Abstract
Distinctive nanoridges on the surface of flowers have puzzled plant biologists ever since their discovery over 75 years ago. Although postulated to help attract insect pollinators, the function, chemical nature, and ontogeny of these surface nanostructures remain uncertain. Studies have been hampered by the fact that no ridgeless mutants have been identified. Here, we describe two mutants lacking nanoridges and define the biosynthetic pathway for 10,16-dihydroxypalmitate, a major cutin monomer in nature. Using gene expression profiling, two candidates for the formation of floral cutin were identified in the model plant Arabidopsis thaliana: the glycerol-3-phosphate acyltransferase 6 (GPAT6) and a member of a cytochrome P450 family with unknown biological function (CYP77A6). Plants carrying null mutations in either gene produced petals with no nanoridges and no cuticle could be observed by either scanning or transmission electron microscopy. A strong reduction in cutin content was found in flowers of both mutants. In planta overexpression suggested GPAT6 preferentially uses palmitate derivatives in cutin synthesis. Comparison of cutin monomer profiles in knockouts for CYP77A6 and the fatty acid ω-hydroxylase CYP86A4 provided genetic evidence that CYP77A6 is an in-chain hydroxylase acting subsequently to CYP86A4 in the synthesis of 10,16-dihydroxypalmitate. Biochemical activity of CYP77A6 was demonstrated by production of dihydroxypalmitates from 16-hydroxypalmitate, using CYP77A6-expressing yeast microsomes. These results define the biosynthetic pathway for an abundant and widespread monomer of the cutin polyester, show that the morphology of floral surfaces depends on the synthesis of cutin, and identify target genes to investigate the function of nanoridges in flower biology.
Keywords: 10,16-dihydroxypalmitic acid; cuticular ridges; CYP77A6; CYP86A4; glycerol-3-phosphate acyltransferase 6
In many flowering plants the surface of epidermal cells from petals and sepals is covered with ridges a few hundred nanometers to a few micrometers wide (1). These ridges, often running parallel, are genuine biological nanostructures and not a result of fixation artifacts because they are observed with environmental scanning electron microcopy (2). In some species, surface ridges are also found on the leaves and fruits. Petal nanoridges are an inspiration for the development of biomimetic surfaces and materials (3), especially because they are responsible for the “petal effect,” a superhydrophobicity state with high adhesive forces for water (4) that is different from the self-cleaning “lotus effect” commonly observed in leaves and stems (3–6).
Nanoridges were described many decades ago on floral organs (7) but their biological function is still poorly known. It is thought that they contribute to the attraction of insect pollinators by holding glistening dew drops (4), creating special patterns of light reflection (8), providing a tactile stimuli for surface recognition or forming a favorable structure to walk on (3). Nanoridges could thus be a factor adding to the known effects of epidermal cell shape and pigment color in attracting pollinator. In addition, they could influence water losses by creating a microcirculation of air at the epidermal surface. A role in rapid epidermis expansion is also suggested by the occurrence in grape berries of tightly appressed nanoridges during early stages of growth and their disappearance in mature fruits (9).
Besides the many hypotheses on the biological function of epidermal nanoridges, basic questions concerning their chemical nature and mechanism of formation remain unanswered to date. Nanoridges are often referred to as cuticular ridges or cuticular folds but they might originate from layers underneath the cuticle (3). The cuticle is an extracellular lipid layer covering and sealing the epidermis of aerial organs (10). It comprises waxes and cutin, a matrix of polymerized acylglycerols (11). Polysaccharides such as pectins might be present in the lower part of the cutin matrix and help anchor it to the cellulose wall. Transmission electron microscopy (TEM) studies indicate that nanoridges are made of sculptures or folds of the polymeric material present at the surface of the outer epidermal wall (reviewed in ref. 3). It is therefore clear that nanoridges do not consist of wax depositions. But whether they primarily originate from the cutin matrix, from massive local thickening of a pectic layer, or from subcuticular deposits of other polysaccharide material has not been demonstrated. It is also not known if the nanoridge pattern is due to mechanical constraints and folding of the wall during cell expansion or is the result of differential polymer synthesis across the epidermal surface. An important question is thus to determine whether the cutin layer is essential to the genesis of the nanopattern itself or whether it is a simple coating on a polysaccharide nanoridge structure. No mutants lacking ridges have been described, which has hampered studies on the function, ontogeny, or composition of these structures.
As a structural polymer of plant cuticles, cutin is one of the most abundant lipid polymers and an important adaptive trait of plants to their terrestrial environment. Cutin has notably been shown to be important for the architecture of stomata (12). Structurally, it is a polymer of fatty hydroxyacids and diacids that are esterified to each other and to glycerol (11, 13, 14). A wide variety of hydroxyacids and other cutin monomers is found in different organs and species but no link has been established yet between specific monomers and the structural and functional features of the corresponding cuticles. The recent use of Arabidopsis as a model for cutin research has allowed rapid progress on the biosynthesis of this polymer. Acylglycerols have been identified as likely basic building blocks for cutin assembly (15) and the network of proteins involved in cutin monomer synthesis has started to be unraveled (16, 17). Much remains to be done, however. For example, the enzymes introducing hydroxyl groups at internal positions of acyl chains to form widespread polyhydroxy fatty acids such as 10,16-dihydroxypalmitate remain unidentified to date.
Here, we report two mutants of the model plant Arabidopsis thaliana that completely lack petal nanoridges. We show that both mutants have a strong reduction in total cutin content and further demonstrate that the two proteins affected are enzymes involved in the cutin biosynthesis pathway. A fatty acid terminal hydroxylase required for cutin synthesis is also identified in this study. These results define in Arabidopsis the biosynthetic pathway of 10,16-dihydroxypalmitate, a major cutin monomer in nature. They also show that cutin biosynthesis is not operating to merely coat a polysaccharide nanoridge structure but on the contrary is essential to the mechanism of nanopattern formation. The identification of genes affecting nanoridges opens the way to future functional studies in other model flowers.
Results
Identification of Arabidopsis Mutants Lacking Flower Nanoridges.
In the last few years, the list of genes of cutin biosynthesis in A. thaliana has grown significantly (12, 18–25). These genes have been shown to affect monomers of stems, leaves, or seeds, but none has been demonstrated to affect the synthesis of the cutin layer in floral organs on the basis of a chemical analysis of cutin. Using gene expression profiles, we have identified two genes, GPAT6 (At2g38110 locus) and CYP77A6 (At3g10570 locus), as strong candidates for the biosynthesis of surface polyesters in flowers. Several of the eight members of the plant-specific family of membrane-bound glycerol-3-phosphate acyltransferases (GPAT) (26) have been previously shown to be required for the synthesis of polyesters (12, 27). GPAT6 showed a distinctively high expression in flowers according to publicly available Arabidopsis microarray data (28). Further analysis of microarray data using the Arabidopsis Coexpression Tool (29) indicated that the gene most highly correlated with GPAT6 was CYP77A6, a gene belonging to a family of cytochrome P450-dependent monooxygenases with no known biological function. CYP77A6 was found to be highly expressed in flowers (28) and moderately so in other organs. In stems, expression of CYP77A6 was 2.5-fold higher in epidermis than in whole stems (30), which was consistent with a putative role in cutin synthesis.
Two independent T-DNA insertional mutant lines homozygous for GPAT6 (gpat6–1 and gpat6–2) and two others for CYP77A6 (cyp77a6–1 and cyp77a6–2) were obtained by PCR screening. Silencing of gene transcript in mutant flowers was verified by RT-PCR (Fig. S1). The gpat6 and cyp77a6 mutants showed normal vegetative growth and development under standard growth conditions. However, consistent with the high expression of the two genes in flowers, the reproductive structures of both mutant lines displayed several prominent phenotypes. Both gpat6 and cyp77a6 mutant lines produced abnormal flowers, most of which displayed organ fusions and did not open. Also, the petals of the mutants were readily stained when immersed in a solution of toluidine blue (Fig. 1A). By contrast, rosette leaves of the mutants were not permeable to the dye. An increase in permeability to toluidine blue is indicative of cuticular defects (31). The presence in the dye solution of 0.01% Tween 20, a mild detergent, was used to minimize the surface effects of ridges and thus ensure a similar wettability between wild type (WT) and mutants.
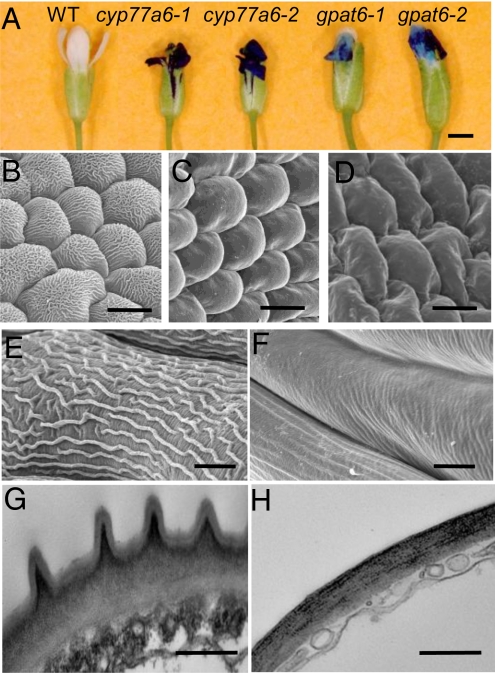
Fig. 1.
Flower morphology and surface characteristics of WT and mutants (gpat6 and cyp77a6). (A) Permeability to toluidine blue of the epidermis of flowers of WT and T-DNA insertional mutants for CYP77A6 and GPAT6. (B–D) Examination of petal abaxial epidermis using SEM (B, WT; C, gpat6–1; and D, cyp77a6–1). (E and F) Higher magnification of sepal surface showing a close-up of nanoridges under SEM (E, WT; F, gpat6–1). (G and H) Cross section of petals viewed under TEM (G, WT; H, gpat6–1). (Scale bars: A, 0.5 mm; B–D, 10 μm; E and F, 5 μm; G and H, 0.5 μm.) Similar SEM observations were made also for cyp77a6–2 and gpat6–2 mutants (see Fig. S2).
Further evidence of cuticular changes in the mutants was observed by detailed scanning electron microscopy (SEM) studies carried out on sepals and petals of mature flowers. Examination of petals and sepals of Arabidopsis by conventional SEM revealed that the cuticular ridges seen in the WT were absent in the mutants from the epidermal cells of petals (Fig. 1 B–D) and sepals (Fig. 1 E and F). SEM images for additional mutant lines are presented in Fig. S2. The absence of cuticular ridges was also evidenced by TEM (Fig. 1 G and H). Strikingly, no sculptures present at the surface of the outer epidermal wall in the abaxial or adaxial epidermis of WT petals were left in the mutants. The electron-lucent outermost layer, which is usually interpreted as the matrix of the cuticle proper, was absent in the mutants. Some electron-dense material, which was arranged as ridges beneath the cuticle proper in the WT, could still be seen in the mutants but the cell wall surface was smooth.
GPAT6 Provides Palmitate-Based Monomers for Cutin Synthesis.
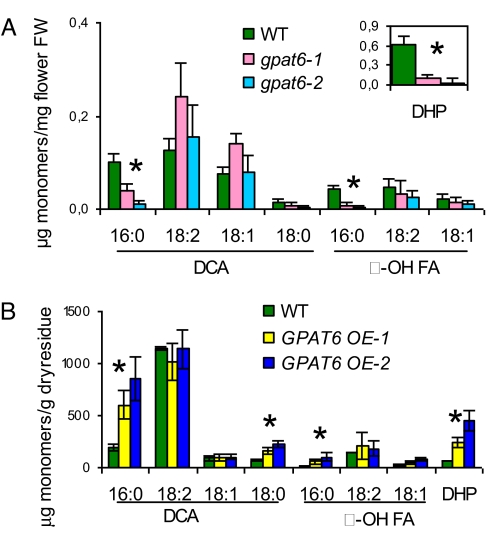
To check that GPAT6 knockout mutants had reduced cutin content in sepals and petals, we analyzed the monomer composition and load of lipid polyesters in the mature flowers of gpat6 mutants and WT. In Arabidopsis, unlike leaves and stems that have an unusual cutin composition with a high content in unsaturated dicarboxylic acids (32, 33), whole flowers have a classical cutin composition (27), with 80% of 10,16-dihydroxypalmitate, a major cutin monomer in fruits and leaves of many species. Analysis of polyesters on dissected sepals and petals confirmed that 10,16-dihydroxypalmitate was the major component in both organs (Fig. S3). When analyzing whole flowers of the gpat6 mutants, we found a reduction of 58% in total monomer load, with 90% reduction in the major monomer 10,16-dihydroxypalmitate (Fig. 2A). Significant reductions were also found in flowers of gpat6 mutants with all other C16 monomers, but not with C18 monomers. By contrast, surface waxes and fatty acids from intracellular soluble lipids were not significantly changed, ruling out a role of GPAT6 in lipid secretion or in providing the bulk of acyl chains in cells. Furthermore, a consistent reduction in all C16 monomers but almost no change in the more abundant C18 monomers was also observed in the cutin of rosette leaves from the gpat6 mutants (Fig. S4A). The low reduction in total cutin content in rosette leaves (<15%) was consistent with the low expression level of GPAT6 (28) and the absence of strong toluidine blue permeability phenotype in this organ.
Fig. 2.
GPAT6 provides C16-based monomers for polyester synthesis. (A) Polyester monomer content in flowers of WT and gpat6–1 and gpat6–2 mutants. (B) GPAT6 overexpression in Arabidopsis stems increases C16 monomers. Data are mean with 95% CI (n = 9). Asterisks denote statistically significant difference between WT and both mutant lines (P < 0.001, t test). DHP: 10,16-dihydroxypalmitate. DCA, α, ω-dicarboxylic acids; FA, fatty acids.
Ectopic overexpression of GPAT6 in Arabidopsis under the 35S promoter resulted in a three- to eightfold increase in C16 monomers of stem polyesters whereas most C18 monomers did not vary (Fig. 2B). This observation was in clear contrast with the increase in both C18 and C16 chains seen in GPAT4 or GPAT8 overexpressors (12). Taken together, these results strongly supported the view that GPAT6 is an acyltransferase of the polyester biosynthesis pathway with a preference for C16 chains.
CYP77A6 Is the In-Chain Hydroxylase of 10,16-Dihydroxypalmitate Biosynthesis.
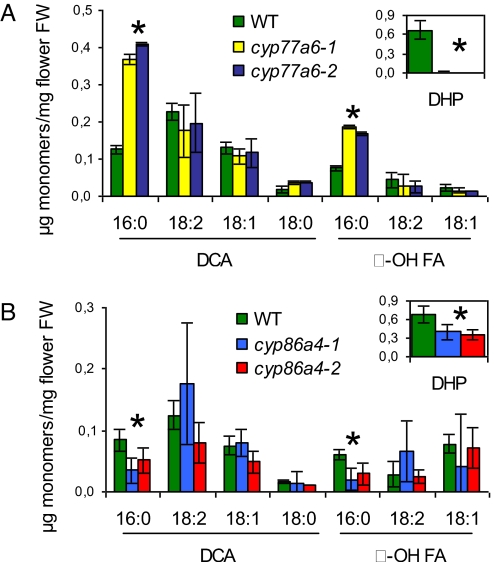
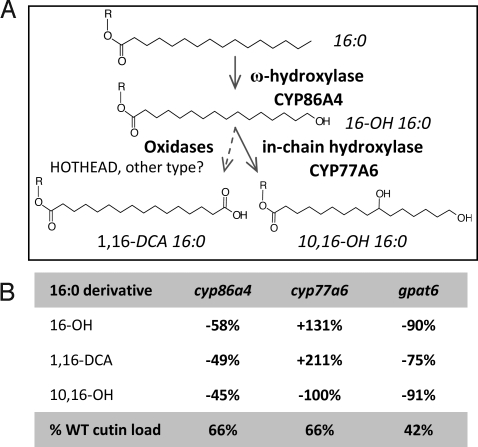
The polyester monomer analysis of cyp77a6–1 and cyp77a6–2 mutant flowers showed a smaller reduction in total cutin content (34%) than for gpat6 mutants. But the monomer profile was very different. It revealed a complete reduction in 10,16-dihydroxypalmitate (and its minor 9,16 and 8,16 isomers) concomitant with a two- to threefold increase in 16-hydroxypalmitate and 1,16-hexadecanedioic acid (Fig. 3A). This pattern of change in cutin monomers was also observed in stems (Fig. S4B). These results strongly suggested that CYP77A6 encoded an in-chain fatty acid hydroxylase acting on 16-hydroxypalmitate to form dihydroxypalmitates.
Fig. 3.
Polyester monomer profile for flowers of cyp77a6 and cyp86a4 mutants. (A) Polyester monomer content in WT and cyp77a6–1 and cyp77a6–2 mutants. (B) Polyester monomer content in WT and cyp86a4–1 and cyp86a4–2 mutants. Data are mean with 95% CI (n = 4 for cyp86a4 mutants). Asterisks denote statistically significant difference between WT and both mutant lines (P < 0.001, t test). DHP: 10,16-dihydroxypalmitate. DCA, α, ω-dicarboxylic acids; FA, fatty acids.
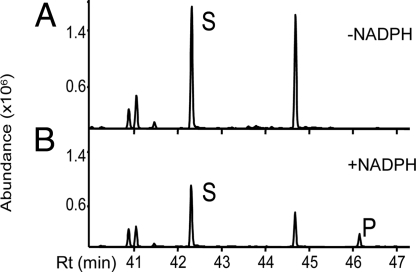
Biochemical activity of CYP77A6 was demonstrated by incubating microsomes of yeast (Saccharomyces cerevisiae) expressing CYP77A6 cDNA with free fatty acids. Microsomes were first incubated with radiolabeled laurate (C12), a generic substrate for many fatty acid hydroxylases. Reaction products were analyzed by TLC. These products, which were strictly NADPH dependent, were identified as in-chain hydroxylaurates on the basis of TLC migration. Identification as 8-, 9-, 10-, and 11-hydroxylaurate was achieved by GC-MS using nonradiolabeled laurate as substrate. Enzyme activity was linear over 10 min and was proportional to the amount of microsomal P450 added. CYP77A6 was then incubated with C16 aliphatic chains, which are more relevant to cutin synthesis. Incubation of CYP77A6-expressing yeast microsomes with 16-hydroxypalmitic acid resulted in the production of a new peak that was NADPH dependent as seen by GC-MS analysis (Fig. 4). MS analysis revealed this peak was a mixture of dihydroxypalmitates with positional isomers 8,16 and 9,16 as the major isomers and 10,16 as a minor one (Fig. S5). By contrast, no metabolite formation could be detected upon incubation of radiolabeled palmitic acid. These results obtained in vitro demonstrated that CYP77A6 indeed can catalyze in-chain hydroxylase activity as inferred by the genetic analysis (Fig. 3A) and furthermore that it could discriminate between ω-hydroxylated and nonhydroxylated palmitate chains.
Fig. 4.
Production of dihydroxypalmitate isomers by CYP77A6-expressing yeasts. Microsomes from CYP77A6-expressing Saccharomyces cerevisiae were incubated with 16-hydroxypalmitic acid (S), in the absence (A) or the presence (B) of NADPH. The mixture of dihydroxypalmitates (P), which included the 10,16-dihydroxy isomer, was produced only in the presence of NADPH.
To provide confirmation that the ω-hydroxylation was upstream of the in-chain hydroxylation in the pathway, a candidate gene for the ω-hydroxylation of palmitate in flowers was sought. Among the top three genes correlated with GPAT6 and CYP77A6 that were found with the Arabidopsis Coexpression Tool was a member of the CYP86A family (CYP86A4, At1g01600 locus). Two independent lines homozygous for a T-DNA insertion in CYP86A4, cyp86a4–1 and cyp86a4–2, were isolated by PCR and gene silencing was checked by RT-PCR (Fig. S1C). No morphological phenotypes were observed in the cyp86a4 knockout mutants. However, analysis of flower polyesters showed that they had a clear biochemical phenotype consisting of a 45–58% reduction in 16-hydroxypalmitate, 10,16-dihydroxypalmitate, and 1,16-hexadecanedioic acid (Fig. 3B). This result was consistent with the in vitro fatty acid ω-hydroxylase activity of CYP86A4 (34), its high expression in flowers (28), and its regulation by WIN1, a transcription factor of cutin biosynthesis (35).
Taken together, the results on CYP77A6 and CYP86A4 identify the encoded proteins as in-chain hydroxylase and ω-hydroxylase, respectively. They also show CYP77A6 acts after CYP86A4 in the biosynthesis of 10,16-dihydroxypalmitate (Fig. 5).
Fig. 5.
The biosynthetic pathway of 10,16-dihydroxypalmitate in Arabidopsis. (A) Enzymes. CYP86A4 and CYP77A6 are identified in this study. The HOTHEAD oxidase has been shown to be involved in dicarboxylic acid formation (22). Diacid formation could also be catalyzed by a single cytochrome P450 (36). GPAT6 and long-chain acyl-CoA synthetases could act upstream or downstream of fatty acid oxidases (17), so R is unknown and may be H or SCoA or glycerolipid. (B) Changes in C16 cutin monomers and total cutin content in the knockout mutants.
Discussion
Biosynthetic Pathway of Dihydroxypalmitates in Arabidopsis.
The cutin monomer 10,16-dihydroxypalmitate is an abundant and widespread component of cutins in various organs of many plant species. It accounts, for example, for 80% of cutin in leaves of Vicia faba (37) and in fruits of tomato (38). The cutin monomer profiles of the Arabidopsis insertional mutants for CYP86A4 and CYP77A6 presented here indicate that the ω-hydroxylase and in-chain hydroxylase for the biosynthesis of 10,16-dihydroxypalmitate, which were studied by Kolattukudy and coworkers 30 years ago using biochemical assays in V. faba (39, 40), have now been identified in Arabidopsis. Furthermore, we have confirmed that the reaction sequence of ω-hydroxylation followed by in-chain hydroxylation that was proposed on the basis of in vitro studies occurs in vivo (Fig. 5). Moreover, the fact that in yeast assays CYP77A6 is active on 16-hydroxypalmitate and not on palmitate provides a clear basis for the sequential order of the two hydroxylation reactions and thus for the absence of 10-hydroxypalmitate in Arabidopsis cutins. Interestingly, the major monomer formed in vitro by CYP77A6 was not 10,16-dihydroxypalmitate but the 8,16 and 9,16 isomers. This contrasts with the Arabidopsis cutin composition in flowers and stems where the latter are minor components of polyesters compared to the former. It is thus clear that other factors exist in vivo that are critical for a preferential hydroxylation at the 10 position of the palmitate chain. Interaction of CYP77A6 with other proteins of a multienzyme protein complex or the structural difference between the substrates used in vitro (free fatty acids) and the in vivo substrates (possibly acyl-CoAs, glycerolipids, etc.), as well as posttranslational modifications, are among the potential factors.
The complete reduction observed in 10(9)(8),16-dihydroxypalmitate in flowers and stems seems to rule out that one of the four other members of the CYP77 family (Fig. S6) is functionally redundant with CYP77A6. It is, however, likely that some of the other CYP77 proteins are involved in the synthesis of the various C18 in-chain oxygenated fatty acids found in Arabidopsis lipid polyesters (32, 33, 41). The recent finding that CYP77A4 has in vitro an epoxygenase activity on unsaturated C18 fatty acids (42) lends support to this hypothesis. The involvement of other CYP77 P450s in polyester biosynthesis is under current investigation. In addition to CYP86A4, several other fatty acid ω-hydroxylases (CYP86A family) are also expressed in various floral organs and could explain why 16-hydroxypalmitate was not 100% reduced in polyesters of whole flowers of cyp86a4 knockout lines.
Recent work has identified sets of P450 and acyltransferase genes associated with the biosynthesis and deposition of specific aliphatic polyesters in plants. In the case of Arabidopsis the P450 CYP86A2 (ATT1) (20), LACS2 (19, 24), and a pair of redundant acyltransferases, GPAT4 and GPAT8 (12), control most of the accumulation of the dicarboxylic acids in leaf and stem cutin, including the dominant yet unusual monomer octadecadiene-1,18-dioic acid. In the case of suberin, a suberin-associated acyltransferase, GPAT5, and two suberin-correlated P450s, CYP86A1 and CYP86B1, are required for the synthesis of suberin-like monomers in cutin of transgenic Arabidopsis (12, 43–45). This general pattern is now observed with the identification of the acyltransferase GPAT6 and the P450 CYP86A4, which are required for the accumulation of C16 cutin monomers. Several lines of evidence indicate that the acyltransferase GPAT6 is required to provide C16 aliphatic chains for the synthesis of the cutin polymer. On the basis of the gain- and loss-of-function mutants presented here, it is, however, not possible to determine whether the hydroxylases act downstream or upstream of GPAT6. The main possibilities regarding the order of reaction of GPATs, fatty acid oxidases, and acyl-activating enzymes have been presented in a recent review (17). The GPAT6 acyltransferase could act at any step of the C16 cutin monomer pathway (Fig. 5). Resolving the uncertainty will require definitive biochemical characterization.
Nanoridge Formation and Cutin Assembly.
Through study of cutin mutants (gpat6 and cyp77a6) in Arabidopsis, we have provided solid evidence that the formation of flower nanoridges requires the synthesis of the cutin polymer. The fact that in the gpat6 and cyp77a6 mutants the cell wall surface is smooth (Fig. 1 F and H) shows that the ridges of Arabidopsis flowers are not likely caused by ridges of the underlying cell wall that are simply coated with cutin as seems to occur in some ridges (3). Our results show on the contrary that the synthesis of the cutin polymer is instrumental to the formation of flower ridges in Arabidopsis. Whether the mechanism involves mechanical folding of the cutin matrix or differential synthesis of the cutin polymer across the cell wall remains to be determined.
The mutants presented here as well as other mutants affected in stomatal ledge formation (12, 46) highlight the ability of epidermal cell types to regulate the morphology of the plant surface by controlling the synthesis of their outermost polymer layer. Cuticular ridges are present at the surface of the 10,16-dihydroxypalmitate-rich cutins of Arabidopsis petals and sepals but are not observed on TEM sections of dicarboxylic acid-rich cutins of stems and leaves. This comparison supports a hypothesis that 10,16-dihydroxypalmitate may be one component specifically required during cutin assembly to give rise to the nanoridges that are prevalent in flowers. The absence of cuticular ridges in the cyp77a6 mutants that are lacking 10,16-dihydroxypalmitate but that still have a significant cutin load (66% of WT) supports this view. However, it is clear that a high content in this monomer alone is not sufficient because, as judged by TEM, the cuticle of developing tomato fruits has a surface free of these nanostructures (38). A more complex view may be required than that of monomer load. Factors required for nanoridge formation may include the fraction of cutin monomers that give only linear polyester chains (16-hydroxypalmitate), the relative contributions of primary and secondary ester bonds to give dendrimeric structures (10,16-dihydroxypalmitate), and amount of cross-linking, for example, to polysaccharides or via dicarboxylic acid (hexadecane-1,16-dioic acid) esterification with glycerol (17). In this context, the view of membrane lipid structure where lipid molecules of specific shapes are associated with flat, convex, and concave surfaces may be instructive (47). Perhaps the (still unknown) distribution of cutin polymer domains is also a critical aspect for ridge formation?
Whether cuticular ridges of Arabidopsis are a residual feature of evolution or whether they have another function remains to be determined. Such questions can now be addressed by studies of mutants. Despite the significant gaps in our understanding of how the structures of plant surfaces relate to biological functions, some properties of plant surfaces have already inspired the development of biomimetic surfaces and materials (3). Further studies of the properties and inferred functions of cuticular nanostructures are expected to give rise to more applications. An additional biotechnological goal will be the production of either polyesters or their respective monomers in recombinant plant or microbial systems for the production of specialty chemical feedstocks (48). Genes such as CYP86A4, CYP77A6, and GPAT6 may enable the production of high amounts of polyhydroxy fatty acids with in-chain hydroxyl groups at defined positions.
Materials and Methods
Plant Growth and Maintenance.
A. thaliana ecotype Col-0 is used throughout this study. Plants were germinated and maintained in standard growth conditions as described previously (49). Transgenic plants were selected on MS agar plates containing 30 μg/mL hygromycin before transfer to soil.
T-DNA Insertional Mutant Identification and Isolation.
Expression profiles of Arabidopsis genes based on publicly available microarray data were visualized using the eFP Browser tool (50) and coexpression analysis was done with the Arabidopsis Coexpression Tool (29). Two independent T-DNA insertions were identified for each gene: for CYP77A6, lines SALK_019080C and SALK_023926C; for GPAT6, lines SALK_136675 and SALK_146013; and for CYP86A4, lines SALK_073078 and SALK_077857. Homozygous plants were isolated by PCR (for primers, see Table S1). Mutant identification, isolation, and gene expression analysis by RT-PCR are all described in SI Materials and Methods.
Microscopy.
Flower epidermal surfaces were examined under SEM. Sample preparation is as described for leaves (12). Images were taken with a JEOL scanning electron microscope. For TEM, freshly harvested fully open flowers (stage 15) were vacuum infiltrated for 30 min with fixation solution (2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer), followed by overnight fixation in the same solution at 4 °C. Flower tissue was prepared as described for leaves (12). Images were taken with a JEOL 100CX transmission electron microscope. For epidermal permeability, whole flowers were dipped in a 0.05% (wt/vol) toluidine blue-O solution (with Tween 20 added at a concentration of 0.01% wt/vol) for 5 min, rinsed in distilled water, and observed under a light microscope (Leica MZ 12.5) coupled with a digital camera.
Polyester Monomer Analyses.
Typically 15 open flowers (stage 15) were harvested with sharp tweezers and immediately quenched in isopropanol at 85 °C for 10 min. Polyester analysis was performed on delipidated tissues using NaOMe depolymerization and GC-MS analysis (23). Petals and sepals were dissected with a pair of sharp tweezers under a dissecting microscope.
GPAT6 Expression Construct.
GPAT6 genomic DNA was amplified as an NcoI–SpeI fragment and inserted into the plant binary vector pCAMBIA1302 (CAMBIA) for transformation of WT Arabidopsis plants (for primers, see Table S1). Cloning procedure and plant transformation are described in SI Materials and Methods.
CYP77A6 Expression and in Vitro Activity Assay.
The coding sequence of CYP77A6 was cloned by PCR with primers CYP77A6-cF and CYP77A6-cR (for primers, see Table S1). The PCR product was cloned as a BglII–KpnI fragment into the yeast expression vector pYeDP60. Details of the PCR cloning procedure are described in SI Materials and Methods. For heterologous expression of CYP77A6 in yeast, a system developed for the expression of P450 enzymes and consisting of plasmid pYeDP60 and Saccharomyces cerevisiae WAT11 strain (51) was used. Yeast culture maintenance, transformation, and microsome preparation are described in SI Materials and Methods. The cytochrome P450 content was measured by the method of Omura and Sato (52). Radiolabeled lauric acid ([1-14C], 45 Ci/mol; Commissariat à l'Energie Atomique, Gif sur Yvette, France) and radiolabeled palmitic acid ([1-14C], 54 Ci/mol, Perkin–Elmer Life Sciences) were dissolved in ethanol, which was evaporated onto the glass tube before the addition of microsomes. Resolubilization of the substrate was confirmed by measuring the radioactivity of the incubation media. Enzymatic activities of CYP77A6 from transformed yeast were determined in standard assay (0.1 mL) containing 20 mM sodium phosphate (pH 7.4), 1 mM NADPH, and radiolabeled lauric or palmitic acid (100 μM) or nonradiolabeled 16-hydroxypalmitic acid (100 μM). The reaction was initiated by the addition of NADPH and was stopped by the addition of 20 μL acetonitrile (containing 0.2% acetic acid). For palmitic acid the reaction products were resolved by TLC (see SI Materials and Methods). For 16-hydroxypalmitic acid incubation, metabolites were extracted from incubation media and subjected to GC-MS analysis as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Alicia Pastor and Ewa Danielewics (Michigan State University Advanced Microscopy Center) for assisting with TEM and SEM analyses, respectively. This work was supported in part by US Department of Agriculture Grant 2005-35318-15419, National Science Foundation Grant MCB-0615563, and the 7th European Community Framework Program through Marie Curie International Reintegration Grant 224941.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909090106/DCSupplemental.
References
- 1.Jeffree CE. The fine structure of the plant cuticle. In: Riederer M, Müller C, editors. Biology of the Plant Cuticle. Oxford: Blackwell; 2006. pp. 11–125. [Google Scholar]
- 2.Kolosova N, Sherman D, Karlson D, Dudareva N. Cellular and subcellular localization of S-adenosyl-l-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methylbenzoate in snapdragon flowers. Plant Physiol. 2001;126:956–964. doi: 10.1104/pp.126.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch K, Bhushan B, Barthlott W. Diversity of structure, morphology and wetting of plant surfaces. Soft Matter. 2008;4:1943–1963. [Google Scholar]
- 4.Feng L, et al. Petal effect: A superhydrophobic state with high adhesive force. Langmuir. 2008;24:4114–4119. doi: 10.1021/la703821h. [DOI] [PubMed] [Google Scholar]
- 5.Barthlott W, Neinhuis C. Purity of the sacred lotus or escape from contamination in biological surfaces. Planta. 1997;202:1–8. [Google Scholar]
- 6.Bargel H, Koch K, Cerman Z, Neinhuis C. Structure-function relationships of the plant cuticle and cuticular waxes- a smart material? Funct Plant Biol. 2006;33:893–910. doi: 10.1071/FP06139. [DOI] [PubMed] [Google Scholar]
- 7.Martens P. Origin and role of surface foldings in the epidermis of petals (in French) C R Acad Sci Paris. 1933;197:785–787. [Google Scholar]
- 8.Whitney HM, et al. Floral iridescence, produced by diffractive optics, acts as a cue for animal pollinators. Science. 2009;323:130–133. doi: 10.1126/science.1166256. [DOI] [PubMed] [Google Scholar]
- 9.Casado CG, Heredia A. Ultrastructure of the cuticle during growth of the grape berry (Vitis vinifera L.) Physiol Plant. 2001;111:220–224. [Google Scholar]
- 10.Samuels L, Kunst L, Jetter R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 11.Kolattukudy PE. Polyesters in higher plants. Adv Biochem Eng/Biotechnol. 2001;71:1–49. doi: 10.1007/3-540-40021-4_1. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, et al. Identification of acyltransferases required for cutin synthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graça J, Schreiber L, Rodrigues J, Pereira H. Glycerol and glyceryl esters of ω-hydroxyacids in cutins. Phytochemistry. 2002;61:205–215. doi: 10.1016/s0031-9422(02)00212-1. [DOI] [PubMed] [Google Scholar]
- 14.Ray AK, Chen ZJ, Stark RE. Chemical depolymerization studies of the molecular architecture of lime fruit cuticle. Phytochemistry. 1998;49:65–70. [Google Scholar]
- 15.Li Y, Beisson F, Ohlrogge J, Pollard M. Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol. 2007;144:1267–1277. doi: 10.1104/pp.107.099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nawrath C. Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol. 2006;9:281–287. doi: 10.1016/j.pbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plants Sci. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Wellesen K, et al. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc Natl Acad Sci USA. 2001;98:9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao F, et al. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004;23:2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurdyukov S, et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell. 2006;18:321–339. doi: 10.1105/tpc.105.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurdyukov S, et al. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- 23.Molina I, Ohlrogge JB, Pollard M. Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 2008;53:437–449. doi: 10.1111/j.1365-313X.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- 24.Bessire M, et al. A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 2007;26:2158–2168. doi: 10.1038/sj.emboj.7601658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lü S, et al. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) and has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009;59:553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, et al. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 2003;15:1872–1887. doi: 10.1105/tpc.012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 29.Manfield IW, et al. Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suh MC, et al. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 2004;37:139–146. doi: 10.1046/j.1365-313x.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- 32.Bonaventure G, Beisson F, Ohlrogge J, Pollard M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 2004;40:920–930. doi: 10.1111/j.1365-313X.2004.02258.x. [DOI] [PubMed] [Google Scholar]
- 33.Franke R, et al. Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 34.Rupasinghe SG, Duan H, Schuler MA. Molecular definitions of fatty acid hydroxylases in Arabidopsis thaliana. Proteins. 2007;68:279–293. doi: 10.1002/prot.21335. [DOI] [PubMed] [Google Scholar]
- 35.Kannangara R, et al. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell. 2007;19:1278–1294. doi: 10.1105/tpc.106.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandel S, et al. Characterization of a methyl jasmonate and wounding responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS J. 2007;274:5116–5127. doi: 10.1111/j.1742-4658.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 37.Kolattukudy PE, Croteau R, Walton TJ. Biosynthesis of cutin: Enzymatic conversion of omega-hydroxy fatty acids to dicarboxylic acids by cell-free extracts of Vicia faba epidermis. Plant Physiol. 1975;55:875–880. doi: 10.1104/pp.55.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saladié M, et al. A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 2007;144:1012–1028. doi: 10.1104/pp.107.097477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soliday CL, Kolattukudy PE. Biosynthesis of cutin: Omega-hydroxylation of fatty acids by a microsomal preparation from germinating Vicia faba. Plant Physiol. 1977;59:1116–1121. doi: 10.1104/pp.59.6.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soliday CL, Kolattukudy PE. Midchain hydroxylation of 16-hydroxypalmitic acid by the endoplasmic reticulum fraction from germinating Vicia faba. Arch Biochem Biophys. 1978;188:338–347. doi: 10.1016/s0003-9861(78)80018-6. [DOI] [PubMed] [Google Scholar]
- 41.Molina I, Bonaventure G, Ohlrogge J, Pollard M. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry. 2006;67:2597–2610. doi: 10.1016/j.phytochem.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Sauveplane V, et al. Arabidopsis thaliana CYP77A4 is the first cytochrome P450 able to catalyze the epoxidation of free fatty acids in plants. FEBS J. 2009;276:719–735. doi: 10.1111/j.1742-4658.2008.06819.x. [DOI] [PubMed] [Google Scholar]
- 43.Höfer R, et al. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot. 2008;59:2347–2360. doi: 10.1093/jxb/ern101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Compagnon V, et al. CYP86B1 is required for very long chain omega-hydroxyacid and alpha,omega-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009;150:1831–1843. doi: 10.1104/pp.109.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. Identification of an Arabidopsis feruloyl-CoA transferase required for suberin synthesis. Plant Physiol. 2009;151:1317–1328. doi: 10.1104/pp.109.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macgregor DR, Deak KI, Ingram PA, Malamy JE. Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell. 2008;20:2643–2660. doi: 10.1105/tpc.107.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cullis PR, Hope MJ, Tilcock CPS. Lipid polymorphism and the roles of lipids in membranes. Chem Phys Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Beisson F. The biosynthesis of cutin and suberin as an alternative source of enzymes for the production of bio-based chemicals and materials. Biochimie. 2009;91:685–691. doi: 10.1016/j.biochi.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Beisson F, Pollard M, Ohlrogge J. Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67:904–915. doi: 10.1016/j.phytochem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Winter D, et al. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 52.Omura T, Sato R. The carbon mononoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.