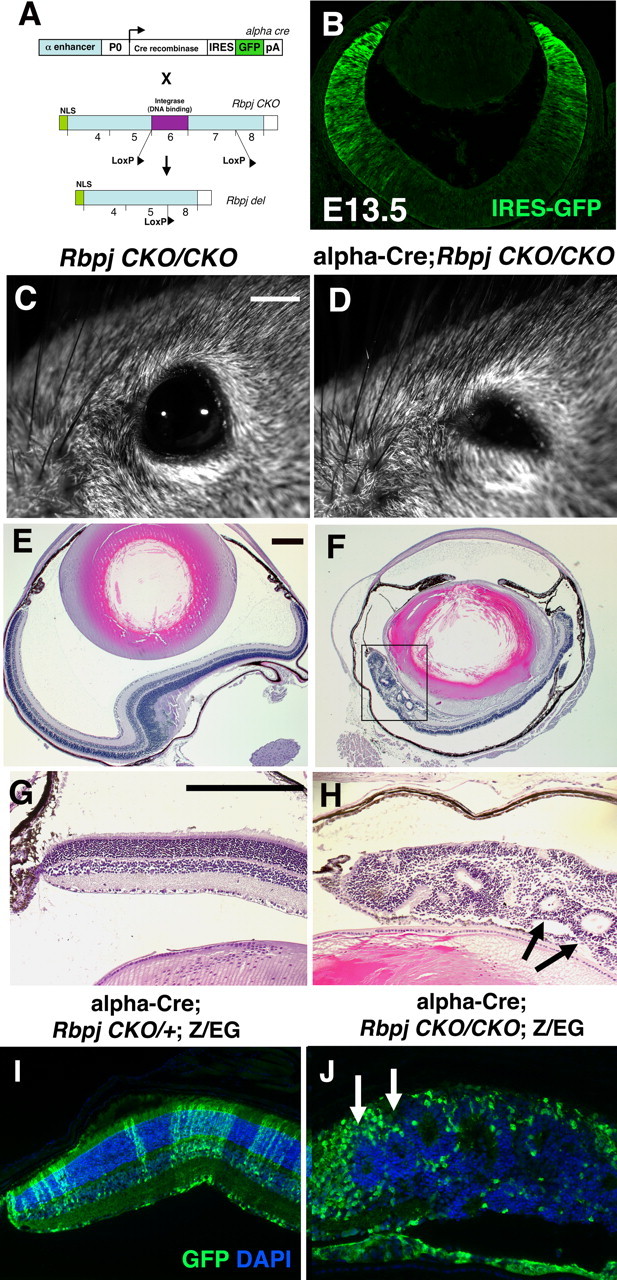
Adult phenotypes of α-Cre;RbpjCKO/CKO conditionally mutant eyes. A, B, Conditional deletion strategy for α-Cre; RbpjCKO/CKO retinal mutants. The α-Cre transgene has an IRES-GFP expression cassette that marks peripheral retinal cells with Cre activity (Marquardt et al., 2001), shown by anti-GFP labeling in B. C, D, α-Cre;RbpjCKO/CKO adult eyes were abnormally small (microphthalmic) compared to RbpjCKO/CKO or α-Cre;RbpjCKO/+ littermates. No Rbpj heterozygous phenotypes were observed, grossly or by histology (n ≥ 4 per genotype). E–G, Histologic sections of P21 eyes further highlighted the small size of mutant eyes (F), and severe mispatterning in the distal retina (boxed area). H is a higher magnification of boxed area in F, with arrows pointing to two retinal rosettes. I, J, Anti-GFP/DAPI double label of α-Cre;RbpjCKO/+;Z/EG and α-Cre;RbpjCKO/CKO;Z/EG P21 retinal sections. Normally this GFP+ lineage contains all retinal cell classes (I). But, Rbpj−/−GFP+ cells exhibited an abnormal, rounded morphology, and were excluded from rosettes (J, arrows). Distal is up in B, E, F; rostral is left in C, D; in G–J vitreal is down. Scale bars: 500 μm in C, D; 5 μm in E, F; 20 μm in G–J, B. α, Pax6 intronic α enhancer; P0, Pax6 promoter; pA, poly A sequence; NLS, nuclear localization sequence.
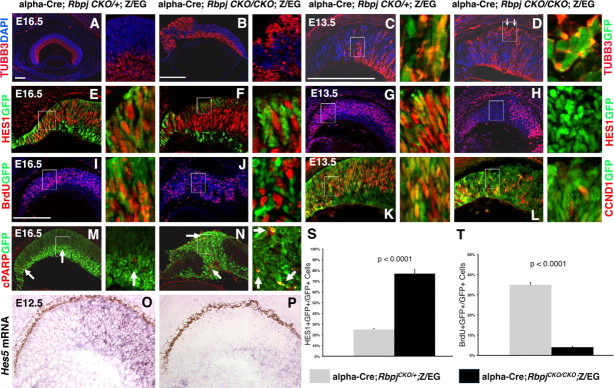
Figure 2.
Embryonic Rbpj mutant eyes are mispatterned, with segregation of proliferating and nonproliferating retinal populations. In C–N, the boxed area is to the right at a higher magnification. A–D, Anti-TUBB3 (red), anti-GFP (green in insets), DAPI triple labels of retinal sections. In E16.5 Rbpj retinal mutants, excess differentiated neurons surrounded forming retinal rosettes (B). Due to a dorsal-ventral gradient of α-Cre expression (Bäumer et al., 2002; Yaron et al., 2006), some sections contained distal mutant tissue on only one side. Note magnification difference between A and B. C, D, Upon removal of Rbpj, differentiating retinal neurons became mispatterned as early as E13.5. Arrows in D point to the early, cell autonomous appearance neurons on the wrong side of the retina. E, F, Control E16.5 α-Cre RPCs (GFP+) coexpress HES1 (E), but without Rbpj function, they autonomously lose HES1 (F). A high degree of GFP and HES1 coexpression also occurred at E13.5, but essentially no HES1 expression could be detected in Rbpj−/−GFP+ cells at this age (H). I, J, BrdU and GFP are normally coexpressed in E16.5 RPCs in the α-Cre lineage, but in Rbpj retinal mutants, mostly BrdU+GFP-neg cells were observed. K, L, In E13.5 control eyes, CCND1 is broadly coexpressed with GFP. However, many Rbpj−/−GFP+ cells autonomously downregulated this marker (L). M, N, Arrows point to cPARP+ apoptotic cells that were autonomously increased in Rbpj mutant tissue. O, P, Hes5 mRNA expression was greatly diminished in the distal optic cup of α-Cre;RbpjCKO/CKO eyes. S, Profound cell autonomous loss of HES1 within E16.5 Rbpj−/− cells. T, The percentage of BrdU+GFP+ cells was also significantly decreased in Rbpj mutant cells. Scale bars: 100 μm in A–C, E; insets magnified 8- or tenfold. For each marker, n ≥ 3 embryos per age and genotype.
The assessment of cell autonomous gene function has been an instrumental tool in the fruit fly eye for deciphering multiple roles for Notch signaling (for review, see Baker, 2000, 2001; Lai, 2002). Because this pathway is inherently more complex in the mouse retina, we wished to approach the genetic rigor of the Drosophila experiments. Moreover, many Cre transgenes (including α-Cre) are mosaically expressed, thereby complicating phenotypic analyses. To address these issues, we integrated the Z/EG transgene in our mouse breeding scheme, to mark and follow Rbpj−/− retinal cells (Novak et al., 2000). In retinal progenitor cells (RPCs) with α-Cre activity, GFP expression was permanently activated by removal of a flox-stop cassette within the Z/EG transgene, simultaneous with Cre-mediated deletion of Rbpj (Fig. 1A) (Novak et al., 2000; Han et al., 2002). Figure 1, I and J, show the distribution of α-Cre lineage (GFP+ cells), within the distal retina of adult control and Rbpj mutant eyes. Normally, α-Cre+ RPCs produce all seven cell classes that span across the laminated retina (Figs. 3–5; supplemental Fig. 2, available at www.jneurosci.org as supplemental material). However, all distal optic cup cells do not express GFP, due to mosaic transgene expression (Marquardt et al., 2001; Yaron et al., 2006; Riesenberg et al., 2009). Interestingly, when α-Cre cells (GFP+) were mutant for Rbpj, they were inappropriately separated from Rbpj+/+GFP-negative (neg; wild-type) cells, such that wild-type cells largely resided in the retinal rosettes (Fig. 1J, arrows), and Rbpj−/−GFP+ cells had an abnormal, rounded morphology.
Figure 3.
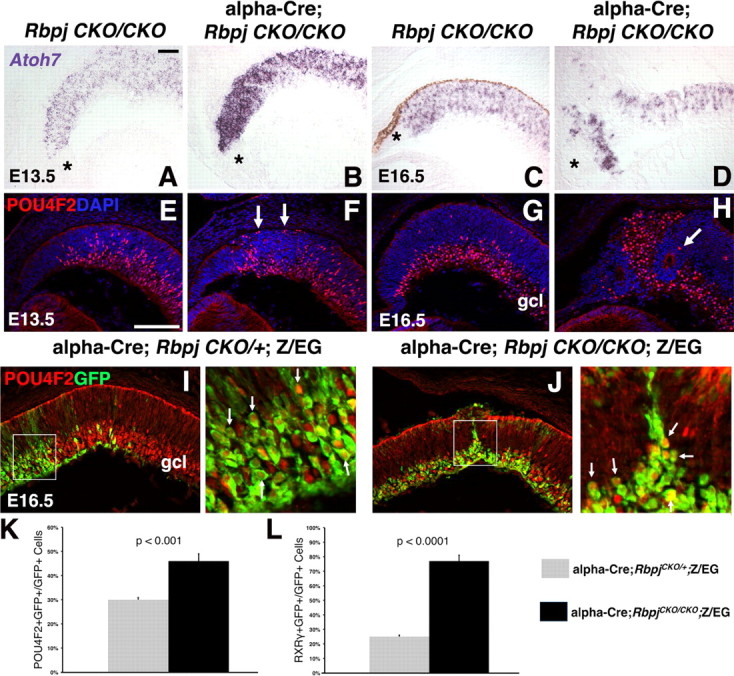
Rbpj cell autonomous suppression of RGC formation. A–D, In situ hybridization for Atoh7 mRNA. At E13.5, the Atoh7-expression domain was expanded in the distal optic cup Rbpj conditionally mutants (B). By E16.5, forming retinal rosettes largely lacked Atoh7 mRNA (upper left in D). Asterisks mark the forming ciliary body. E, F, Without Rbpj at E13.5, the POU4F2 expression domain was mispatterned (compare red nuclei in E, F). Arrows in F point to POU4F2+ RGCs inappropriately at the outer optic cup. G, H, POU4F2/DAPI double labels showed E16.5 Rbpj conditional mutant rosettes devoid of POU4F2+ RGCs (arrow in H points to a labeled cell next to a rosette). I, J, At E16.5, both Rbpj control and mutant eyes had extensive POU4F2 and GFP coexpression in the distal retina. Boxed areas at the right are a higher magnification, where arrows point to GFP+ cell bodies with red nuclei. K, The percentage of POU4F2+ RGCs significantly increased in Rbpj−/− cells. L, Both RXRγ+ RGCs and cones were significantly increased in Rbpj mutant retinal cells. Scale bars: 100 μm in A, E; vitreal is down in all panels; n = 3/3 embryos per age and genotype.
Figure 4.
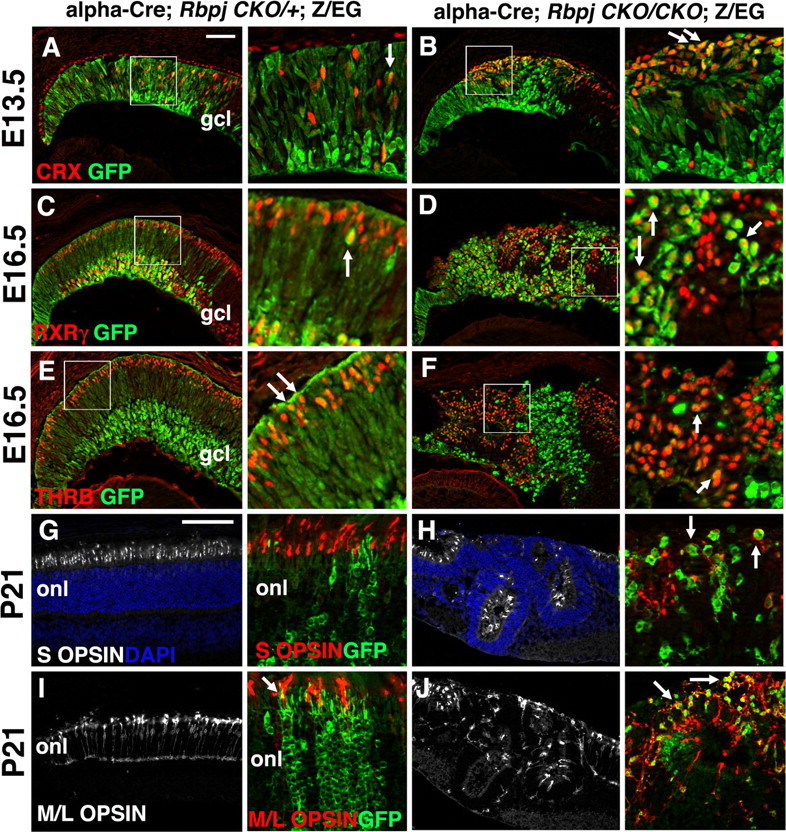
Rbpj represses cone photoreceptor cell development. Boxed areas in A–F are shown at higher magnification immediately to the right. A, B, CRX-GFP double labeled retinal sections showing derepression and mispatterning of CRX (red) in photoreceptor precursors, autonomously in E13.5 Rbpj−/−GFP+ cells (B). C, D, At E16.5, RXRγ+ expression (red) in RGCs within the gcl, and cones within the outer retina, was disorganized in Rbpj retinal mutants. Retinal rosettes had formed in the outer retina containing RXRγ+ cones, while RXRγ+ RGCs in the gcl surrounded the rosettes (D). Mutant cones sometimes displayed lower levels of GFP expression than inner retinal mutant cells. E, F, THRB+ cone photoreceptors were also increased autonomously in Rbpj−/−GFP+ cells. G, H, Adult retinal sections colabeled with S OPSIN (white) and DAPI. I, J, M/L OPSIN (white) labeling of distal retinal sections. Both types of cone photoreceptors were present in the rosettes of Rbpj conditional mutants. G–J, Higher magnifications. The adult α-Cre lineage (GFP+) normally contains mostly M/L cones (compare I versus G insets). When Rbpj was removed from this lineage, excess M/L OPSIN+ cones appeared (arrows in H,J insets point to coexpressing yellow cells). Mutant rosettes also contained cone photoreceptors derived from wild-type RPCs (red only cells). onl, Outer nuclear layer; vitreal is down in all panels. Scale bars: 100 μm in A, G; insets 8× magnified; n ≥ 3 animals per age and genotype.
Figure 5.
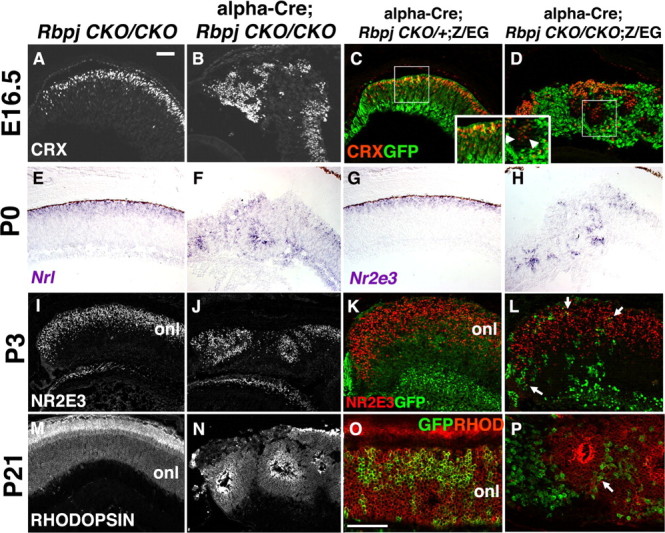
Conditional loss of Rbpj alters rod photoreceptor differentiation. A, B, E16.5 CRX+ photoreceptor precursor cells (white nuclei) in the forming rosettes of Rbpj retinal mutants. C, D, In controls, CRX+GFP+ cells were obvious in the outer retina. In Rbpj mutants, both Crx+GFP+ and CRX+GFP-neg cells were in the rosettes. E–H, Nrl and Nr2e3 mRNA expression at P0 showed rod precursor cells in rosettes. I, J, Nuclear NR2E3 protein expression at P3 further highlighted the abundance of rods in retinal rosettes. K, L, Normally, NR2E3 and GFP were extensively coexpressed in the P3 α-Cre lineage, but only very limited double-positive cells were present in Rbpj mutants (L, arrows). M, N, RHODOPSIN expression (white) in the adult eye indicated differentiated rods in Rbpj conditional mutant rosettes. O, P, Although the α-Cre lineage included rods, without Rbpj, only rare GFP+RHODOPSIN+ rods were found (arrow in P). onl, Outer nuclear layer; vitreal is down in all panels; scale bars: 100 μm in A, O; n ≥ 3 animals per age and genotype.
Rbpj promotes RPC proliferation and suppresses differentiation
To understand when and how the severely mispatterned retinas of Rbpj mutants arose, we surveyed embryonic eyes from E13.5–E16.5. By E16.5, removal of Rbpj had already disrupted retinal lamination and caused excess differentiated TUBB3+ (βIII Tubulin+) neurons, which surrounded the rosetted areas (Fig. 2A,B) (data not shown). In younger, E13.5 α-Cre;RbpjCKO/CKO eyes, TUBB3+ neurons were already disorganized and mispositioned at the outer retina (Fig. 2, compare C, D). Mitotically active RPCs were also examined by BrdU pulse labeling and CCND1/CyclinD1 expression. In E16.5 control eyes, RPCs expressed these markers broadly (Fig. 2I), but in Rbpj mutants there was an eightfold loss of S-phase Rbpj−/−GFP+ cells, with the remaining proliferative RPCs mislocalized to the periphery of forming rosettes (Fig. 2J,T) (control 35 ± 1.5%, n = 6758 GFP+ cells from 3 eyes; Rbpj mutants, 4.2 ± 0.2%, n = 4345 GFP+ cells from 3 eyes; p < 0.0001). Earlier at E13.5, we also observed fewer Rbpj−/−GFP+BrdU+ (data not shown) or Rbpj−/−GFP+CCND1+ RPCs, compared to littermate control eyes (Fig. 2K,L). This simultaneous loss of proliferating RPCs and increased neuronal differentiation in the absence of Rbpj were consistent with the phenotypes of Notch1 conditional mutants (Jadhav et al., 2006b; Yaron et al., 2006).
Next we asked how the removal of Rbpj affected the expression of Hes1 and Hes5, two known transcriptional targets of the NOTCH-ICD/MAML/RBPJ complex (Kageyama and Ohtsuka, 1999; Kopan, 2002). Normally, many E16.5 GFP+ RPCs coexpress HES1 (Fig. 2E,S), but in Rbpj mutants there was an 11-fold loss of GFP+HES1+ cells (Fig. 2F,S) (control, 59 ± 0.3%, n = 4586 GFP+ cells from 3 animals; Rbpj mutants, 5 ± 0.2%, n = 3401 GFP+ cells from 3 mutants; p < 0.0001). Moreover, an Rbpj-dependent reduction in HES1+ cells was already evident by E13.5 (Fig. 2G,H). We also examined Hes5 mRNA expression, and found it downregulated in E12.5-E16.5 α-Cre;RbpjCKO/CKO peripheral retinas (Fig. 2O,P) (data not shown).
We then tested whether Rbpj mutant cells undergo apoptosis, by double labeling E13.5-P3 retinal sections with anti-GFP and anti-cPARP. Apoptotic cells are normally very rare in E13.5-E16.5 control retinas (Fig. 2M, arrows). Although there was no significant difference in cPARP+ cells between E13.5 control and Rbpj conditional mutant eyes (n = 3 animals/per age and genotype, data not shown), by E16.5 apoptosis was cell autonomously increased in the Rbpj−/−GFP+ population (Fig. 2N, arrows) (controls, 0.2 ± 0.04%, n = 4970 GFP+ cells from 3 animals; Rbpj−/−, 0.9 ± 0.1%, n = 3311 GFP+ cells from 3 animals; p = 0.006). At P3, excess cPARP+GFP+ cells were still obvious in α-Cre;RbpjCKO/CKO eyes (data not shown). We conclude that without Rbpj function, embryonic RPCs autonomously downregulate Hes1 and Hes5, prematurely exit the cell cycle and differentiate. These defects are accompanied by disorganization of the developing retinal architecture and cell autonomous death of a portion of the α-Cre;Rbpj−/− retinal lineage. Reduced RPC proliferation, increased differentiation, and to a lesser extent increased apoptosis, all contribute to a significant reduction in the GFP+Rbpj mutant lineage by E16.5 (Fig. 6A).
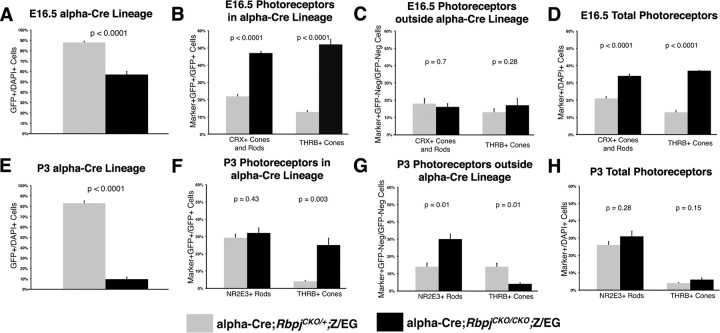
Figure 6.
Cell autonomous increase in cones, and postnatal non-autonomous correction of the total photoreceptor population in Rbpj mutants. A, Significant reduction in E16.5 α-Cre lineage cells without Rbpj. B, The proportion of CRX+GFP+ photoreceptor precursors and THRB+GFP+ cones increased autonomously at E16.5 in the absence of Rbpj. C, Both CRX+GFP-neg and THRB+GFP-neg cell populations were unaffected in Rbpj conditional mutants. D, The overall percentages of both photoreceptor precursors and nascent cones are significantly increased in E16.5 Rbpj conditional mutants. E, At P3, the α-Cre lineage was drastically smaller without Rbpj function. F, The proportion of NR2E3+GFP+ rods was the same between genotypes, THRB+GFP+ cones increased in an Rbpj-dependent manner. G, P3 NR2E3+GFP-neg wild-type rods (outside the α-Cre lineage) were significantly increased, at the same time that THRB+GFP-neg cones were reduced, in α-Cre;RbpjCKO/CKO;Z/EG eyes. H, The shifts in rod and cone populations within and outside of the α-Cre lineage resulted in the correct overall ratio for each cell type.
Rbpj cell autonomously blocks RGC formation
Because embryonic retinal neurogenesis is derepressed in the absence of Rbpj, we were interested to learn how RGC would be affected, since this cell class differentiates first. Therefore, we tested the expression of two genes critical for RGCs formation, Atoh7/Math5 and Pou4f2/Brn3b (Gan et al., 1999; Brown et al., 2001; Wang et al., 2001). The bHLH gene Atoh7 is required for RGC genesis, presumably because it activates Pou4f2 and additional RGC factors, although paradoxically terminally mitotic Atoh7+ RPCs give rise to all seven retinal cell fates (Brown et al., 2001; Hutcheson and Vetter, 2001; Liu et al., 2001; Wang et al., 2001; Yang et al., 2003; Brzezinski, 2005). In E13.5 Rbpj mutants, Atoh7 expression was upregulated (Fig. 3, compare A, B), along with an analogous increase in Pou4f2+ RGCs (Fig. 3E,F) (data not shown). At E16.5, the Atoh7 expression pattern was extremely disrupted in Rbpj conditional mutants, with distal-most RPCs displaying an intense area of Atoh7 expression (Fig. 3C,D). Derepression of Atoh7 in Rbpj mutants correlated with the loss of Hes1 (Fig. 2F,H), which normally represses Atoh7 activation (Brown et al., 1998; Takatsuka et al., 2004; Lee et al., 2005).
POU4F2 encodes a POU-domain transcription factor that is expressed by the majority of differentiating RGCs (Erkman et al., 1996; Gan et al., 1996). Without Rbpj function, POU4F2+ cells were moderately expanded at E13.5 and increased by 1.5-fold at E16.5 (Fig. 3E–K, arrows in J) (control, 30.3 ± 0.9%, n = 1060 GFP+ cells from 3 animals; Rbpj−/−, 46.4 ± 2.7%, n = 828 GFP+ cells from 3 animals; p < 0.001). Like TUBB3+ neurons, ectopic POU4F2+ RGCs were excluded from forming rosettes (Fig. 3H, arrow). While some Rbpj mutant RGCs were mispositioned at the outer retina (Fig. 3F, arrows), others correctly migrated to the ganglion cell layer (gcl) (Fig. 3G–J), implying that the initiation of lamination does not require Rbpj activity. However, the subsequent mispatterning of α-Cre;RbpjCKO/CKO retinas ultimately affected the gcl, since it was bent around the forming rosettes in mutant eyes (Fig. 3H,J). Despite the formation of ectopic embryonic RGCs from E13.5-E16.5, very few POU4F2+ RGCs were found in P21 α-Cre;RbpjCKO/CKO eyes, which had thinner optic nerves (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). The loss of RGCs was correlative with increased apoptosis of Rbpj−/− GFP+ cells (Fig. 2M,N).
Rbpj suppresses cone photoreceptor fates autonomously
Next, we wished to understand to what extent Rbpj regulates cone photoreceptor fates. The orphan retinoic acid receptor RXRγ and its heterodimeric partner thyroid hormone receptor, THRB/TRβ2, are two of the earliest markers of cone photoreceptors (Hoover et al., 1998; Mori et al., 2001; Roberts et al., 2005; Ng et al., 2009). RXRγ is also expressed by prenatal RGCs, but RGCs and cones are easily distinguishable by their locations on opposite sides of the optic cup and distinct morphologies (Fig. 4A,I). In E13.5 Rbpj mutants, the RGC and cone populations were properly located at the inner and outer retina respectively, with some disorganization distally (data not shown). At E16.5, we quantified the percentage of GFP+RXRγ+ cells in control and Rbpj conditional mutants, and found a threefold, autonomous increase in RXRγ+ Rbpj−/− cells (Figs. 3L, 4C,D) (controls, 25 ± 1.1%, n = 6381 GFP+ cells from 3 animals; Rbpj−/−, 77 ± 3.8%, n = 4261 GFP+ cells from 3 animals; p < 0.0001). This represented a 3.2-fold increase in embryonic RGCs (controls 13.5% versus Rbpj−/− 43.5%) and 2.9-fold increase of embryonic cones in the outer retina (controls 11.5% versus Rbpj−/− 33.5%). Importantly, the RXRγ+ cones were always abnormally clustered in the center of forming rosettes within α-Cre;RbpjCKO/CKO;Z/EG eyes (Fig. 4C,D). To verify that mispositioned RGCs had not skewed our RXRγ+ cone quantification, we also assayed E16.5 THRB+ cones, which displayed a fourfold increase in the absence of Rbpj (Figs. 4E,F, 6B) (controls 13 ± 0.7%, n = 2234 GFP+ cells from 3 animals; Rbpj−/− 52 ± 3%, n = 1320 GFP+ cells from 3 animals).
Cones and rods are thought to originate from CRX+ bipotential precursor cells (Furukawa et al., 1997; Chen et al., 2002). The decision of a CRX+ precursor to adopt either a cone or rod fate is partly regulated by the rod genes Nrl and Nr2e3 (Swaroop et al., 1992; Liu et al., 1996; Chen et al., 1999; Kobayashi et al., 1999; Mears et al., 2001). In the distal optic cup of E13.5 Rbpj conditional mutants, CRX+GFP+ cells (Fig. 4A,B) were disorganized and expanded, just like RXRγ+ cones. The percentage of CRX+GFP+ in E16.5 control and Rbpj mutant retinas was also determined (Figs. 5A–D, 6B) (data not shown). Here, CRX+ postmitotic photoreceptors were increased by 2.1-fold, autonomously within the Rbpj−/− population (Figs. 5A–D, 6B) (controls, 22.3 ± 1.1%, n = 5616 GFP+ cells from 3 eyes; Rbpj−/−, 47.3 ± 1.1%, n = 5278 GFP+ cells from 3 eyes). We also observed this same outcome for the marker OTX2 (data not shown), which acts upstream of CRX during retinal development (Nishida et al., 2003). Therefore, we assume that Rbpj is genetically required for cone genesis at the level of Otx2/Crx gene activation. To understand how the loss of Rbpj affected mature cone photoreceptors, S OPSIN (blue) and M/L OPSIN (red/green) expression were examined in adult eyes (Fig. 4G–J) (Zhu and Craft, 2000; Zhu et al., 2006). Both types of cones were present in the rosettes of Rbpj mutants but, we were unable to score their cell autonomy, due to severe mispatterning and aberrant outer segment morphologies (Fig. 4H,J). However, in Rbpj retinal mutants M/L OPSIN+ cones were more prevalent than S OPSIN+ cones (Fig. 4I,J, arrows), which differed from Notch1 conditional mutants, where the S cones outnumbered M/L cones (Jadhav et al., 2006b). Overall, we conclude that Rbpj normally acts cell autonomously within embryonic RPCs to block cone photoreceptor formation.
Rod photoreceptor development in Rbpj conditional mutants
At E16.5, cone differentiation has peaked within the CRX+ retinal cell population, while the apex of rod genesis is still days away. Thus, at this age the CRX population might be expected to contain more cones than rods, but because rods are the largest retinal cell class in rodents, they already outnumber cones (Cepko et al., 1996; Rapaport et al., 2004). The expanded CRX-expressing population of Rbpj conditional mutants prompted us to ask whether rod development was also affected, for which we foresaw several possible mechanisms. First, the removal of Rbpj autonomously depleted the RPC pool (Figs. 2S,T, 6A), resulting in fewer RPCs to adopt a rod fate. Complicating this idea is the potential for the remaining RPC population to autonomously require Rbpj to block rod development, resulting in most (or all) of the Rbpj−/− RPCs becoming rods, at the expense of bipolar and Müller glial fates. To explore this idea further, the mRNA expression patterns of Nrl and Nr2e3, two transcription factors critical for rod differentiation, were examined (Swaroop et al., 1992; Liu et al., 1996; Chen et al., 1999; Kobayashi et al., 1999; Mears et al., 2001). At P0, Nrl and Nr2e3 are normally present in postmitotic rods within the outer nuclear layer (Fig. 5E,G). In Rbpj conditional mutants both genes were expressed by a large proportion of the rosetted cells (Fig. 5F,H). At E13.5 and E16.5 neither Nrl nor Nr2e3 mRNAs were precociously expressed in the absence of Rbpj (data not shown). Thus, although Nrl+ and Nr2e3+ rods were mispositioned within the rosettes of Rbpj mutants, it was not obvious whether the number of rods produced was abnormal.
The other retinal cell classes were also surveyed in the α-Cre lineage of adult Rbpj conditional mutants (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). There were no noticeable alterations in amacrines, using the markers CALBINDIN (marks horizontals and a subset of amacrines; supplemental Fig. 2A–C, available at www.jneurosci.org as supplemental material) and CALRETININ (marks A2 amacrines; supplemental Fig. 2J–L, available at www.jneurosci.org as supplemental material). Calbindin+ horizontal neurons were also unaffected in Rbpj conditionally mutant eyes (right arrows in supplemental Fig. 2C,available at www.jneurosci.org as supplemental material). In addition, CHX10+ bipolars (supplemental Fig. 2D–F, available at www.jneurosci.org as supplemental material) and SOX 9+ or CRALBP+ Müller glia (supplemental Fig. 2G–I, available at www.jneurosci.org as supplemental material) (data not shown) were compared in control versus Rbpj conditionally mutant eyes. Here, we observed fewer CHX10+GFP+ or SOX9+GFP+ cells within the Rbpj mutant α-Cre lineage (supplemental Fig. 2D–I, available at www.jneurosci.org as supplemental material). While this supported the idea that postnatal RPCs erroneously adopt an rod fate, it was also consistent with a role for Rbpj in autonomously promoting Müller glial fates, like Notch1 regulation of this cell class (Furukawa et al., 2000; Bernardos et al., 2005; Roesch et al., 2008). Therefore, we concluded it was not possible to discriminate among these possibilities further here, since the autonomy of potential Rbpj mutant bipolar and Müller glia phenotypes must be examined by conditional deletion of Rbpj specifically after RGC and cone genesis is completed.
Wild-type cells compensate for shifts in Rbpj mutant photoreceptor cell populations
Yet another potential mechanism by which rod fates could be affected in Rbpj−/− retinas might occur if postmitotic cones non-autonomously signal RPCs to adopt a rod fate. Intriguingly, we noticed that in E16.5 Rbpj conditional mutants, CRX+GFP-neg cells usually resided next to CRX+GFP+ cells, particularly around the periphery of forming rosettes (Fig. 5D inset, arrowheads). However, the E16.5 CRX+GFP-neg population was not significantly increased (Fig. 6C), and expansion of CRX+ retinal cells was solely attributable to the cell autonomous increase in THRB+ cones (Fig. 6B–D). Nevertheless, Rbpj−/− cones may non-autonomously influence the fate that later RPCs adopt, for which the outcome is not immediately evident. To test this possibility, we quantified nascent THRB+ cones and NR2E3+ rods in early postnatal control and Rbpj conditionally mutant eyes (Figs. 5I–L, 6E–H) (data not shown). At P3, the α-Cre lineage normally encompasses 80% of the distal retina (Fig. 6E), of which 30% are NR2E3+ rods and 4% are THRB+ cones (Fig. 6F). In contrast, the Rbpj mutant lineage was reduced eightfold (Fig. 6E), but with a sixfold increase in THRB+ cones (control 4 ± 0.4%, n = 15,441 GFP+ cells from 3 animals; Rbpj−/− 25 ± 4%, n = 818 GFP+ cells from 3 animals; p = 0.01). Remarkably, although the mutant lineage was much smaller (Fig. 6E), it contained the correct proportion of rods (Fig. 6F) (control, 29.3 ± 0.2%, n = 18,459 GFP+ cells from 3 animals; Rbpj−/−, 32 ± 0.3%, n = 15,560 GFP+ cells from 3 animals; p = 0.43). Furthermore, the total population of rods and cones was the same between controls and Rbpj conditional mutant eyes (Fig. 6H), suggesting that the percentages of cones and rods were adjusted within the wild-type population, to correct for abnormal numbers of each cell type within the α Cre;Rbpj−/− lineage. Consistent with this hypothesis, we observed that NR2E3+ rods were significantly increased, and THRB+ cones significantly reduced, only in the GFP-neg population (Fig. 6G). We conclude that wild-type RPCs compensated for two simultaneous abnormalities in the α-Cre;Rbpj−/− lineage: (1) cell autonomous overproduction of cones that began around E13.5, and (2) dramatic loss of the postnatal α-Cre lineage, which contained the correct ratio, but not the proper number of GFP+ rods (Fig. 6E–H).
Finally, differentiated rods were examined by comparing the expression of the terminal differentiation marker RHODOPSIN at P10 and P21, in both α-Cre;RbpjCKO/+;Z/EG and α-Cre;RbpjCKO/CKO;Z/EG eyes (Fig. 5M–P) (data not shown). Although control retinas had an abundance of RHODOPSIN+GFP+ rods (Fig. 5O), only rare RHODOPSIN+GFP+ rods were observable in Rbpj conditional mutants (Fig. 5P, arrow), indicating that most of the rosettes were almost entirely comprised of wild-type rods. Importantly, the predominantly rod-filled rosettes of Rbpj mutants (Fig. 5P) drastically differed from Notch1 retinal mutants, whose rosettes contained mostly cones (Yaron et al., 2006), implying that Rbpj might act either outside of the Notch pathway, or complexes with a different activated receptor, to control photoreceptor cell population dynamics.
Broad removal of Rbpj with Chx10-Cre causes analogous embryonic retinal phenotypes
To independently verify each Rbpj retinal phenotype, we also deleted Rbpj with a different retinal Cre driver, Chx10-Cre, which is expressed by the vast majority of embryonic RPCs, albeit with some mosaicism (Rowan and Cepko, 2004; Jadhav et al., 2006a). The Z/EG transgene was also included in these experiments, to demonstrate the cell autonomy of each retinal cell class. At E13.5 and E16.5, we observed identical shifts in the expression of HES1 (supplemental Fig. 3A,B), POU4F2 (supplemental Fig. 3C,D), CRX (supplemental Fig. 3E,F), and RXRγ (supplemental Fig. 3G,H, all available at www.jneurosci.org as supplemental material) in Rbpj−/−GFP+ cells. Thus, HES1+ RPCs decreased cell autonomously while RGC and cone differentiation also increased cell autonomously. These shifts were found as early as E13.5 (data not shown). We also tested NR2E3 expression at P3, and found that the rod precursor cells were overwhelmingly GFP-neg, in CHX10-Cre; RbpjCKO/CKO;Z/EG eyes (supplemental Fig. 3I,J, available at www.jneurosci.org as supplemental material).
Notch3 and Hes1 suppress RGC, but not cone development
Notch1 and Rbpj each suppress cone formation, yet their mutant phenotypes suggest they perform distinct functions during RGC genesis (Jadhav et al., 2006b; Yaron et al., 2006). Therefore, we hypothesized that distinct combinations of Notch pathway genes regulate each retinal cell type. To address this idea, we compared the functions of Rbpj to those of the Notch3 receptor and downstream effector, Hes1. First, we better defined the expression pattern of Notch3, which was already known to be present in the prenatal rodent retina (Lindsell et al., 1996), and then searched for RGC or photoreceptor phenotypes in Notch3 mutants. Here, we took advantage of a Notch3 gene trap mutant allele, in which an in-frame βgal-neo insertion into the Notch3 coding region causes a 99% loss of mRNA in homozygotes (Leighton et al., 2001; Mitchell et al., 2001). Moreover, the resulting Notch3-βgal fusion protein is localized to the endoplasmic reticulum and secretory vesicles of Notch3-expressing cells (Fig. 7A–I) (Mitchell et al., 2001). We observed Notch3LacZ in E11.5, E13.5 and E16.5 retinal progenitor cells and nascent RGCs, although expression in the latter cell class was downregulated at E16.5 (Fig. 7A–C). The Notch3LacZ expression pattern in E11.5–E13.5 RPCs is consistent with the published mRNA expression pattern (Lindsell et al., 1996), although additional expression in nascent RGCs presumably reflects βgal perdurance. Indeed, there was extensive Notch3LacZ expression within POU4F2+ RGCs at E13.5 (Fig. 7E), although βgal+HES1+ RPCs (Fig. 7D) and a few βgal+CRX+ photoreceptor precursors (Fig. 7F) were also evident. Interestingly, by E16.5 only the youngest RGC cell bodies at the GCL-neuroblastic boundary expressed Notch3LacZ (Fig. 7H, arrows), along with some residual βgal expression in RGC axons (Fig. 7C,H). At this older age, there were numerous βgal+HES1+ progenitor cells (Fig. 7G), but only a few, scattered βgal+CRX+ photoreceptors (Fig. 7I). By comparing E16.5 wild-type and Notch3 mutant retinas, we found a 1.3-fold increase in POU4F2+ RGCs (Fig. 7K) (control 16.8 ± 0.9%, n = 1930 cells from 3 animals; Notch3−/− 21.2 ± 0.6%, n = 1405 cells from 3 animals; p = 0.01), which correlated with the 1.1-fold decrease in HES1+ progenitor cells (Fig. 7J) (control 48.4 ± 1.1%, n = 1785 cells from 3 animals; Notch3−/− 43.9 ± 1%, n = 1152 cells from 3 animals; p < 0.01). Interestingly, the loss of Notch3 did not affect CRX+ photoreceptor precursors (Fig. 7L) (control 25.2 ± 0.7%, n = 1756 cells from 3 animals; Notch3−/− 23.3 ± 1.1%, n = 1371 cells from 3 animals; p = 0.11). We conclude that Notch3 normally suppresses RGC, but not cone formation.
Figure 7.
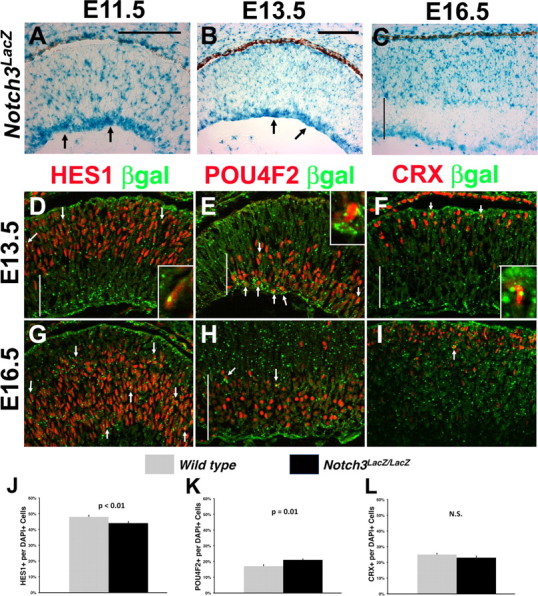
Notch3 expression and mutant phenotypes during embryonic RGC and cone development. A–C, LacZ expression from an in-frame β-geo insertion in the Notch3 gene during early retinogenesis. From E11.5 to E13.5, many cells express Notch3LacZ predominantly at the inner optic cup, where differentiating RGC neurons accumulate (arrows in A, B). By E16.5, Notch3LacZ expression was restricted to the outer retina, although some RGC axons exhibited retained expression. D–I, Colabeling of E13.5 and E16.5 retinal sections for βgal with HES1 (progenitors, D, G), POU4F2 (RGCs, E, H), or CRX+ (photoreceptors, F, I). Arrows point to cells coexpressing βgal in vesicles, and marker of interest in nucleus. At E13.5 there were some colabeled cells for each marker, but extensive colabeling of βgal in differentiated RGCs (E). Insets in D–F show colabeled cells at high magnification. G, H, At E16.5, there were many more βgal+HES1+ retinal progenitors, but markedly fewer RGCs expressing Notch3LacZ in the GCL, except for a few colabeled cells at the border with the neuroblastic layer (arrows). I, Rare Notch3LacZ+CRX+ cells were also found at this age (arrow). The vertical line in C–F, H shows the width of the GCL. J–L, Percentage of HES1+, POU4F2+ or CRX+ cells in the central retina of E16.5 Notch3 controls and mutants. There was a significant loss of progenitors and increase in RGCs in the absence of Notch3. n = 6 sections from 3 embryos per genotype; N.S., Not significant. Scale bars: 100 μm in A, B.
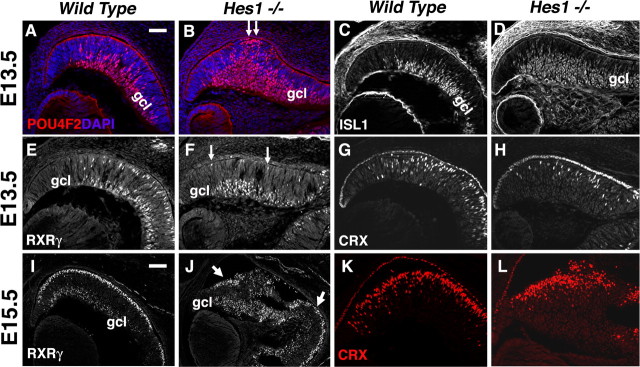
Next, we surveyed RGC and early photoreceptor development in Hes1 germline mutants (Tomita et al., 1996). Hes1 was previously shown to promote RPC proliferation, repress RGC, horizontal and rod neurogenesis and promote Müller glia differentiation (Tomita et al., 1996; Furukawa et al., 2000). In addition, Hes1−/− retinas are mispatterned by E15.5, with retinal rosettes that contain too many rod photoreceptors (Tomita et al., 1996; Takatsuka et al., 2004). Somewhat surprisingly, cone development has not been examined in Hes1 mutants. Therefore, we assayed the prenatal RGC and photoreceptor phenotypes of Hes1 mutants. Previous reports showed both precocious and an expanded domain of TUBB3+ neurons in Hes1−/− embryonic retinas, during RGC genesis (Takatsuka et al., 2004; Lee et al., 2005). We revisited this by comparing the RGC markers POU4F2, ISL1 and RXRγ in E13.5 and E15.5 Hes1 control and mutant eyes (Fig. 8A–D) (data not shown). Indeed, all three RGC markers were greatly expanded in the absence of Hes1 (Fig. 8B,D,F,J). In some embryos, we also found RGCs inappropriately located at the outer optic cup (Fig. 8B, arrows), like Rbpj conditional mutants (Figs. 2D, 3F). Unexpectedly, the increase in RGCs present in Hes1 mutants was accompanied by a loss of outer RXRγ+ cones (Fig. 8F, arrows). We also found a 1.5-fold reduction in E13.5 CRX+ cells in Hes1 mutants, further confirming the loss of cones (control, 11.9 ± 0.007%, n = 5300 DAPI+ cells from 3 embryo eyes; Hes1−/−, 8.2 ± 0.01%, n = 5712 DAPI+ cells from 3 embryo eyes; p = 0.02). Interestingly, by E15.5, the proportion of CRX+ cells in Hes1−/− eyes appeared to rebound to that of controls, although this is likely due to the precocious onset of rod development as previously reported (Tomita et al., 1996). Unfortunately Hes1 germline mutant embryos could not be recovered beyond E15. We conclude that RGC neurogenesis is normally suppressed by Notch3, Rbpj and Hes1, while cone formation is regulated by Notch1-Rbpj signaling through a different downstream effector, instead of Hes1.
Figure 8.
Hes1 simultaneously suppresses RGC and promotes cone photoreceptor fates. A–D, POU4F2+ (red in A, B) and ISL1+ (white in C, D) RGC nuclei were dramatically increased in E13.5 Hes1−/− retinas. Arrows in B point to mispatterned POU4F2+ RGCs in the outer optic cup. Mouse anti-ISL1 nonspecifically labeled the surrounding mesenchyme, hyaloid vasculature and forming lens capsule. E, F, RXRγ expression in RGCs in the gcl, and cones in the outer optic cup, highlighted the simultaneous increase in RGCs and decrease in cones (F, arrows). G, H, E13.5 CRX expression was also reduced in Hes1−/− eyes. Anti-CRX also labeled RPE nuclei, due to cross-reactivity with OTX2 (Zhu and Craft, 2000). I, J, In E15.5 Hes1−/− eyes, the outer retinal domain of RXRγ was disrupted. Arrows in J point to two regions containing cone cell nuclei, disrupted by the expansion of RGCs. K, L, CRX+ cells in E15.5 Hes1 mutants appeared were mispatterned but present in nearly normal proportions. Vitreal is down, distal left in all panels; scale bar: 100 μm; n = 3 embryos per age and genotype.
Discussion
Although the Notch pathway clearly has multiple roles in the vertebrate retina, the requirements for each gene remain largely unknown, and the cell autonomy of those gene functions already investigated has not been very well determined. Nevertheless, loss of Delta-Notch signaling results in excess embryonic RGCs and cone photoreceptors; while overexpression of Delta1, activated Notch or Hes1 prolongs the mitotic activity of RPCs (Austin et al., 1995; Dorsky et al., 1995; Henrique et al., 1995; Tomita et al., 1996; Ahmad et al., 1997; Dorsky et al., 1997; Schneider et al., 2001; Silva et al., 2003; Takatsuka et al., 2004; Jadhav et al., 2006b; Yaron et al., 2006). In this paper, we demonstrate that the Notch signal integrator Rbpj autonomously promotes progenitor cell growth, and suppresses RGC and cone photoreceptor development. In the embryonic retina, both Rbpj−/− RGCs and cones were overproduced and mispatterned, including mislocalization of RGCs at the outer retina, and cones in the center of forming rosettes. But, only the ectopic cones fully persisted into adulthood in Rbpj conditionally mutant eyes. An increase in apoptotic Rbpj−/− cells occurred where mutant RGC cell bodies reside, during the period of normal RGC axon outgrowth and connectivity, which correlated with the loss of RGCs and thinner optic nerves in Rbpj−/− adult eyes. Although Notch signaling can regulate cell survival (Oishi et al., 2004), we suggest the additional possibility that Rbpj−/− RGCs die potentially during the normal corrective process of RGC overproduction. Alternatively, the ectopic RGCs may produce misrouted axons that fail to reach the optic nerve.
Our interest in RGC cell fate specification drew us to the mechanism of how Notch signaling controls the timing of RGC differentiation. Notch regulation of RGC neurogenesis is less complex, since these cells initiate differentiation in the absence of extrinsic signals from other neurons (Waid and McLoon, 1998; Silva et al., 2003; Dakubo and Wallace, 2004; Liu et al., 2006). Based on previous work in the vertebrate and Drosophila eye, the prevailing model holds that an ‘equivalence group’ of mitotic RPCs coexpressing DELTA and NOTCH, subsequently undergo lateral inhibition to produce one or more postmitotic cells, which downregulate Notch/Rbpj/Hes activity and upregulate bHLH proneural expression thereby controlling the sequential onset of each retinal neuron class (Cepko, 1999; Kageyama and Ohtsuka, 1999; Kageyama et al., 2008). However, Notch1, Notch3, Rbpj, and Hes1 mutant mice exhibited separate and overlapping retinal phenotypes, provoking the question of which combinations of ligands, receptors and downstream effectors regulate RGC versus cone formation. Here, we tested the embryonic roles of Rbpj, which integrates input from all combinations of Notch ligand and receptors. We found that Rbpj represses RGC fates, consistent with the function of Hes1, as well as a Notch-mediated blockade of RGC formation in other vertebrate eyes. But, Notch1 conditional mutants had reduced numbers of RGC marker+ cells (Jadhav et al., 2006b; Yaron et al., 2006), and although Notch1 and Rbpj each block cone fates, neither Notch3 nor Hes1 participate in this process. Therefore, we delineated two branch points in the Notch pathway through which Rbpj regulates RGC and cone formation simultaneously, namely variable receptor input, and/or the activation of different downstream effectors (Fig. 9). The 1.3-fold increase in POU4F2+ RGCs in Notch3 mutants is similar to the 1.5-fold RGC increase that occurred without Rbpj. However, because conditional deletion of Rbpj underestimates its full requirements during retinal cell type specification, it remains plausible that Notch1 and Notch3 act synergistically, or cross-regulate one another, during RGC formation. Thus, the total requirement for Notch receptors during RGC neurogenesis should become evident through simultaneous removal of both receptors. We are optimistic that future Notch1;Notch3 mutant analyses will finally unify the Notch RGC phenotypes among different vertebrate model organisms.
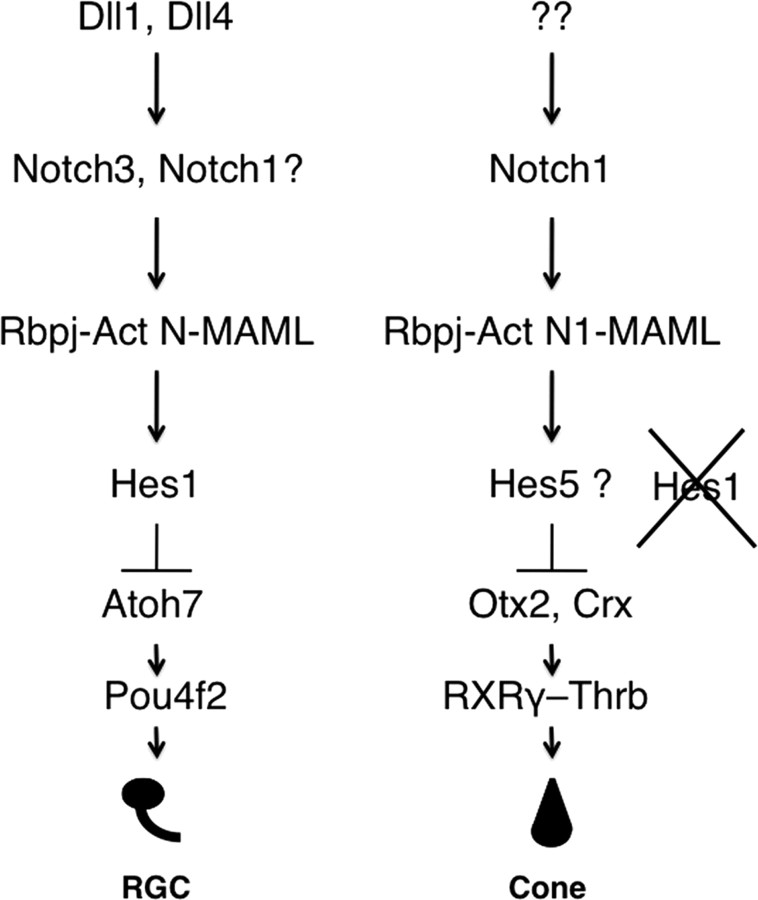
Figure 9.
Model for differential Notch pathway regulation of RGC versus cone fates. Distinct combinations of Notch pathway genes regulate the development of particular retinal cell types.
Because Rbpj can activate either Hes1 or Hes5, Hes5 is the obvious candidate to respond to Notch1-Rbpj during cone photoreceptor genesis. This raises interesting questions about the spatial and temporal overlap of HES1 and HES5 expression and resulting retinal lineages, about the influences of other signaling pathways, such as shh, in modulating Hes1 or Hes5 gene activity (Wall et al., 2009), and whether these transcriptional repressors act separately, or with partially overlapping cell autonomous functions. Because cone differentiation was reduced in Hes1 mutants, Hes1 may normally repress a negative regulator of cone fates. Interestingly, Hes1 and Hes5 genetically repress one another in particular contexts (Hatakeyama et al., 2004). Alternatively, Rbpj may regulate the timing of cone precursor cell formation directly, or act cell autonomously through another transcriptional target (Iso et al., 2003). Future experiments that establish the cell autonomy of Deltalike1, Notch3, Hes1 and Hes5 gene functions will distinguish among these possibilities. Interestingly, yet another level of Notch signal complexity is likely to exist, since the requirement for Deltalike1 and Deltalike4 ligands was recently suggested for RGC development (Rocha et al., 2009) (Fig. 9).
In comparing photoreceptor development between marked Rbpj−/− and control retinal lineages, we obtained clear evidence that retinal cells adjust their production of rods and cones, when confronted with population shifts in a neighboring lineage. Therefore, the developing retina monitors both the overall production of photoreceptors to non-photoreceptors, and the correct proportion of rods to cones. Previous in vitro studies with mixed-age retinal cultures showed that rod precursors can induce nearby embryonic RPCs to differentiate as rods (Wantanabe and Raff, 1990; Reh, 1992). In mixed pellet cultures of embryonic and postnatal retinal cells, the embryonic RPCs had a higher propensity to differentiate as rods (Wantanabe and Raff, 1990). In the second study, embryonic RPCs were introduced to retinal monolayers containing photoreceptor-filled rosettes. Here too, the embryonic RPCs were induced to adopt the rod fate, especially when situated next to rod-containing rosettes (Reh, 1992). Exogenous growth factor addition could influence the rate of rod production, but not the fate chosen by RPCs. In addition, embryonic RPCs could not be induced to become rods when likewise cultured with a monolayer of cortical cells. Together, these studies suggested that a local cue, emanating from closely situated rod precursors, directs prenatal RPCs to adopt the rod fate.
Our discovery of non-autonomous compensation by wild-type retinal cells for the shifts in rod and cone numbers in Rbpj conditionally mutant retinas, raises the obvious question of whether Rbpj regulates some, or all, aspects photoreceptor homeostasis during development. RBPJ, either within or outside of the context of Notch signaling, for example in a complex with PTF1A (Masui et al., 2007; Hori et al., 2008), could non-autonomously influence the choice of bipotential CRX+ cells through a mechanism that maintains the balance of rod to non-rods, cones to non-cones and/or total photoreceptor to non-photoreceptor populations. Theoretically, such a signal might be transduced from cell-to-cell in a subsequent round of Notch signaling, or use other signaling pathways. Importantly, Notch regulates tissue homeostasis in different organs of the body, although it does so by controlling a variety of physiologic processes (Lin and Kopan, 2003; Lewis, 2008; Okuyama et al., 2008; Robinson, 2008; Brabletz et al., 2009). In addition, Notch is a key regulator of normal tissue growth, and Notch activity is inappropriately upregulated during tumor cell overgrowth (for review, see Kopan, 2002; Gridley, 2003; Lasky and Wu, 2005; Sjölund et al., 2005; Louvi and Artavanis-Tsakonas, 2006). If the photoreceptor homeostasis highlighted in our Rbpj conditional mutant analysis is Notch-dependent, it might act through a different receptor, since Notch1−/− cells autonomously overproduced cones, but without an analogous appearance of rod photoreceptors within the forming rosettes (Jadhav et al., 2006a; Yaron et al., 2006). On the other hand, we observed that Rbpj−/−GFP+ cells autonomously maintained the correct ratio of rods (Fig. 6F), despite a profound loss of the α-Cre lineage. Furthermore, we found a loss of cone photoreceptors in Hes1 germline mutants, at the same age that RGC development was both precocious and expanded. This suggests that since all the retinal cells lacked Hes1 activity, at least some RPCs were shunted away from the cone fates to maintain the correct overall number of photoreceptors, perhaps because rod fates are expanded in this mutant background. At present our data implicate but do not clearly demonstrate whether Notch signaling regulates photoreceptor cell population dynamics. Alternatively, the quantification of cell autonomy for each mutant phenotype, coupled with the reduced mutant RPC pool in this mutant, may have identified an Rbpj-independent retinal process for regulating photoreceptor cell numbers. To understand the genetic hierarchy that controls this important process, future experiments will compare both the cell biological characteristics and gene profiles of the wild-type and Rbpj−/− marked cell populations, within the period cone and rod development examined here (E16-P3).
The ability of tissues to sense and regulate their overall size, and the proportion of each cell type, was first hypothesized more than two decades ago (Gurdon, 1988). Both characteristics are critical for normal development, and presumably are affected during tumor formation. These elusive homeostatic mechanisms are still intensely investigated, with multiple signaling pathways implicated as the inducers of this process (Gurdon et al., 1998, 1999; Standley et al., 2001; Piddini and Vincent, 2009). Here, we demonstrate that during a critical developmental period the mammalian retina keeps track of, and can correct, the size of its photoreceptor populations. This finding is directly relevant for embryonic stem cell or retinal progenitor cell therapies, which aim to restore reduced or missing vision (MacLaren et al., 2006; Lamba et al., 2009). Although much progress has been made in this area, several significant hurdles remain, including the ability to produce pure populations of photoreceptor precursors for reintroduction and improving their efficiency of tissue integration. It is exciting to speculate that the future identification of molecular pathways that monitor photoreceptor population dynamics will contribute beneficially toward these unresolved cell therapy issues.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants GM55479, DK66408, and HD44056 to R.K. and NIH Grants EY13612 and EY18097 to N.L.B. We thank Tasuku Honjo and Tom Gridley for RbpjCKO mice; Ruth Ashery-Padan and Peter Gruss for alpha-Cre transgenic mice; Anand Swaroop, Tom Glaser, Cheryl Craft, and Doug Forrest for antibody reagents; Kenny Campbell and Alan Mears for cDNA plasmids; Ashley Riesenberg, Tien Le, and Kevin Conley for technical support and Valerie Wallace, Tiffany Cook, Masato Nakafuku, Kenny Campbell, Rob Hufnagel, and Tom Glaser for valuable discussion or critical evaluation of this manuscript.
References
- Ahmad I, Dooley CM, Polk DL. Delta-1 is a regulator of neurogenesis in the vertebrate retina. Dev Biol. 1997;185:92–103. doi: 10.1006/dbio.1997.8546. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Baker NE. Notch signaling in the nervous system. Pieces still missing from the puzzle. Bioessays. 2000;22:264–273. doi: 10.1002/(SICI)1521-1878(200003)22:3<264::AID-BIES8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Baker NE. Notch and the patterning of ommatidial founder cells in the developing Drosophila eye. In: Moses K, editor. Results and problems in cell differentiation: Drosophila eye development. Heidelberg: Springer; 2001. pp. 35–58. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumer N, Marquardt T, Stoykova A, Ashery-Padan R, Chowdhury K, Gruss P. Pax6 is required for establishing naso-temporal and dorsal characteristics of the optic vesicle. Development. 2002;129:4535–4545. doi: 10.1242/dev.129.19.4535. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-Delta signaling is required for spatial patterning and Muller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Brabletz S, Schmalhofer O, Brabletz T. Gastrointestinal stem cells in development and cancer. J Pathol. 2009;217:307–317. doi: 10.1002/path.2475. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA. Ann Arbor, MI: Department of Human Genetics, University of Michigan; 2005. The Role of Math5 in retinal development. [Google Scholar]
- Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Figueroa DJ, Marmorstein AD, Zhang Q, Petrukhin K, Caskey CT, Austin CP. Retina-specific nuclear receptor: A potential regulator of cellular retinaldehyde-binding protein expressed in retinal pigment epithelium and Muller glial cells. Proc Natl Acad Sci U S A. 1999;96:15149–15154. doi: 10.1073/pnas.96.26.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Xu S, Liu I, Li LY, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet. 2002;11:873–884. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Wallace VA. Hedgehogs and retinal ganglion cells: organizers of the mammalian retina. Neuroreport. 2004;15:479–482. doi: 10.1097/00001756-200403010-00019. [DOI] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Rapaport DH, Harris WA. Xotch inhibits cell differentiation in the Xenopus retina. Neuron. 1995;14:487–496. doi: 10.1016/0896-6273(95)90305-4. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O'Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. Pou domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet 12 Spec No. 2003;1:R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Gurdon JB. A community effect in animal development. Nature. 1988;336:772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Dyson S, St Johnston D. Cells' perception of position in a concentration gradient. Cell. 1998;95:159–162. doi: 10.1016/s0092-8674(00)81747-x. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Standley H, Dyson S, Butler K, Langon T, Ryan K, Stennard F, Shimizu K, Zorn A. Single cells can sense their position in a morphogen gradient. Development. 1999;126:5309–5317. doi: 10.1242/dev.126.23.5309. [DOI] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquié O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- Hoover F, Seleiro EA, Kielland A, Brickell PM, Glover JC. Retinoid X receptor gamma gene transcripts are expressed by a subset of early generated retinal cells and eventually restricted to photoreceptors. J Comp Neurol. 1998;391:204–213. [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, Magnuson MA, Macdonald RJ, Birchmeier C, Johnson JE. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH Factors Xath5 and XNeuroD Can Upregulate the Expression of XBrn3d, a POU-Homeodomain Transcription Factor. Dev Biol. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-lop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Cho SH, Cepko CL. Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci U S A. 2006a;103:18998–19003. doi: 10.1073/pnas.0608155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006b;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci U S A. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba DA, Gust J, Reh TA. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell. 2009;4:73–79. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;57:104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- Lewis J. From signals to patterns: space, time, and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- Lin MH, Kopan R. Long-range, nonautonomous effects of activated Notch1 on tissue homeostasis in the nail. Dev Biol. 2003;263:343–359. doi: 10.1016/j.ydbio.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Liu H, Thurig S, Mohamed O, Dufort D, Wallace VA. Mapping canonical Wnt signaling in the developing and adult retina. Invest Ophthalmol Vis Sci. 2006;47:5088–5097. doi: 10.1167/iovs.06-0403. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene. 1996;12:207–211. [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci U S A. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lesson from the retina. Nature Rev. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- MacLaren RE, Pearson RA, MacNeil A, Douglas RH, Salt TE, Akimoto M, Swaroop A, Sowden JC, Ali RR. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ, Tate P, Dill K, Pangilinan E, Wakenight P, Tessier-Lavigne M, Skarnes WC. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- Mori M, Ghyselinck NB, Chambon P, Mark M. Systematic immunolocalization of retinoid receptors in developing and adult mouse eyes. Invest Ophthalmol Vis Sci. 2001;42:1312–1318. [PubMed] [Google Scholar]
- Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci. 2006;28:128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- Ng L, Ma M, Curran T, Forrest D. Developmental expression of thyroid hormone receptor beta2 protein in cone photoreceptors in the mouse. Neuroreport. 2009;20:627–631. doi: 10.1097/WNR.0b013e32832a2c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Oishi K, Kamakura S, Isazawa Y, Yoshimatsu T, Kuida K, Nakafuku M, Masuyama N, Gotoh Y. Notch promotes survival of neural precursor cells via mechanisms distinct from those regulating neurogenesis. Dev Biol. 2004;276:172–184. doi: 10.1016/j.ydbio.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Okuyama R, Tagami H, Aiba S. Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J Dermatol Sci. 2008;49:187–194. doi: 10.1016/j.jdermsci.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, Kopan R. gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Perron M, Harris WA. Retinal stem cells in vertebrates. Bioessays. 2000;22:685–688. doi: 10.1002/1521-1878(200008)22:8<685::AID-BIES1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Piddini E, Vincent JP. Interpretation of the wingless gradient requires signaling-induced self-inhibition. Cell. 2009;136:296–307. doi: 10.1016/j.cell.2008.11.036. [DOI] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Reh TA. Cellular interactions determine neuronal phenotypes in rodent retinal cultures. J Neurobiol. 1992;23:1067–1083. doi: 10.1002/neu.480230811. [DOI] [PubMed] [Google Scholar]
- Riesenberg AN, Le TT, Willardsen MI, Blackburn DC, Vetter ML, Brown NL. Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis. 2009;47:175–187. doi: 10.1002/dvg.20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- Robinson GW. Using notches to track mammary epithelial cell homeostasis. Cell Stem Cell. 2008;3:359–360. doi: 10.1016/j.stem.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Rocha SF, Lopes SS, Gossler A, Henrique D. Dll1 and Dll4 function sequentially in the retina and pV2 domain of the spinal cord to regulate neurogenesis and create cell diversity. Dev Biol. 2009;328:54–65. doi: 10.1016/j.ydbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Schneider ML, Turner DL, Vetter ML. Notch signaling can inhibit Xath5 function in the neural plate and developing retina. Mol Cell Neurosci. 2001;18:458–472. doi: 10.1006/mcne.2001.1040. [DOI] [PubMed] [Google Scholar]
- Silva AO, Ercole CE, McLoon SC. Regulation of ganglion cell production by Notch signaling during retinal development. J Neurobiol. 2003;54:511–524. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- Sjölund J, Manetopoulos C, Stockhausen MT, Axelson H. The Notch pathway in cancer: differentiation gone awry. Eur J Cancer. 2005;41:2620–2629. doi: 10.1016/j.ejca.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Standley HJ, Zorn AM, Gurdon JB. eFGF and its mode of action in the community effect during Xenopus myogenesis. Development. 2001;128:1347–1357. doi: 10.1242/dev.128.8.1347. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka K, Hatakeyama J, Bessho Y, Kageyama R. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004:148–155. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Ganglion cells influence the fate of dividing retinal cells in culture. Development. 1998;125:1059–1066. doi: 10.1242/dev.125.6.1059. [DOI] [PubMed] [Google Scholar]
- Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, Kageyama R, Wallace VA. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wantanabe T, Raff MC. Rod photoreceptor development in vitro: Intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron. 1990;2:461–467. doi: 10.1016/0896-6273(90)90058-n. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Zhu X, Craft CM. Modulation of CRX transactivation activity by phosducin isoforms. Mol Cell Biol. 2000;20:5216–5226. doi: 10.1128/mcb.20.14.5216-5226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J Neurosci. 2003;23:6152–6160. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, Guillemot F, Kageyama R, Rosenfeld MG. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. doi: 10.1101/gad.1444706. [DOI] [PMC free article] [PubMed] [Google Scholar]