Abstract
Nuclear movement is critical during neurogenesis and neuronal migration that are fundamental for mammalian brain development. While dynein, Lis1 and other cytoplasmic proteins are known for their roles in connecting microtubules to the nucleus during interkinetic nuclear migration (INM) and nucleokinesis, the factors connecting dynein/Lis1 to the nuclear envelope (NE) remain to be determined. We report here that the SUN-domain proteins SUN1 and SUN2 and the KASH-domain proteins Syne-1/Nesprin-1 and Syne-2/Nesprin-2 play critical roles in neurogenesis and neuronal migration in mice. We show that SUN1 and SUN2 redundantly form complexes with Syne-2 to mediate the centrosome-nucleus coupling during both INM and radial neuronal migration in the cerebral cortex. Syne-2 is connected to the centrosome through interactions with both dynein/dynactin and kinesin complexes. Syne-2 mutants also display severe defects in learning and memory. These results fill an important gap in our understanding of the mechanism of nuclear movement during brain development.
Keywords: KASH domain, UNC-84, nuclear envelope, nucleokinesis, interkinetic nuclear movement, learning and memory
Introduction
Neuronal migration is a fundamental process for the development of laminary structures in the mammalian brain, including the cortex, hippocampus, midbrain and hindbrain. Defects in neuronal migration are responsible for human diseases such as lissencephaly and double-cortex syndromes. In the cerebral cortex, the majority of neurons are born in the germinal zone along the lateral ventricles and migrate toward the pial surface in an inside-out fashion. Interkinetic nuclear migration (INM) and nucleokinesis are two distinct processes in which the nucleus undergoes dramatic movement which is necessary for proper brain development (Feng and Walsh, 2001; Tsai et al., 2007; Tsai and Gleeson, 2005; Wynshaw-Boris, 2007). INM refers to the nuclear movement of neural epithelial stem cells (NESC) and radial glial progenitor cells (RGPC). From E8 to E11, the nuclei of NESCs migrate between the apical and basal surfaces of the neural epithelium in conjunction with the progression of the cell cycle. When neurogenesis begins at E11, the NESCs differentiate into RGPCs, and their nuclei continue to move within the ventricular zone (VZ).
Nucleokinesis describes the nuclear movement during cell migration. After the transition of young neurons from multipolar to bipolar shape in the subventricular zone (SVZ), they migrate along the radial glial fibers across a long distance toward the pial surface. These projection neurons reiterate a four-step process during radial migration. First, the leading edge of the neuron extends along the radial glial fiber, and a swelling of the plasma membrane forms in the leading process. The microtubule network is then pulled forward and the centrosome moves steadily into the swelling. Next, the nucleus is pulled by microtubules toward the centrosome. Finally, the trailing cytoplasmic region follows the nucleus and finishes soma translocation (Tsai et al., 2007; Tsai and Gleeson, 2005).
How the large nucleus moves during INM and nucleokinesis remains an important and fascinating question. Microtubules have been shown to exert pulling forces during granule cell migration in the cerebellum and radial neuronal migration in the cortex (Keays et al., 2007; Solecki et al., 2004; Xie et al., 2003). Factors including LIS1, Ndel1, DCX and the dynein complex have been shown to have critical roles in coupling microtubules and the nucleus (Feng et al., 2000; Sasaki et al., 2000; Shu et al., 2004; Tanaka et al., 2004; Tsai et al., 2007). In the prevailing model, one group of cytoplasmic dynein proteins is anchored on the plasma membrane and pulls the microtubule and centrosome forward, while another group of cytoplasmic dynein proteins is anchored on the nuclear envelope (NE) by some unknown factors and transmits the force to the nucleus (Morris, 2003; Reinsch and Gonczy, 1998; Tsai et al., 2007).
During INM, the centrosome is maintained along the apical surface, and the nucleus moves up and down (Higginbotham and Gleeson, 2007). Interestingly, disruption of either cytoplasmic dynein, Lis1 or centrosome proteins in rodents results in failure of INM (Tsai et al., 2005; Xie et al., 2007). Lis1 is also required for the symmetric division of murine NESC and RGPC cells, where it attaches the microtubule plus end to the cell cortex (Yingling et al., 2008). These findings suggest that the protein complexes utilized in nucleokinesis are also critical for INM. However, in both cases, the essential link between cytoplasmic dynein and the NE is still undetermined.
Mammalian SUN1 and SUN2 proteins, identified as homologs of C. elegans UNC-84 (Malone et al., 1999), are conserved SUN family proteins that contain transmembrane domains spanning the inner nuclear membrane and a C-terminal SUN domain localizing to the lumen of the NE (Haque et al., 2006; Hodzic et al., 2004; Padmakumar et al., 2005). The N-termini of SUN proteins have been shown to be in the nucleoplasm and to interact with nuclear lamins (Crisp et al., 2006; Fridkin et al., 2004; Haque et al., 2006; Mejat et al., 2009). Mammalian Syne-1/Nesprin-1 and Syne-2/Nesprin-2 proteins belong to a family of giant KASH proteins that are conserved in the worm and fly (Starr and Fischer, 2005; Wilhelmsen et al., 2006). The KASH proteins, which contain conserved 60-residue KASH (Klarsicht/ANC-1/Syne Homology) domains at the C-termini (Starr and Han, 2002), have been shown to be recruited to the outer NE through interactions between the KASH domains and SUN domains in the lumen of NE (Crisp et al., 2006; Malone et al., 2003; McGee et al., 2006; Padmakumar et al., 2005; Starr and Han, 2002; Starr et al., 2001). The roles of SUN and KASH proteins and their functional interactions during nuclear positioning were first uncovered by genetic studies in non-mammalian animal models. In C. elegans, SUN proteins have been shown to recruit KASH proteins to the NE for nuclear migration and anchorage (Malone et al., 1999; Malone et al., 2003; Starr and Han, 2002; Starr et al., 2001). In Drosophila and Zebrafish retina, KASH and SUN proteins play critical roles in nuclear movement (Del Bene et al., 2008; Kracklauer et al., 2007; Mosley-Bishop et al., 1999; Tsujikawa et al., 2007).
We and others have previously shown that the KASH protein Syne-1/Nesprin-1 and Lamin A/C are essential for the anchorage of synaptic and non-synaptic nuclei of skeletal muscle cells in mice (Grady et al., 2005; Mejat et al., 2009; Puckelwartz et al., 2008; Puckelwartz et al., 2009; Zhang et al., 2007) and that SUN1 is essential for gametogenesis (Chi et al., 2009; Ding et al., 2007). We have also determined that SUN1 and SUN2 play critical and redundant roles in recruiting Syne-1 to the NE and in anchoring myonuclei in mice (Lei et al., 2009). However, the neonatal lethality of Syne-1; Syne-2 double KASH deletion (Syne-1/2 DKD or Syne-1-/-; Syne-2-/-) mice and Sun1; Sun2 double knockout mice (Sun1/2 DKO or Sun1-/-; Sun2-/-) indicate that Syne-1/2 and SUN1/2 may have essential functions in developmental processes other than myonuclear anchorage and gametogenesis (Ding et al., 2007; Lei et al., 2009; Luke et al., 2008; Zhang et al., 2007). Consistent with this notion, human SYNE1 has been shown to be associated with cerebellar ataxia in a French-Canadian cohort (Gros-Louis et al., 2007).
In this study, we have examined the central nervous system in Syne-1/2 DKD and Sun1/2 DKO mutants and uncovered the critical functions of these proteins in nucleokinesis and INM during mammalian brain development. Our results indicate that the SUN-KASH complexes mediate the coupling between the nucleus and the centrosome, and provide anchors in the NE for cytoplasmic dynein/dynactin during neuronal migration.
Results
Loss of both SUN1 and SUN2 Lead to Severe Laminary Defects in Mouse Brain
Sun1/2 DKO (Sun1-/-; Sun2-/-) pups failed to breathe and died shortly after birth (Lei et al., 2009). Necropsy showed that their brain size was significantly reduced compared to that of their littermate controls (Figure 1A; data not shown). Histological analysis of the brains of embryonic-day (E) 18.5 Sun1/2 DKO embryo indicated severe defects that include malformed cortices, enlarged lateral ventricles and a smaller corpus callosum (Figure 1B; data not shown). Multiple brain regions of the DKO pups displayed severe laminary defects, including loss of the mitral cell layer in the olfactory bulb, loss of the pyramidal cell layer in the hippocampus, loss of the Purkinje cell layer in the cerebellum, and widespread defects in the midbrain and hindbrain (Figure 1C-H).
Figure 1. Brains of Sun1/2 DKO embryos display severe laminary defects and inverted layers in multiple brain regions.
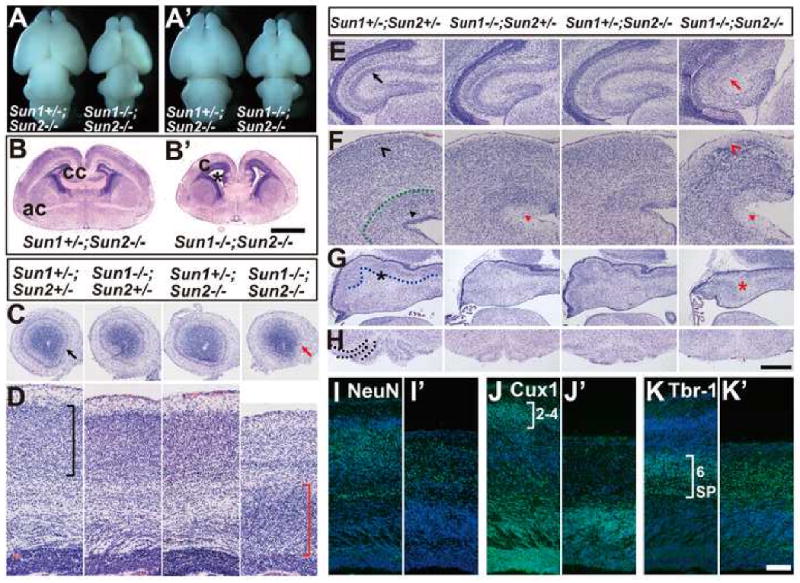
(A-A′) Dorsal (A) and ventral (A′) views of E18.5 brains. The brains of Sun1/2 DKO embryos were significantly smaller than that of their littermates.
(B-B′) Coronal sections of E18.5 brains at anterior commissure (ac) stained with H&E. Compared to the Sun1+/-; Sun2-/- control, the Sun1/2 DKO brain displayed smaller size, significantly enlarged lateral ventricles (asterisk) and abnormal cortical structures (c). cc, corpus callosum. Bar, 1000μm.
(C-H) Coronal sections of E18.5 brains stained with H&E, showing defects in various regions of Sun1/2 DKO brains compared to their littermates. (C) Mitral cell layer was missing in olfactory bulb in the DKO brain (red arrow). (D) Neocortex of the DKO embryo displayed inverted layers (red bracket) compared to the controls (black bracket). (E) Pyramidal cell layer was disrupted in the DKO hippocampus (red arrow). (F) Laminary defects in the DKO midbrain are shown (red arrowheads). (G) Purkinje cell layer (highlighted by a dotted line in the control) was malformed in the DKO cerebellum. (H) The inferior olive (highlighted by dotted lines in the control) was lost in the DKO brain stem. Bar in (H) equals 475μm for (C), 159μm for (D), 365μm for (E), 266μm for (F), and 500μm for (G-H).
(I-K) Coronal sections of E18.5 brains stained with different markers. (I) NeuN staining revealed that the number of cortical neurons (green) was decreased and the sub-plate was poorly formed in Sun1/2 DKO brain (I′), compared to that of Sun1+/-; Sun2-/- control (I). (J-K) Cux-1 and Tbr-1 positive neurons (green) in Sun1/2 DKO brain (J′ and K′) occupied inverted positions compared to that in Sun1+/-; Sun2-/- control (J and K). SP, subplate. Bar, 100 μm.
The cerebral cortex is organized in six layers that were clearly observable in the cortex of Sun1+/-; Sun2-/- mice (Figure 1D). In contrast, the six-layer structure could not be distinguished in the cortex of Sun1/2 DKO mice (Figure 1D). Noticeably, the cell density in the region above the ventricular zone (VZ) was abnormally high, likely due to the failure of radial neuronal migration. Immuno-staining with an antibody recognizing the neuronal specific protein NeuN showed a reduced number of neurons and a poorly developed subplate in the Sun1/2 DKO cortex (Figure 1I). We further examined the laminary structure using an anti-Cux1 antibody that is specific for neurons in the later-born layers 2 and 3, and an anti-Tbr-1 antibody that is specific for neurons in the early-born layer 6 and the subplate (Molyneaux et al., 2007). Although Cux1 and Tbr-1 positive neurons were readily detectible in Sun1/2 DKO cortices, the positions they occupied, referring to the distance from the pial surface, were inverted compared to that in the Sun1+/-; Sun2-/- control (Figure 1J-K). To determine whether the marginal zone was properly developed in the Sun1/2 DKO mutants, we examined the cerebral cortices of E15.5 embryos with an anti-Reelin antibody. Reelin positive neurons were localized beneath the pial surface in the Sun1+/-; Sun2-/- control (Figure S1A-D). In contrast, the number of Reelin positive cells is significantly increased in Sun1/2 DKO embryo brain and these cells occupied deeper positions referring to the Tbr1 positive cells (Figure S1 E-H). These results indicate that the laminary structure of cerebral cortex is inverted in Sun1/2 DKO mice.
Syne-1/2 DKD Mice Phenocopy Sun1/2 DKO Brains
Syne-1 collaborates with SUN1 and SUN2 for proper myonuclear anchorage in mice (Lei et al., 2009), and Syne-1/2 DKD (Syne-1-/-; Syne-2-/-) pups also failed to breathe and died shortly after birth (Zhang, et al., 2007). We thus examined whether Syne-1 and Syne-2 mutants displayed similar neuronal defects as Sun1/2 DKO mice. The brains of E18.5 Syne-1/2 DKD (Syne-1-/-; Syne-2-/-) mice were significantly smaller than those of Syne-1+/-; Syne-2+/-, Syne-1-/-; Syne-2+/-, and Syne-1+/-; Syne-2-/- littermates (Figure S2). Syne-1/2 DKD brains displayed enlarged lateral ventricles, inverted layers in the cerebral cortex, loss of the pyramidal cell layer in hippocampus, loss of the Purkinje cell layer in cerebellum, and laminary defects in midbrain, brain stem and many other brain regions (Figure 2A-E; Figure S2). Cux-1 and Tbr1 positive cells also occupied inverted position in the cerebral cortex of Syne-1/2 DKD mice (data not shown). Interestingly, brains of E18.5 Syne-1+/-; Syne-2-/- mice displayed inverted layers in the cerebral cortex and abnormal pyramidal cell layer in the hippocampus, similar to that seen in Syne-1-/-; Syne-2-/- mice, but had a normal Purkinje cell layer in the cerebellum and normal organization of the brain stem (Figure 2A-E).
Figure 2. Brains of Syne-1/2 DKD mice phenocopy Sun1/2 DKO mutants.
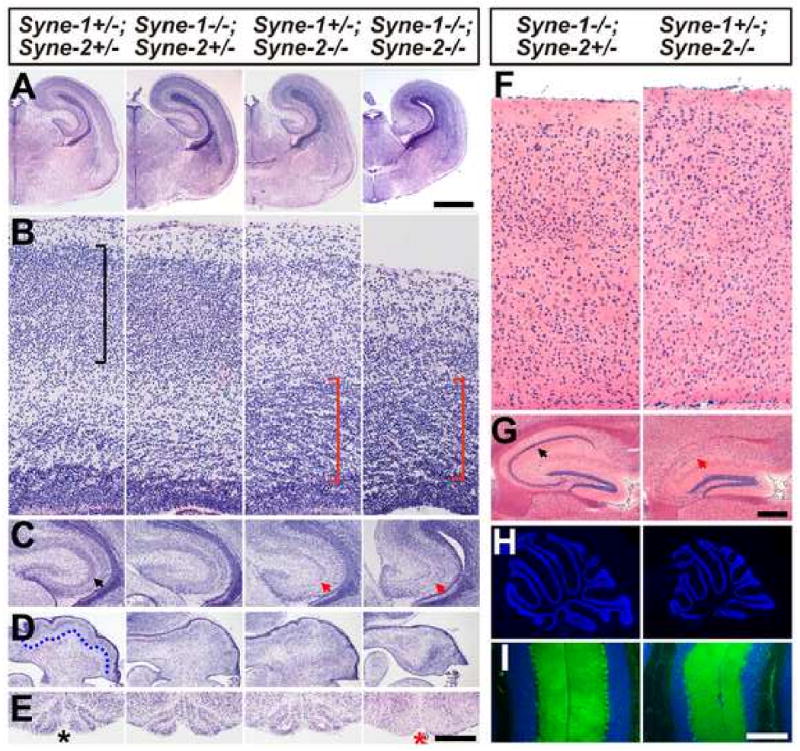
(A-E) Coronal sections of E18.5 brains stained with H&E, showing defects in various regions of the Syne-1-/-; Syne-2-/- brains compared to that of their littermates. (A) Syne-1-/-; Syne-2-/- brain displayed smaller size and enlarged cerebral ventricles. Bar, 1000μm. (B) Both Syne-1+/-; Syne-2-/- and Syne-1-/-; Syne-2-/- brains displayed inverted layers in the cerebral cortex (red brackets). (C) The pyramidal cell layers were malformed in Syne-1+/-; Syne-2-/- and Syne-1-/-; Syne-2-/- hippocampus (red arrows). (D) The Purkinje cell layer (highlighted by a dotted line in a control) was disrupted in Syne-1-/-; Syne-2-/-cerebellum. (E) The inferior olive clearly seen in the controls (asterisk) was missing in Syne-1/2 DKD brain stem (red asterisk). Bar in (E) equals 120μm in (B), 487μm in (C), 635μm in (D) and 500μm in (E).
(F-G) Coronal sections of adult brain stained with H&E. (F) The distinct 6-layer structure could be easily observed in Syne-1-/-; Syne-2+/- neocortex, but was disrupted in Syne-1+/-; Syne-2-/- brain. (G) The pyramidal cell layer seen in controls was severely disrupted in Syne-1+/-; Syne-2-/- hippocampus (red arrow). Bar in (G) equals 60μm in (F) and 250μm in (G)
(H-I) Sagittal sections of adult cerebellums stained with DAPI (blue) and anti-Calbindin antibody (green). (H)The cerebellum of Syne-1+/-; Syne-2-/- mouse displayed a smaller size but normal foliations compared to the control. (I) In the cerebellum of Syne-1+/-; Syne-2-/- mouse, the inner granule cell layer (IGL, blue) is significantly thinner than that of the control, but the Purkinje cells occupied correct positions (green dots lining the IGL). Bar in (I) equals 265μm in (H) and 200μm in (I).
In adult Syne-1+/-; Syne-2-/- but not in Syne-1-/-; Syne-2+/- mice, the distinct laminary structure of the cerebral cortex was disrupted and the pyramidal cell layer in the hippocampus was mostly abolished (Figure 2F&G). In contrast, although the cerebellum was smaller, the Purkinje cells and granule cells occupied correct positions in the Syne-1+/-; Syne-2-/- mice (Figure 2H&I). We also examined Syne-1-/- and Syne-2-/- mice and found that the cerebral cortex and the hippocampus of Syne-2-/- brains displayed impaired laminary structures, similar to those seen in the Syne-1+/-; Syne-2-/- mice, but the brains of Syne-1-/- mutants showed no obvious defects (data not shown). These results indicate that Syne-2 alone is essential for the laminary structure formation in both the cerebral cortex and hippocampus, and that Syne-1 and Syne-2 have redundant roles in the cerebellum, midbrain, brain stem and other brain regions.
SUN1/2 and Syne-2 Are Essential for Radial Neuronal Migration in the Cerebral Cortex
The inverted laminary structure in the cerebral cortex and severe laminary defects in various brain regions are similar to the defects seen in mice with mutations in the Reelin or Disabled gene (D'Arcangelo et al., 1995; Howell et al., 1997; Sheldon et al., 1997), implying the failure of neuronal migration in Sun1/2 DKO and Syne-1/2 DKD brains. To investigate the roles of SUN1/2 in neuronal migration, we intercrossed Sun1+/-; Sun2-/-mice with either Sun1+/-; Sun2-/- or Sun1+/- mice, and injected BrdU intraperitoneally into the pregnant mothers at either E12.5 or E14.5. The BrdU positive cells were examined on coronal sections of E18.5 brains. In the cortex of Sun1+/-; Sun2+/-, Sun1+/-; Sun2-/-, and Sun1-/-; Sun2+/- mice, the E12.5 labeled BrdU positive cells were located in regions close to the intermediate zone (IZ), while the E14.5 labeled cells were close to the pial surface. In contrast, the positions of BrdU positive cells were inverted in Sun1-/-; Sun2-/- mutants (Figure 3A-D). These results indicate that SUN1 and SUN2 are essential and mutually redundant for the normal radial neuronal migration in the cerebral cortex.
Figure 3. Loss of SUN1/2 or Syne-2 disrupts radial neuronal migration in the cerebral cortex.
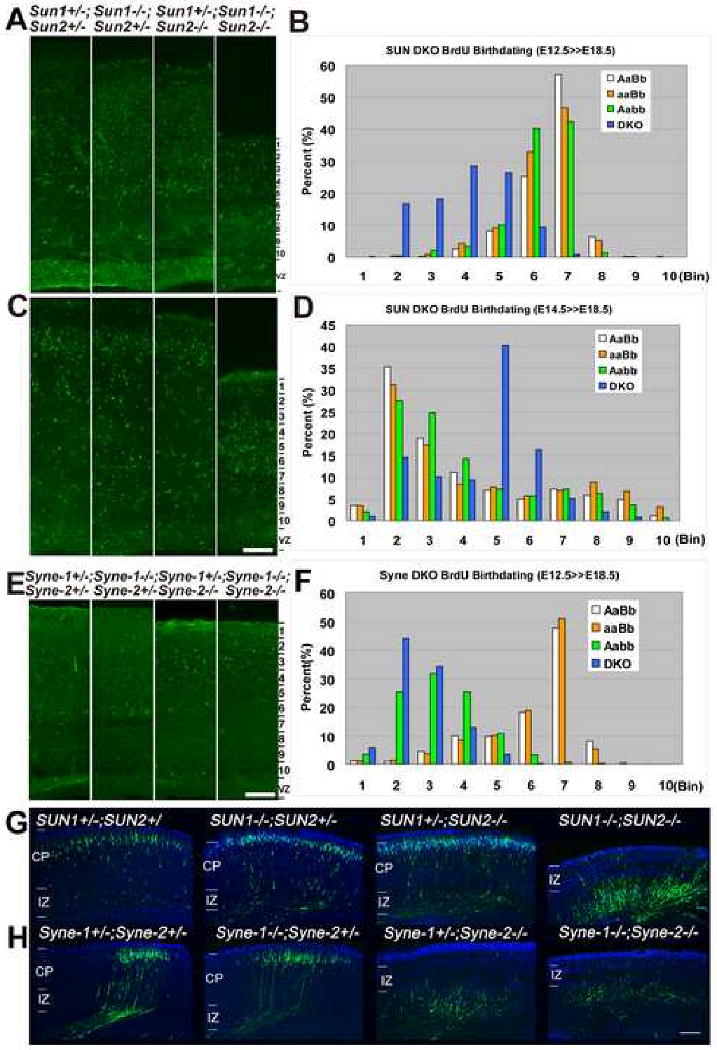
(A-D) BrdU birth-dating assays from E12.5 to E18.5 in Sun1/2 DKO mice. (A) E12.5 labeled BrdU positive cells (green) in Sun1/2 DKO brain were randomly distributed in the cortex (coronal section), while the BrdU positive cells in controls were localized deep in the cortex along the intermediate zone. (C) E14.5 labeled BrdU positive cells were localized deep in the Sun1/2 DKO cortex while most BrdU positive cells in controls were localized in layer 2-3. Bar in (C) equals 100μm for (A)&(C). (B)&(D) The region between pial surface and the VZ was arbitrarily divided into ten bins, and cell numbers in each bin were calculated. Statistical analysis showing the inverted distribution of BrdU positive cells between Sun1/2 DKO (DKO) brain and controls. More than three mice for each genotype and more than 4 sections were analyzed for each brain. AaBb, Sun1+/-; Sun2+/-. aaBb, Sun1-/-; Sun2+/-. Aabb, Sun1+/-; Sun2-/-.
(E-F) BrdU birth-dating assays from E12.5 to E18.5 in Syne-1/2 DKD mice. E12.5 born neurons occupied inverted position in the cortex of Syne-1+/-; Syne-2-/- (Aabb) and Syne-1/2 DKD (DKO) embryos compared to that in the Syne-1+/-; Syne-2+/- (AaBb) and Syne-1-/-; Syne-2+/- (aaBb) controls. Bar, 100μm.
(G-H) Coronal sections of E18.5 brains showing EYFP positive cells labeled by EYFP-expressing plasmids at E14.5. (G) The EYFP positive cells failed to migrate to layer2-3 in Sun1/2 DKO brain compared to those in controls. (H) The EYFP positive cells in Syne-1+/-; Syne-2-/- and Syne-1/2 DKD embryos failed to migrate to layer2-3 compared to those in the controls. CP, cortical plate. IZ, intermediate zone. Bar, 252μm.
The same BrdU birth-dating assay was also applied to Syne-1/2 mutants. Syne-1-/-; Syne-2+/- and Syne-1+/-; Syne-2-/- mice were intercrossed and BrdU was introduced into the pregnant mother at E12.5 or E14.5. When examined at E18.5, the BrdU positive cells occupied inverted position in both the Syne-1/2 DKD and the Syne-1+/-; Syne-2-/- brains, compared to the Syne-1+/-; Syne-2+/- and Syne-1-/-; Syne-2+/- controls at both time points (Figure 3E&F; Figure S3).
Radial neuronal migration in SUN1/2 and Syne-1/2 mutants was also examined after newly-formed neurons were labeled with EYFP through in utero electroporation (Tsai et al., 2007). Plasmids carrying an EYFP expression cassette were introduced into the lateral ventricles of E14.5 embryos, and the distribution of labeled neurons was examined at E18.5. In the control samples, most of the EYFP positive cells had successfully migrated from the VZ to the region right under the pial surface (Figure 3G; Figure S4). In contrast, most EYFP positive cells remained at the IZ in Sun1/2 DKO and Syne-2-/- mice (Figure 3G&H). These results are consistent with BrdU birth-dating results, indicating that the radial migration is disrupted in Sun1/2 DKO and Syne-2-/- mutants.
SUN1 and SUN2 Are Localized to the NE in the Embryonic Cortex and Both Function to Anchor Syne-2 to the NE
By immunofluorescence staining in embryonic brains, we found that Syne-1 and Syne-2 were localized to the nuclear envelope (NE) in certain cell types in the olfactory bulb, midbrain, cerebellum and brain stem (Figure S5; data not shown). In the cerebral cortex (cortex, IZ and VZ) of E13.5 and E15.5 mouse brains, SUN1, SUN2 and Syne-2 co-localized with lamin B at the NE (Figure S6; Figure 4A-C; data not shown), while Syne-1 displayed Lis1-associated pattern (Figure 4D; Figure S7A-D). In primary cultured cortical neurons, SUN1, SUN2 and Syne-2 again displayed NE localization, and Syne-1 showed Lis1-associated punctate distribution around the centrosome (Figure 4E-G; Figure S7E-J). These results are consistent with the findings that Syne-1 and Syne-2 display redundant functions in brain regions other than the cerebral cortex and the hippocampus (Figure 2&3; Figure S3).
Figure 4. SUN1 and SUN2 are required for the NE localization of Syne-2.
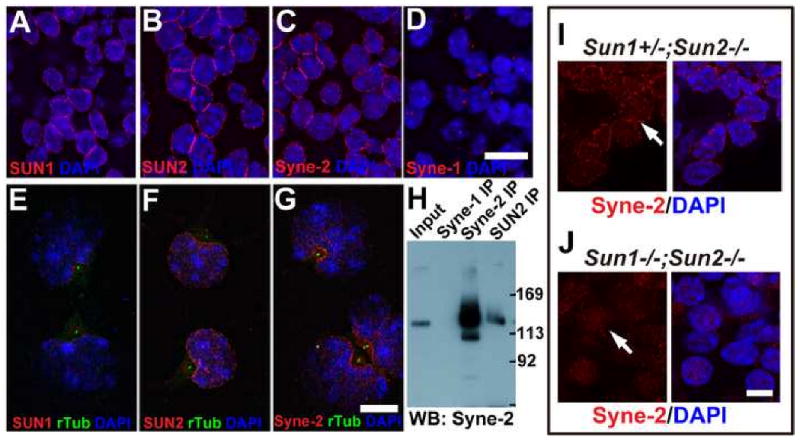
(A-D) Coronal sections of E15.5 wild-type mouse brain stained with antibodies against indicated proteins. SUN1, SUN2 and Syne-2 were localized on the nuclear envelope (NE), and Syne-1 showed bright dots adjacent to the nuclei. Bar, 10μm. The NE localization of these three proteins in brain neurons were confirmed by their co-localization with lamin B (Figure S6).
(E-G) Immuno-staining images of primary cultured E15.5 neurons showing high levels of SUN1, SUN2 and Syne-2 signals (red) at the NE region facing the centrosome (green dots, anti-γ-tubulin antibody staining). Bar, 10μm.
(H) Western blot showing interaction between Syne-2 and SUN2. E17.5 brain lysates were immuno-precipitated with antibodies against Syne-1, Syne-2 and SUN2 (rabbit polyclonal), and immunoblotted with anti-Syne-2 (mouse monoclonal). The two bands of Syne-2 in the input also exist in the anti-Syne-2 and anti-SUN2 immunoprecipitates.
(I-J) Immuno-staining images showing that the NE localization of Syne-2 (red) in E15.5 cortex was disturbed in Sun1/2 DKO brain. Bar, 5μm.
Furthermore, Syne-2 co-immunoprecipitated with SUN2 from both the E15.5 and E17.5 brain lysates (Figure 4H; data not shown) and the NE localization of Syne-2 was disturbed in E15.5 Sun1/2 DKO brains (Figure 4I&J). These results indicate that both SUN1 and SUN2 contribute to the recruitment of Syne-2 to the NE in migrating neurons.
SUN1, SUN2 and Syne-2 are Essential for Nucleokinesis and Centrosome-Nucleus Coupling during Radial Neuronal Migration in the Cerebral Cortex
To examine the nucleokinesis process in Sun1/2 DKO neurons, plasmids encoding Cherry-centrin2 and EGFP-Histone1B were co-injected into the lateral ventricles of E15.5 embryos, and the dynamic positioning of centrosomes and nuclei was time-lapse recorded on E18.5 brain slices. Consistent with previous reports, centrosomes migrated forward at a relatively stable speed and the nucleus followed in a saltatory fashion in wild-type samples (Figure 5A; Supplemental Video 1) (Tsai et al., 2007). However, in Sun1/2 DKO samples, nuclei vibrated around the original position and failed to move toward the pial surface, even though centrosomes were able to migrate forward for long distances (Figure 5B&D; Supplemental Video 2). Furthermore, the leading processes of Sun1/2 DKO neurons were significantly longer compared to that in controls (data not shown), suggesting that the neurites were competent for outgrowth. Similar nucleokinesis defects were also observed in Syne-1+/-; Syne-2-/- mice (Figure 5C&D; Supplemental Video 3). These results indicate that SUN1, SUN2 and Syne-2 play essential roles in nucleokinesis, likely through coupling the centrosome to the nucleus and transducing the pulling force from the microtubule network to the nucleus.
Figure 5. Loss of SUN1/2 or Syne-2 disrupts nucleokinesis during radial neuronal migration.
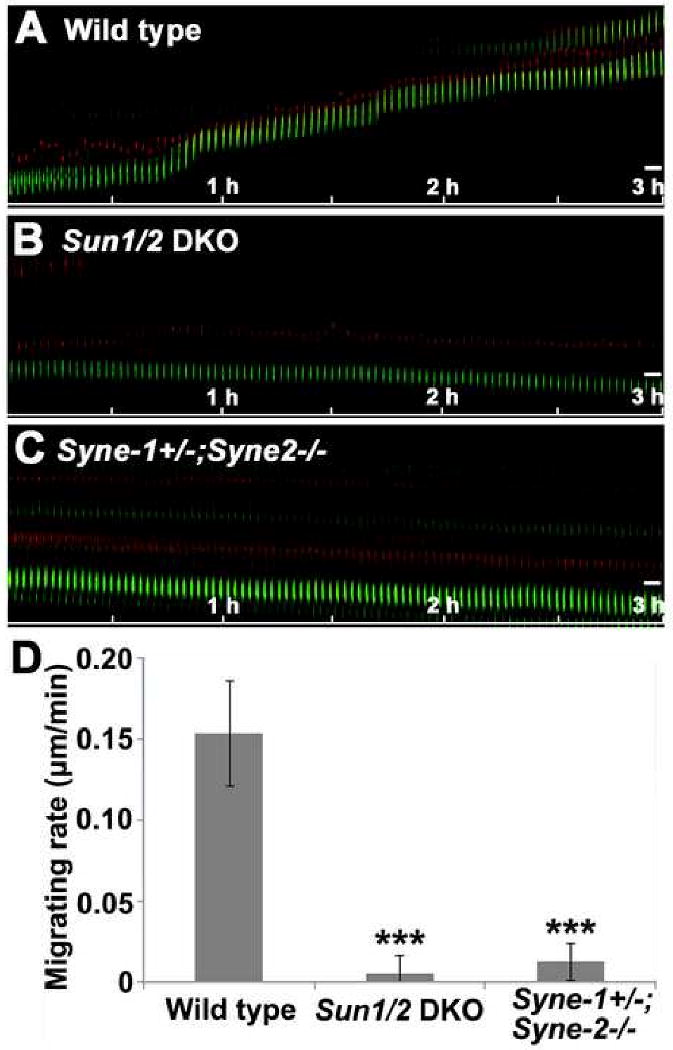
(A-C) Kymographs showing the movement of centrosomes (red) and nuclei (green) on live brain slices. The saltatory movement seen in the wild-type control (A) was disrupted in Sun1/2 DKO (B) and Syne-1+/-; Syne-2-/- brains (C). Bar, 5μm.
(D) Statistical data showing the migration rate of nuclei of brain cells of indicated genotypes. The nuclei in Sun1/2 DKO and Syne-1+/-; Syne-2-/- mice migrate drastically slower than those in the wild-type controls. n>16, p<0.001.
SUN1, SUN2 and Syne-2 are Essential for Centrosome-Nucleus Coupling during Glial Migration in vitro
We further utilized primary cultured glial cells to examine the coupling between the centrosome and the nucleus. Glial cells were isolated from E15.5 cortices and plated on laminin-coated dishes for 10 to 15 days. By this time, most cells were undergoing prominent migration (data not shown). In Sun1/2 DKO and Syne-1/2 DKD glia cells, the distance between the centrosome and the nucleus was drastically increased compared to that in wild-type and heterozygous controls (Figure 6A-D; Figure S8). Syne-1+/-; Syne-2-/- cells also displayed a defect that was similar to but less severe than that in the Syne-1/2 DKD mice (Figure 6C&D). Additionally, the centrosome-nucleus distance in the Sun1/2 DKO and Syne-1/2 DKD mutant glia showed a more randomized distribution pattern, indicating that the centrosome was uncoupled from the nucleus.
Figure 6. The centrosome-nucleus coupling is disrupted in Sun1/2 DKO and Syne-1/2 DKD glia.
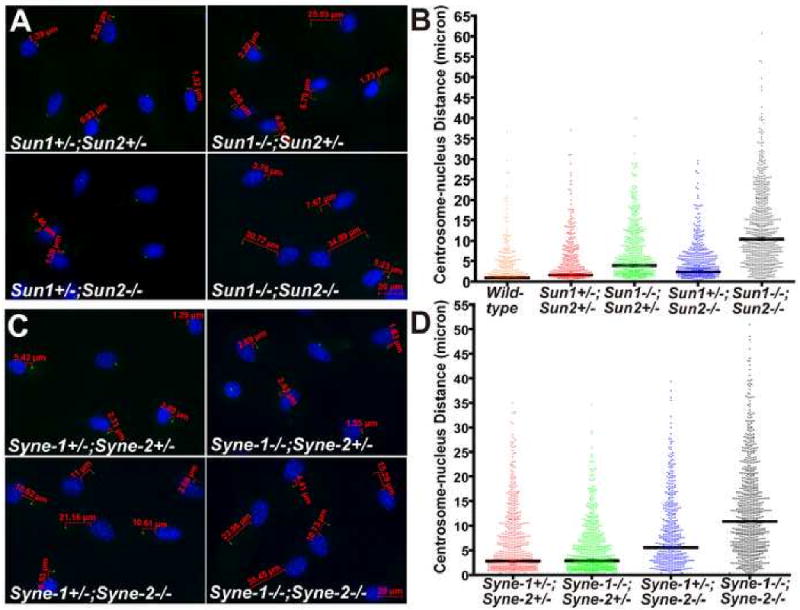
(A-B) Glial cells were stained with DAPI (blue) and anti-γ-tubulin antibody (green). The distance between the nucleus and the centrosome was measured using AxioVision (Carl Zeiss Microimaging, Inc). The centrosome-nucleus distance in Sun1/2 DKO glia was dramatically increased and randomized compared to the controls (B).
(C-D) The centrosome-nucleus distance was significantly increased in Syne-1+/-; Syne-2-/- glial cells, and the defect was exacerbated in Syne-1/2 DKD cells. Over 400 cells were scored for each genotype, and the p-values are shown in the two tables below.
Loss of SUN1/2 or Syne-2 Leads to Progressive Depletion of Neural Progenitors
The smaller brain size could be indicative of defects in neuronal proliferation. We thus examined the proliferation status in cortices of SUN1/2 and Syne-1/2 mutants at E12.5, E15.5 and E17.5 using BrdU pulse labeling. The number of neural progenitors in Sun1/2 DKO or Syne-1/2 DKD cortices was similar to that of controls at E12.5, and the number was slightly decreased at E15.5 (Figure 7A&B; Figure S9A&B). At E17.5, many proliferating cells were identified in the SVZ and IZ in controls, but the numbers of progenitor cells in corresponding regions were dramatically decreased in Sun1/2 DKO, Syne-1+/-; Syne-2-/-, and Syne-1-/-; Syne-2-/- mice (Figure 7C; Figure S9C). We also examined the number of neural progenitors in E15.5 Sun1/2 DKO mouse brain using an anti-Nestin antibody, and the result is consistent with the BrdU pulse labeling data (Figure S10). These results indicate that the loss of either SUN1/2 or Syne-2 caused a progressive depletion of neural progenitors in the cerebral cortices. Specifically, the loss of proliferating cells in the IZ and SVZ at E17.5 in these mutant brains suggests the loss of intermediate progenitor cells (IPC) (Colette Dehay and Henry Kennedy, 2007).
Figure 7. Loss of SUN1/2 or Syne-2 disrupts interkinetic nuclear migration and leads to progressive depletion of neural progenitors.
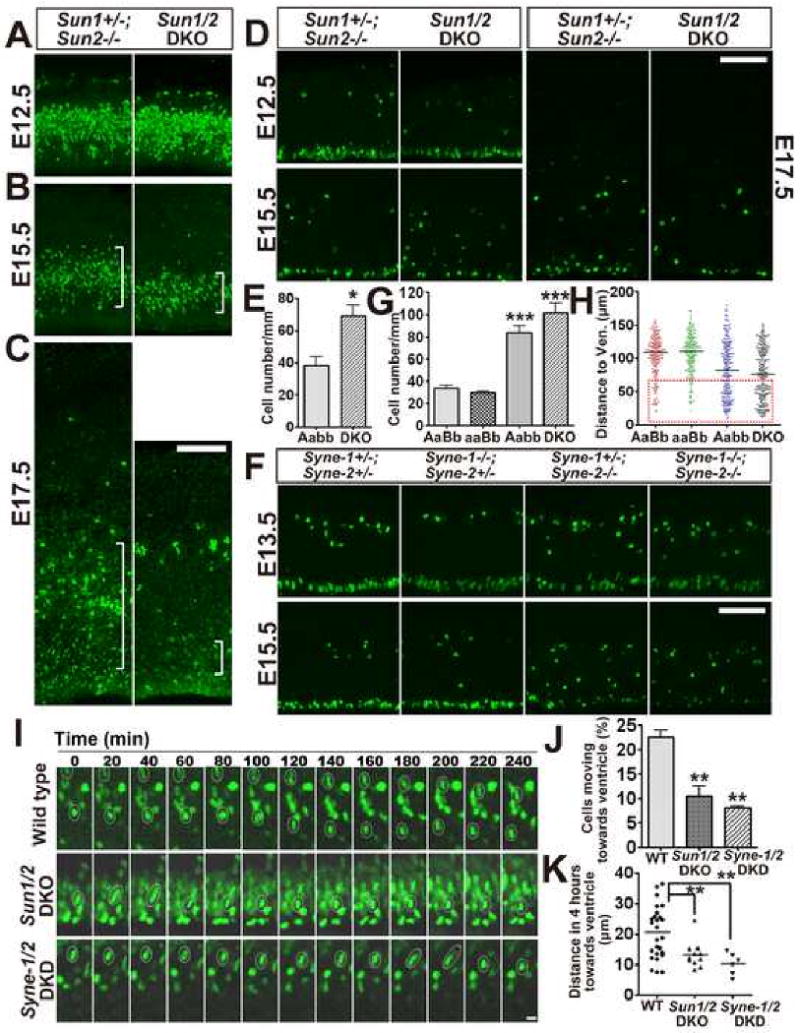
(A-C) Fluorescence images showing proliferating cells pulse labeled with BrdU at E12.5, E15.5 and E17.5 (green). The number of proliferating cells in Sun1/2 DKO brain was progressively decreased compared to the controls. Noticeably, the BrdU positive cells at the IZ (white brackets) of E17.5 Sun1/2 DKO brain were essentially missing compared to that of control brain. Bar, 100 μm.
(D) Immuno-staining images showing phosphorylated-Histone3 (pH3, S10P) positive cells (green) identified at different time points in the ventricular zone (VZ). In Sun1/2 DKO mice, a significant portion of mitotic cells in the VZ was mis-positioned at E15.5 and the total number of mitotic cells at the apical surface was decreased at E17.5. Bar, 100 μm.
(E) Statistical diagrams illustrating mis-positioning of mitotic cells in the VZ of Sun1/2 DKO brains. The average number of mis-positioned pH3 positive cells per 1000 micrometers was analyzed at E15.5, and the number was significantly increased in Sun1/2 DKO brain (DKO). n=3, p<0.05. Aabb, Sun1+/-; Sun2-/-.
(F) Images showing that mitotic cells (green, pH3 positive) in the VZ were dramatically mis-positioned and the number of mitotic cells along the apical surface was significantly decreased in Syne-1+/-; Syne-2-/- and Syne-1/2 DKD brain at E15.5 compared to controls. Bar, 100 μm.
(G-H) Statistical diagrams illustrating mis-positioning of mitotic cells in the VZ of Syne-2-/-brains. (G)Average number of mis-positioned mitotic cells in the VZ was significantly increased in Syne-1+/-; Syne-2-/- (Aabb) and Syne-1/2 DKD (aabb) brains at E15.5 (2 mice for each genotype and over ten sections of each mice were examined, p<0.001 when either Aabb or aabb group was compared with control ones). (H) Y axis represents the distance between the mis-positioned pH3 positive cell and the apical surface of the VZ. 200 cells were counted for each genotype, with obviously more cells were severely mis-positioned in Syne-1/2 DKD brain (red dotted box). AaBb, Syne-1+/-; Syne-2+/-; aaBb, Syne-1-/-; Syne-2+/-.
(I) Time-lapse recording of live brain slices at the VZ showing the movement of soma/nucleus (green). The movement of soma toward the apical surface of the VZ was disrupted in Sun1/2 DKO and Syne-1/2 DKD mice (white circles). Bar, 10 μm.
(J) Bar diagram showing that less than 10% of cells were migrating toward the apical surface of the VZ in Sun1/2 DKO and Syne-1/2 DKD mice, compared to 22.5 % in wild-type mouse brain.
(K) Diagram illustrating the distance an individual nucleus moved toward the apical surface of the VZ within 4 hours. This average distance was significantly decreased in Sun1/2 DKO and Syne-1/2 DKD mouse brain. 2 slices for wild type and Syne-1/2 DKD, and 3 slices for Sun1/2 DKO mouse brains.
To further characterize the proliferation defects in Sun1/2 DKO and Syne-1/2 DKD mutants, we examined the mitotic cells by staining with an S10-phosphorylated Histone3 (pH3) antibody. At E12.5 and E13.5, most pH3 positive cells of all genotypes were located along the apical surface of the VZ, and a small portion of them were located at the basal surface of the VZ. At E15.5, the number of mitotic cells along the ventricle was significantly decreased in the VZ of Sun1/2 DKO, Syne-1+/-; Syne-2-/-and Syne-1/2 DKD mice, but the number of mis-positioned mitotic cells in the VZ was dramatically increased in these mice compared to their littermate controls (Figure 7D-H). These results indicate that SUN1/2 and Syne-2 have essential functions in maintaining mitotic cell divisions of RGPCs at proper positions within the VZ.
SUN1/2 and Syne-2 Are Required for Interkinetic Nuclear Migration in Mouse Cerebra
A possible cause for the mis-positioning of pH3-positive mitotic cells in the VZ of Sun1/2 DKO, Syne-1+/-; Syne-2-/-, and Syne-1/2 DKD mice is the failure of interkinetic nuclear migration (INM). Defective INM may also lengthen the G1 phase of RGPCs and lead to more differentiative mitotic divisions, which are expected to reduce the number of IPCs and progenitors lining the VZ (Colette Dehay and Henry Kennedy, 2007). To elucidate the roles of SUN1/2 and Syne-1/2 in INM, an EGFP-Histone1B expressing cassette was introduced into the embryonic brains at E14.5. After 15-18 hours, acute brain slices were time-lapse recorded and the movement of EGFP positive cells within the VZ was analyzed. In wild-type mice, 22.5% of cells were observed to be migrating toward the apical surface of the VZ, but the number was decreased to less than 10% in either the Sun1/2 DKO or Syne-1/2 DKD brains (Figure 7I&J; Supplemental Video 4-6). Furthermore, the total distance that a single nucleus migrated within 4 hours toward the apical surface of the VZ was significantly reduced in Sun1/2 DKO and Syne-1/2 DKD cells (Figure 7I&K; Supplemental Video 4-6). Therefore, loss of both SUN1 and SUN2, or both Syne-1 and Syne-2, disrupted interkinetic nuclear migration.
SUN-Syne Complexes Co-localize and Interact with the Dynein/dynactin and Kinesin Complexes
In the prevailing model of nucleokinesis, one group of cytoplasmic dynein is proposed to be localized to the NE to mediate the pulling force for nuclear movement (see Introduction). To investigate the potential role of Syne-2 as the NE anchor for cytoplasmic dynein during neuronal migration, we first examined their subcellular localization in the brains of E15.5 embryos. We found that Syne-2 co-localized with dynein intermediate chain (dynein IC, 74.1kd) at the nuclear periphery in the cerebral cortex (Figure 8A; data not shown). Both Syne-2 and dynein IC also displayed asymmetric and partially overlapped localizations in primary cultured cortical neurons (Figure 8B).
Figure 8. Syne-2 interacts with the dynein/dynactin complex at the nuclear envelope.
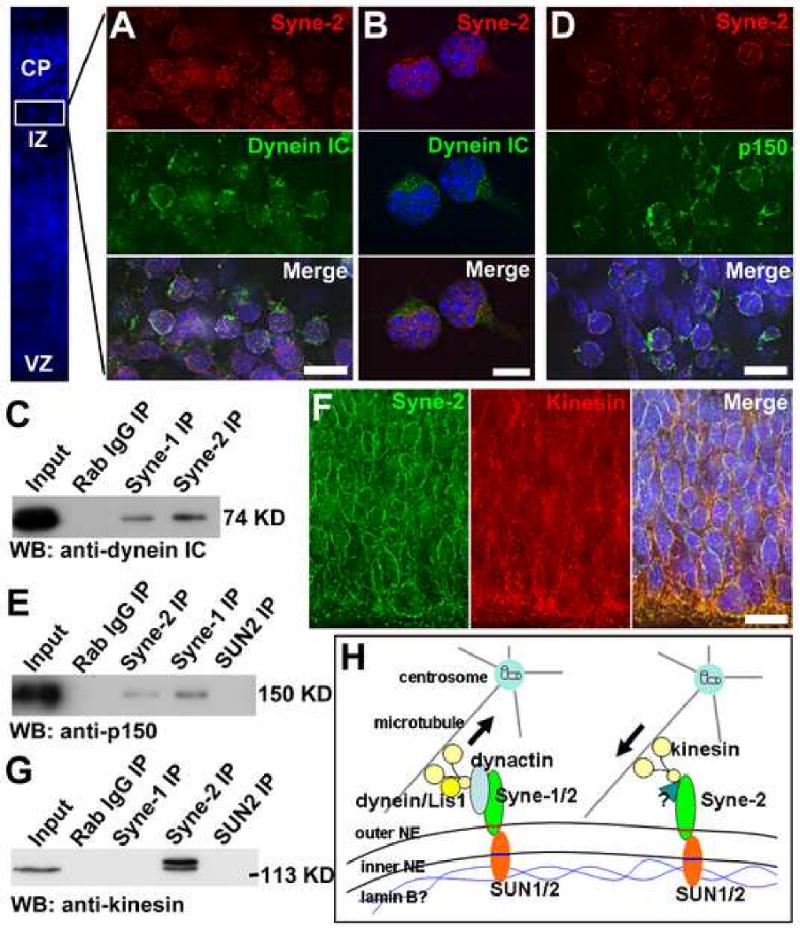
(A-B) Immuno-staining images showing that Syne-2 and dynein intermediate chain (IC) co-localize on the NE of E15.5 mouse cortical cells (A) and primary cultured cortical neurons (B). Bar, 10 μm.
(C) Western blot showing antibodies against Syne-1 and Syne-2 precipitated dynein IC from E17.5 brain lysates.
(D) Immuno-staining images showing that Syne-2 and dynactin (p150) co-localized on the NE of E15.5 mouse cortex. Bar, 10 μm.
(E) Western blot showing that antibodies against Syne-1 and Syne-2 precipitated the p150 subunit of dynactin.
(F) Immuno-staining images showing that kinesin and Syne-2 partially co-localize on the NE in the VZ of E13.5 mouse brain. Bar, 10 μm.
(G) Western blot showing that kinesin was precipitated from E17.5 brain lysate by anti-Syne-2 antibody.
(H) A model illustrating how SUN1/2 and Syne-1/2 mediate the driving force from microtubule to the nucleus. Inner nuclear envelope proteins SUN1/2 interact with Lamin B and form complexes with the outer NE proteins Syne-1 and Syne-2. During INM at G2 phase and nucleokinesis, Syne-1 and Syne-2 interact with cytoplasmic dynein/Lis1 complexes to pull the nucleus toward the centrosome. During INM at G1 phase, Syne-2 forms complexes with kinesin to push the nucleus away from the centrosome.
We further carried out co-immunoprecipitation experiment between dynein and Syne-2 in embryonic brains, using an anti-Syne-2 antibody that recognizes the giant isoform of Syne-2 (over 200kD) in the E15.5 mouse brain (Figure S11A&B). When Syne-2 was immunoprecipitated by the anti-Syne-2 antibody from E17.5 or E18.5 brains, cytoplasmic dynein was readily detected in the precipitates (Figure 8C; Figure S11C). Consistently, an antibody against the dynein intermediate chain was able to precipitate the giant Syne-2 protein from E17.5 brain lysates (Figure S11D). In addition, Syne-1 was localized to the NE in multiple regions of mouse brain except the cerebral cortex and hippocampus (Figure S5), and an anti-Syne-1 antibody recognizing both isoforms was also able to immunoprecipitate cytoplasmic dynein from the whole brain lysates of E17.5 mouse embryos (Figure 8C).
We also examined interactions between Syne-1/2 and dynactin in embryonic brain because dynactin has been shown to be the anchor protein between dynein and its cargo and to localize to the NE in prophase tissue culture cells (Schroer, 2004). First, Syne-2 were found to co-localize with the dynactin subunit p150 in the cerebral cortex of E15.5 mouse brain (Figure 8D). Second, the p150 subunit was consistently precipitated by antibodies against either Syne-1 or Syne-2 from the E17.5 brain lysates (Figure 8E). In addition, an antibody against p150 was capable of pulling down Syne-2 from the E17.5 brain lysates (data not shown). These results indicate that Syne-1/2 interacts with the cytoplasmic dynein/dynactin complex on the NE during brain development.
During INM, the nucleus moves both away from and toward the centrosome. It is conceivable that cytoplasmic dynein motors act to pull the nucleus toward the centrosome during G2, and the kinesin motors function to push the nucleus away from the centrosome in G1. Therefore, we also examined the interaction between Syne-2 and kinesin in the developing mouse brain. We found that Syne-2 co-localized with kinesin on the NE in the VZ at E13.5 (Figure 8F), and that the anti-Syne-2 antibody was able to pull down kinesin from E17.5 brain lysates (Figure 8G). These results suggest that Syne-2 interacts with kinesin motors at the NE during interkinetic nuclear migration.
The microtubule “fork” that associates with the nucleus has been proposed to be essential for the coupling between the centrosome and the nucleus during neuronal migration (Xie et al., 2003). To further address whether the function of SUN1, SUN2 and Syne-2 is associated with microtubule during neuronal migration, we double stained E15.5 mouse brain slices with anti-α-Tubulin and anti-SUN1, anti-SUN2 or anti Syne-2 antibody. We found that α-Tubulin co-localized with SUN1, SUN2 and Syne-2 not only at the NE but also on the “fork” structures (Figure S12 A-C), suggesting that SUN1/2 and Syne-2 function to connect microtubule to the nuclear envelope. We then stained the cortex of E15.5 SUN DKO embryos with anti-α-Tubulin, but found the NE localization of microtubule did not show significant difference when compared to the controls (Figure S12 D-E). This might be due to the small volume of cytosol of cortical neurons and the limitation of the light microscope used in this analysis.
Working Memory Is Impaired in Syne-2 KASH Deletion Mice
The laminary structures of the cerebral cortex and the hippocampus, known to be important for learning and memory, were severely disrupted in Syne-1+/-; Syne-2-/-, and Syne-1+/+; Syne-2-/- mice (Figure 2F&G; data not shown). We thus back-crossed Syne-2-/- mice to the C57BL/6J background for six generations and subjected them to several behavior tests. These back-crossed Syne-2-/- mice had a growth rate that was indistinguishable from their littermate controls.
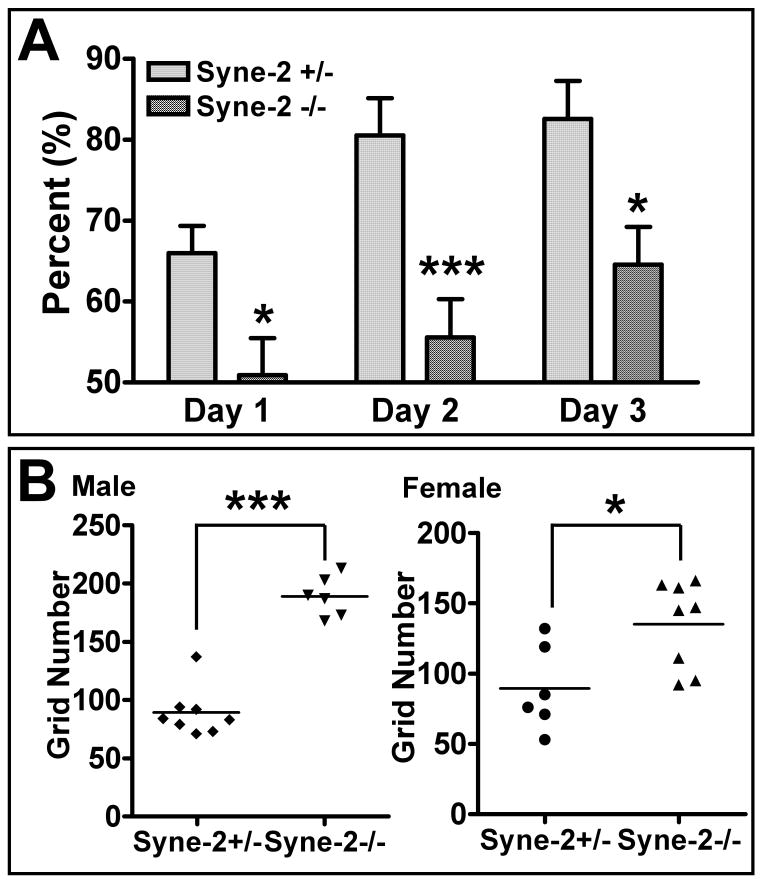
T-maze alternation has been commonly used to test the “working memory” of rodents (Deacon, 2006). In a rewarded T-maze alternation test, Syne-2+/- mice successfully reached 80% correct responses within 3 days, but Syne-2-/- mice learned significantly slower than controls (Figure 9A). This result indicates that working memory is significantly impaired in Syne-2-/- mice. Secondly, in a novel-open-field test, Syne-2-/- mice entered significantly more grids than their littermate controls during a five-minute period (Figure 9B), indicating that Syne-2-/- mice have abnormally active responses to a new environment. Finally, when tested on an accelerating rotating rod or a 0.9cm diameter balance beam, the Syne-2-/- mice performed as well as their littermate wild-type and Syne-2+/- controls (data not shown), indicating that these Syne-2-/- mice have no significant defects in balance or locomotion abilities. Given that the cerebral cortex and hippocampus are severely malformed in the Syne-2-/- mouse brain, it is currently unclear what specific structural defects in the mutant brain lead to the defects in learning and memory.
Figure 9. Loss of KASH-domain containing Syne-2 disrupts working memory in mice.
(A) Bar diagram showing the defect of Syne-2-/- mice in a rewarded T-maze alternation assay. Syne-2-/- mice showed significantly worse performance than their littermate controls. Syne-2+/- n=15, Syne-2-/- n=12.
(B) Diagram showing the results of an open-field assay, indicating that both female and male Syne-2-/- mice entered significantly more grids than controls in an open field.
Discussion
In this study, we uncover for the first time the essential roles of SUN1/2 and Syne-1/2 (Nesprin-1/2) in the nuclear migration process during neurogenesis and neuronal migration in mammals. We show that SUN1 and SUN2 recruit Syne-2 to the nuclear envelope (NE) and that Syne-2 interacts with both dynein/dynactin and kinesin complexes at the NE. These protein complexes are crucial for the coupling between the centrosome and the nucleus during neuronal migration, and the integrity of the SUN-Syne function is important for learning and memory in mice. These results may significantly advance our understanding of the mechanism of nucleokinesis and interkinetic nuclear movement during mammalian brain development (Figure 8H).
SUN1/2 and Syne-1/2 Are Essential for Radial Neuronal Migration and Interkinetic Nuclear Migration
In this study, we showed that loss of either SUN1/2 or Syne-1/2 led to severe defects in the laminary structures of many brain regions. Our analysis of the neocortex revealed the radial neuronal migration failure in both Sun1/2 DKO and Syne-1/2 DKD mice. The laminary defects in other brain regions of these DKO mutants are also likely caused by the disruption of radial neuronal migration processes. For example, pyramidal cells of the hippocampus arise in the ventricular zone (VZ) and migrate outward along radial glial cells (Hatten, 1999). Failure of pyramidal cell migration will directly result in the laminary defects in the hippocampus in Sun1/2 DKO and Syne-1/2 DKD mice. For another example, postmitotic Purkinje cells in the cerebellum migrate outward along radial glial fibers and settle down beneath the external granule cell layer (Hatten, 1999). The loss of Purkinje cell layer in Sun1/2 DKO and Syne-1/2 DKD brain (Figure 1&2) is thus likely to be caused by the radial migration failure. The widespread laminary defects in Sun1/2 DKO and Syne-1/2 DKD brains suggest that the SUN-KASH complexes are broadly utilized during radial neuronal migration.
We also investigated the roles of SUN1 and SUN2 in tangential migration during brain development. Interneurons labeled by anti-Calbindin antibodies were readily observed in cerebral cortices of E18.5 Sun1/2 DKO mice (Figure S13), suggesting that tangential migration of Calbindin positive interneurons from LGE/MGE to the cerebral cortex was not blocked in the Sun1/2 DKO mice. This may further suggest a mechanistic difference between radial and tangential migration in the brain.
Syne and SUN Proteins Play Critical Roles in both Nuclear Anchorage and Nuclear Movement
Our previous genetic studies have shown that the Syne-1 protein was essential for the stable anchorage and even distribution of myonuclei along the cell membrane in muscle cells (Zhang et al. 2007). Syne-1 is speculated to connect the NE of myonuclei to cortical actin through its N-terminal actin binding (ABD or CH) domains and thus keep those nuclei at fixed periphery locations during drastic muscle contraction. Our current study uncovers a somewhat surprising function of Syne-1 and Syne-2 in connecting the NE to the microtubule network during radial neuronal migration and interkinetic nuclear movement. Detailed analysis indicated that Syne-2 interacts with the dynein/dynactin complex, likely for pulling the nucleus towards the centrosome, and with kinesin, likely for pushing the nucleus away from the centrosome. These results have expanded our understanding of how KASH-domain proteins mediate interactions between the NE and cytoskeleton.
In C. elegans and Drosophila, KASH domain proteins have been shown to function in nuclear anchorage and migrations. ANC-1, the only ortholog of Syne-1/2 in C. elegans, is known for its specific function in nuclear anchorage in syncytial cells, likely through interacting with actin network (Starr and Han, 2002). The much smaller molecules UNC-83 and ZYG-12 of C. elegans, as well as Klarsicht of Drosophila, are involved in nuclear movement, likely through interacting with the microtubule system (Malone et al., 2003; Mosley-Bishop et al., 1999; Starr et al., 2001). The Syne-2 protein was recently found to play a role in retina cell movement in Zebrafish and such a function was speculated to be mediated by the dynein/dynactin complex based on the results from the shorter KASH proteins of the invertebrate systems (Del Bene et al., 2008; Tsujikawa et al., 2007).
Like ANC-1, both Syne-1 and Syne-2 contain both the KASH and the ABD domains and are giant proteins with long spectrin repeats (Starr and Han, 2002; Zhen et al., 2002). In comparison, the other known mammalian KASH proteins, Nesprin-3 and Nesprin-4, are smaller proteins that are potential orthologs of Klarsicht in Drosophila, and UNC-83 or ZYG-12 in C. elegans (Roux et al., 2009; Wilhelmsen et al., 2005). Therefore, it is interesting to learn that Syne proteins have the ability to interact with both F-actin and microtubules for their functions in both nuclear anchorage and migration.
Two models may explain the functional diversity of Syne-1 and Syne-2 in mouse brain development. In the first possibility, the ABD of Syne-2 may interact with a subunit of the dynein/dynactin complex. Specifically, the Arp1 subunit of dynactin has been shown to form actin-filament-like octamers and interact with various kinds of spectrin-domain proteins (Holleran et al., 1996; Schroer, 2004), and βIII spectrin has also been shown to directly interact with Arp1 through its CH domains (Holleran et al., 2001). Using an antibody against the ABD domain of Syne-2 (Luke et al., 2008), we have detected high-level expression of the ABD-containing Syne-2 at the NE of brain cells in mouse embryos (Figure S14). The ABD domains of Syne-2 are also highly conserved compared to the CH domains of βIII spectrin (data not shown). Although we are so far unable to detect a direct interaction between the ABD/CH domains of Syne-2 and Arp-1 (data not shown), this model remains a strong possibility. The second possibility is that a shorter Syne-2 isoform without the ABD domain performs the function in neuronal migration. In our IP experiments using an anti-Syne-2 KASH antibody, we detected both the giant isoform and a ∼120 kD shorter isoform of Syne-2 (Figure 4H). Additional genetic experiments are required to distinguish between these two models.
We have also analyzed the mouse Nesprin-3 gene for its potential function during brain cell migration. However, we neither detected any obvious neuronal migration defects in Nesprin-3 deletion mice, nor observed evenly distributed NE localization of the Nesprin-3 protein in the cerebral cortex (data not shown).
SUN1 and SUN2 have been shown to be inner NE proteins and interact with laminA/C (Crisp et al., 2006; Haque et al., 2006; Hodzic et al., 2004; Padmakumar et al., 2005; Zhang et al., 2005). Loss of laminA/C in mouse skeletal muscle cells has been shown to disrupt the myonuclear anchorage and the NE localization of SUN2 (Mejat et al., 2009). In the developing mouse brain, we found lamin B, but not lamin A/C was expressed on the NE (data not shown). Lamin B mutant mice die shortly after birth and show a flattened skull (Vergnes et al., 2004), phenotypes similar to Sun1/2 DKO and Syne-1/2 DKD mice. Therefore, it is possible that lamin B, instead of lamin A/C, interacts with SUN1/2 in the developing mouse brain for neurogenesis and neuronal migration (Figure 8K).
The Redundancy of Two SUN Proteins and Two Syne Proteins in Brain Development
Our studies using single and double mutants indicated that SUN1 and SUN2 proteins act redundantly for much of their roles during mouse development (Lei et al., 2009) (this study). The lethality of Syne-1/2 DKD mice indicates that the KASH-domain-containing Syne-1 and Syne-2 function redundantly to provide an essential function, similar to the functions of SUN1 and SUN2. However, the redundancy between Syne-1 and Syne-2, and between SUN1 and SUN2, is variable from tissue to tissue. While Syne-1 alone plays an essential function in anchoring nuclei in skeletal muscle cells, the SUN1 and SUN2 functions in this tissue are largely redundant even though the function of SUN1 is slightly more prominent than that of SUN2 (Lei et al., 2009). However, their functions during brain development present a different combination: on one hand, Syne-2 alone is essential for neuronal migration in the cerebral cortex and the hippocampus while SUN1 and SUN2 act redundantly in this process; on the other hand, Syne-1 and Syne-2 act redundantly in the midbrain, the cerebellum and the hindbrain, and SUN1 itself seems to be essential for specific brain regions of the midbrain (Figure 1&2). In the case of SUN proteins, the difference in their functions is to a large extend due to the difference in their expression patterns, a notion also supported by the finding that SUN1 but not SUN2 is expressed in germ cells during meiosis (Ding et al., 2007; Lei et al., 2009).
It is interesting that Syne-1 and Syne-2 showed different subcellular localizations in the cerebral cortex and the hippocampus (Figure 4). We also found that Syne-1 was localized to the NE of certain cell types in the olfactory bulb, midbrain, cerebellum, brain stem, but not in the cerebral cortex or the hippocampus in E15.5 and P0 mouse brain (Figure S5; data not shown). In the Co-IP studies using whole brain lysates, it's not feasible to distinguish which isoform of Syne-1 interacts with dynein/dynactin. The subcellular localization of Syne-1 was associated with Lis1 both in the cerebral cortex of E15.5 embryo and in primary cultured cortical neurons (Figure S7). We further showed that the asymmetrical distribution of Syne-1 is dependent on microtubules, not F-actin (Figure S15). The NE-disassociated localization of Syne-1 in the cerebral cortex and hippocampus suggests a possibility that a KASH domain-lacking isoform is expressed during early brain development. Such an isoform could have a function that is distinct from that of Syne-2. Since the expression of this isoform would not likely be affected in our Syne-1 KASH deletion mice, the function of this potential KASH domain-lacking Syne-1 and its relationship with Lis1 need to be explored by further genetic studies.
Mutations in human SYNE1 has been show to be associated with recessive cerebellar ataxia (Gros-Louis et al., 2007). Our data that the Syne-2-/- mice have severe defects in learning and memory but not in growth rate indicate a possibility that Syne-2 is also associated with human neurological deceases. However, at the this stage, what specific defects in the Syne-2-/- mouse brain is the direct cause of the behavior problem remain unclear and would be an attractive question for future studies.
Experimental Procedures
Mouse Procedure
Mouse breeding and animal related experiments were carried out following the general guidelines published by The Association for Assessment and Accreditation of Laboratory Animal Care. Animal-related procedures were reviewed and approved by the Institute of Developmental Biology and Molecular Medicine Institutional Animal Care and Use Committee. Sun1/2 DKO and Sun1+/-Sun2-/- mice were generated by Sun1+/-Sun2-/- intercrosses. Sun1+/- Sun2+/- and Sun1-/-Sun2+/- mice were generated by crossing Sun1+/- mice with Sun1+/-Sun2-/- mice. Syne-1/2 DKD animals were generated by intercross between Syne-1+/-Syne-2-/- and Syne-1-/-Syne-2+/- mice. Noon of the day of virginal plug detection was defined as embryonic day 0.5 (E0.5) in timed mating.
BrdU Birth-dating and Proliferation Assays
For neuronal birth-dating, BrdU (Sigma, 10mg/ml) dissolved in normal saline or PBS was injected intraperitoneally at 50μg/g body weight into E12.5 or E14.5 pregnant mothers, and brains of the E18.5 embryos were collected, fixed in paraformaldehyde and embedded (Chae et al., 1997). For proliferation assay, BrdU solutions were injected intraperitoneally into pregnant mother. Embryos were collected 30 minutes later and subjected to fixation (Feng and Walsh, 2004).
Histology and Immunofluorescence Staining
Frozen sectioning, paraffin sectioning and IF staining were carried out as previously described (Zhang et al., 2007). Before frozen sectioning, brains of E17.5 and E18.5 embryos and heads of E12.5, E13.5 and E15.5 embryos were fixed in either 1% or 4% paraformaldehyde and dehydrated in 20% sucrose overnight at 4°C. Antibodies against the following proteins/antigens were used: Rabbit antibodies against Syne-1(1:100), anti-Syne-2(1:100), anti-SUN1(1:100) and anti-SUN2(1:100) were described previously (Ding et al., 2007; Zhang et al., 2007). Other antibodies against the following proteins were used: Nesprin-2 (K56-386, gift from A. Noegel, University of Cologne), NeuN (Chemicon, 1:100), Cux1 (Santa Cruz, 1:100), Tbr1 (Abcam, 1:1000), Reelin (Chemicon, 1:500, gift from Jinbo deng, Henan University), BrdU (Sigma, 1:1000, Chemicon, 1:100), Nestin (Santa Cruz, 1:100), γ-Tubulin (Abcam, 1:200), Lis1 (D. Smith, University of South Carolina; Santa Cruz, 1:200), Dynein IC (Chemicon, 1:200), Kinesin (Chemicon, 1:500), Histone3-S10P (Upstate) and α-Tubulin (Sigma, 1:1000).
Neuronal Cell Culture
To obtain cortical neurons or hippocampal cells, cortices or hippocampi were dissected from E15.5 mouse embryos and digested by trypsin. Dissociated cells were plated on cover glasses coated with poly-D-Lysine (Sigma) and laminin (Sigma), and cultured in Neurobasal Medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 5% FBS, 1% penicillin/streptomycin and 1% L-Glutamine (Shi et al., 2003). To obtain glial cells, cells isolated from cortices were plated at lower cell density (2×104/3.5cm dish) and cultured for two to three weeks (Osmani et al., 2006).
Nocodazole or lautranculin A (Sigma) were added to the medium at 10μm/ml 36-48 hours after plating, and incubated for 2-3hours(Tanaka et al., 2004). After 36-48 hours in culture, cells were fixed in methanol at -20°C for 10 minutes and subjected to immuno-staining.
In Utero Electroporation and Brain Slice Live Imaging
In utero electroporation and time-lapse imaging on live brain slices were carried out according to standard protocols (Chen et al., 2008; Saito, 2006). Plasmids carrying different expression cassettes were injected into the lateral ventricles of embryos at indicated ages (0.5μg/μl for Cherry-centrin2 and 1.5μg/μl for EGFP-Histone1B). Embryos were collected either at E18.5 for the observation of nucleokinesis or at E15.5 for the observation of interkinetic nuclear migration. Images were collected by Olympus FW1000 microscopes and were further disposed by Image-Pro Plus (MediaCybernetics) and Adobe Photoshop.
Co-Immunoprecipitation (Co-IP) and Western Blot
Brain samples were homogenized in TNP buffer (50mM Tris-HCl, pH7.4, 150mM NaCl, and 0.4% NP-40) or TNT buffer (50mM Tris-HCl, pH7.4, 150mM NaCl, and 0.5% Triton X-100) and was extracted for 30 min on ice. After centrifugation (18,000g, 20 min at 4°C), the supernatant was precleared with proteinA/G agarose (PIERCE) at 4°C for 1 hour and was then subjected to centrifugation again (18,000g, 15min at 4°C). The precleared supernatant was divided into 600μl aliquots, and each aliquot was mixed with the antibody or control IgG and rotated end-over-end overnight at 4°C. Aliquots were further centrifuged (18,000g, 15min at 4°C), and the supernatant of each aliquot was incubated with protein A/G agarose at 4°C for 2-4 hours. The beads were then collected at 1,000g and were washed thoroughly in TNP buffer for 5 times. Finally, the beads were boiled in loading buffer and subjected to SDS-PAGE. SDS-PAGE (5%-10%) and Western blot analysis were carried out according to standard protocols (Joseph Sambrook, 2001) and the manufacturer's instructions (Pierce and Santa Cruz).
Behavior Test
Syne-2+/- mice were back crossed to C57BL/6J for six generations and intercrossed to obtain the Syne-2-/- mice that were subjected to behavior tests. The Rewarded T-Maze was carried out according to standard protocols (Deacon, 2006). For the open-field assay, a 33cm × 55cm field was divided into fifteen 11cm × 11cm grids. One mouse at each time was dropped onto the center of this field from 25cm above, and the total number of grids that a mouse entered in 5 minutes was recorded. The Rotating Rod experiment was carried out as previously described (Ying et al., 2006).
Statistical Analysis
Statistical analyses were carried out using Microsoft Excel 2003 and GraphPad Prism 4. The results are presented as mean +/- SEM, and significant differences are indicated by single asterisk (*) when p<0.05, double asterisk (**) when p<0.01 and triple asterisk (***) when p<0.001.
Supplementary Material
Acknowledgments
We thank Angelika A. Noegel (University of Cologne), Yuanyi Feng (Northwestern University), Deanna Smith (University of South Carolina), Jinbo Deng (Henan University) and Li-Huei Tsai (MIT) for gifts of antibodies; Xu Ding, Diana Ronai, Aileen Sewell, Max Cohen, Brian Kudlow, Jing Cui for valuable comments and discussions; Chuantao Zhao, Di Xia, Xiaoping Huang, Boying Tan, Yanling Yang, and Xiaoqian Jin and others in X. Yuan's lab and at IDM for assistance; and Beibei Ying, Wufan Tao, Kejing Deng, Ling Sun and Yun Zheng for advices. Kai Lei held the funding of National Talent Training Fund in Basic Research of China (No. J0630643). This work was supported by grants from the Ministry of Science and Technology (973 program), National Natural Science Foundation of China, Ministry of Education of China (211 program), and Shanghai Municipal Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chae T, Kwon YT, Bronson R, Dikkes P, Li E, Tsai LH. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron. 1997;18:29–42. doi: 10.1016/s0896-6273(01)80044-1. [DOI] [PubMed] [Google Scholar]
- Chen G, Sima J, Jin M, Wang KY, Xue XJ, Zheng W, Ding YQ, Yuan XB. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11:36–44. doi: 10.1038/nn2018. [DOI] [PubMed] [Google Scholar]
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, Faucette L, Wang JY, Jeang KT. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Deacon RM. Appetitive position discrimination in the T-maze. Nat Protoc. 2006;1:13–15. doi: 10.1038/nprot.2006.3. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Protein-protein interactions, cytoskeletal regulation and neuronal migration. Nat Rev Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci U S A. 2004;101:6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci U S A. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard JP, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Joseph Sambrook DWR. Molecular Cloning: A Laboratory Manual. Third Edition. New York: Cold Spring Habor Laboratory Press; 2001. [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid Encodes a SUN Domain Protein Required for Klarsicht Localization to the Nuclear Envelope and Nuclear Migration in the Eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812037106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, et al. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121:1887–1898. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 Is a KASH Protein Required for Nuclear Migration and Is Recruited to the Outer Nuclear Membrane by a Physical Interaction with the SUN Protein UNC-84. Mol Biol Cell. 2006 doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejat A, Decostre V, Li J, Renou L, Kesari A, Hantai D, Stewart CL, Xiao X, Hoffman E, Bonne G, et al. Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J Cell Biol. 2009 doi: 10.1083/jcb.200811035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr Opin Cell Biol. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, et al. Disruption of nesprin-1 produces an Emery Dreifuss Muscular Dystrophy-like phenotype in mice. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Natl Acad Sci U S A. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci U S A. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Ying M, Xu R, Wu X, Zhu H, Zhuang Y, Han M, Xu T. Sodium butyrate ameliorates histone hypoacetylation and neurodegenerative phenotypes in a mouse model for DRPLA. J Biol Chem. 2006;281:12580–12586. doi: 10.1074/jbc.M511677200. [DOI] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.