Summary
Selective degeneration of midbrain dopaminergic (mDA) neurons is associated with Parkinson’s disease (PD), and thus an in-depth understanding of molecular pathways underlying mDA development will be crucial for optimal bioassays and cell replacement therapy for PD. In this study, we identified a novel Wnt1-Lmx1a autoregulatory loop during mDA differentiation of ES cells, and confirmed its in vivo presence during embryonic development. We found that the Wnt1-Lmx1a autoregulatory loop directly regulates Otx2 through the β-catenin complex and Nurr1 and Pitx3 through Lmx1a. We also found that Lmx1a and Lmx1b co-operatively regulate mDA differentiation with overlapping and cross-regulatory functions. Furthermore, co-activation of both Wnt1 and SHH pathways by exogenous expression of Lmx1a, Otx2 and FoxA2 synergistically enhanced the differentiation of ES cells to mDA neurons. Together with previous works, this study shows that two regulatory loops (Wnt1-Lmx1a and SHH-FoxA2) critically link extrinsic signals to cell-intrinsic factors and cooperatively regulate mDA neuron development.
Introduction
During early brain development, mDA neurons originate from the ventral midline of the mesencephalon. The initial event of mDA neuron development was shown to depend on Sonic hedgehog (SHH), fibroblast growth factor 8 (FGF8), and Wnt1, setting up the initial field for mDA progenitors (McMahon and Bradley, 1990; Prakash et al., 2006; Ye et al., 1998). Among these, Wnt1 and FGF8 are expressed from Isthmus and they cross regulate each other (Chi et al., 2003; Lee et al., 1997; Liu and Joyner, 2001; Matsunaga et al., 2002). Recent studies showing that FGF8 failed to induce ectopic DA neurons in Wnt1 mutant embryos (Prakash et al., 2006) suggest that Wnt1, which can be induced by FGF8, is a more direct regulator of initiation of mDA fields. Furthermore, a recent study established that compound FGFR mutant mice show that FGF8 regulates mDA neuronal precursors (NP) proliferation rather than mDA identity, the latter being more critically mediated by SHH and Wnt1 (Saarimaki-Vire et al., 2007). SHH expressed from the notochord has been shown to directly induce FoxA2 expression in ventral mesencephalon (VM) through Gli binding sites in the FoxA2 gene (Sasaki et al., 1997). FoxA2, in turn, directly induces VM SHH expression through well-conserved FoxA2 binding sites in the SHH gene (Jeong and Epstein, 2003). FoxA2 regulates mDA development by inhibiting an alternate fate (Nkx2.2+ cells), inducing neurogenesis through Ngn2, and regulating Nurr1 and DA phenotype genes (Ferri et al., 2007) as well as regulating survival/maintenance of mDA neurons (Arenas, 2008; Kittappa et al., 2007), strongly suggesting that FoxA2 is the main mediator of SHH signaling in mDA development. Taken together, SHH and Wnt1 are two major extrinsic signals that play critical roles in mDA development. However, for Wnt1, it is less clear what are its direct downstream target genes. These extrinsic signals are thought to initiate the regulatory cascades leading to mDA development by inducing key transcription factors. Indeed, downstream from these initial signaling molecules numerous transcription factors have been implicated, including FoxA2, Otx2, Lmx1a, Lmx1b, Nurr1, and Pitx3 (Ang, 2006; Smidt and Burbach, 2007). How these extrinsic signals and intrinsic transcription factors interact with each other is of utmost importance not only for our understanding of the regulatory network of mDA development but also for optimal stem cell engineering for cell replacement therapy of PD.
Recently, an important transcriptional pathway involving the homeodomain protein Lmx1a has been identified; Lmx1a is expressed in early DA progenitors and induces another homeodomain factor, Msx1, which then suppresses alternative cell fates by suppressing the Nkx6.1 gene and induces neurogenesis by activating the proneural gene, Ngn2 (Andersson et al., 2006b). This study showed that Lmx1a is important for mDA development by gain- and loss-of-function analyses in chick embryos. Interestingly, dreher mutant mice carrying a mutation in the Lmx1a locus (Millonig et al., 2000) showed only modest developmental defect of mDA neurons (Ono et al., 2007), suggesting that there may be differences in mDA developmental mechanisms between the chick and mammalian systems. While these elegant studies shed important insights into mDA development, how Lmx1a interacts with critical extrinsic signals and how it regulates key mDA factors remain largely unknown.
In this study, we identified a Wnt1-Lmx1a autoregulatory loop during mDA differentiation of ES cells and confirmed that such a regulatory loop is also functional in vivo during mouse embryonic development. We found that this Wnt1-Lmx1a autoregulatory loop directly regulates Otx2 gene expression (through the canonical Wnt signaling pathway) and Nurr1 and Pitx3 gene expression (through Lmx1a). We also found that Lmx1a has overlapping function with Lmx1b in regulating downstream target genes and that they cross-regulate each other during mDA differentiation. Furthermore, forced expression of key targets of the Wnt1 pathway (Otx2 and Lmx1a) and a key target of the SHH pathway (FoxA2) synergistically induced mDA differentiation of ES cells, showing the importance of understanding mDA developmental mechanisms in optimal differentiation of ES cells into mDA neurons.
Results
Wnt1 directly regulates the expression of Lmx1a and Otx2 during mDA differentiation
FoxA2 is a direct downstream target of the SHH signaling pathway (Sasaki et al., 1997) but the direct downstream targets of the Wnt1 signaling pathway during mDA development are still unclear (Fig. 1a). To address this question, J1 ES cells were differentiated in vitro and infected with empty or Wnt1-expressing retrovirus at the NP stage (Suppl. Fig. 1). To clearly see the effect of transgene expression without masking their effect by culture conditions, we used suboptimal condition without any DA-inducing factors. Cells were further differentiated and analyzed at day 3 of the neuronal differentiation (ND) stage (termed ND3 in the text). This is the time point of active mDA neurogenesis and differentiation in this stem cell culture bioassay, thus optimal to analyze the expression of potential mDA regulators/targets. Quantitative real time PCR (qPCR) analysis revealed that forced Wnt1 expression significantly increased mRNA levels of Otx2, Pitx3 and to a greater extent Lmx1a (Fig. 1b), but not those of FoxA2, Nurr1 or Msx1. The lack of an effect on the expression of these genes at this early time point suggests that they are not the direct targets of Wnt1 signaling, though they may be regulated by genes further downstream. Consistent with this mRNA analysis, immunocytochemisty analysis revealed an increased number of Lmx1a+ cells after Wnt1 overexpression (Fig. 1c–d) from 7.05±0.78 to 16.20±1.11 (%Lmx1a+ cells/Hoechst+ cells; P<0.05). In addition, Otx2+ or Pitx3+ cell numbers, but not Nurr1+ cell numbers, were significantly increased after Wnt1 overexpression (Suppl. Fig. 2).
Fig. 1.
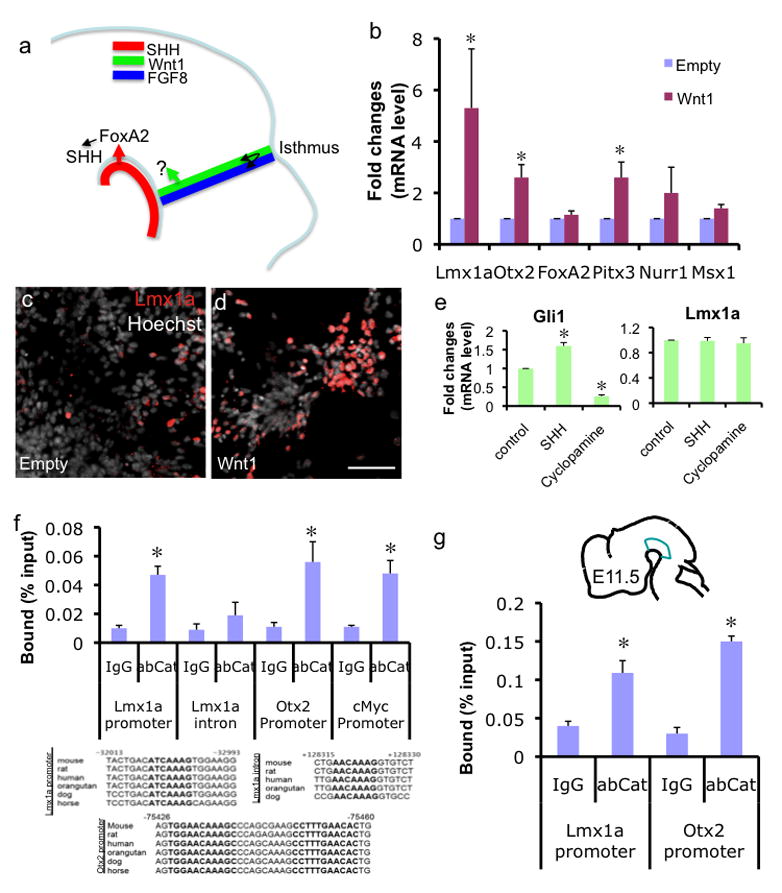
Wnt1 directly regulates Lmx1a and Otx2 through the β-catenin complex. a. Two major signaling molecules involved in mDA differentiation are SHH from notochord and Wnt1 from Isthmus. FoxA2 is shown to be a direct downstream target of the SHH signaling pathway and then FoxA2 in turn induces VM SHH expression. The direct downstream target of the Wnt1 signaling pathway remains elusive. FGF8 from the hindbrain side of Isthmus and Wnt1 from the midbrain side of Isthmus cross-regulate each other, shown by black arrow. b. qPCR analysis of DA regulator expression on in vitro differentiated cells transduced with empty or Wnt1-expressing retrovirus (ND3; n=4, p<0.05, data are represented as mean±SEM throughout this study). c–d. Immunocytochemistry analysis on the same cells. Scale bar represents 50μm. e. NP stage cells were treated with 500ng/ml of SHH or 1μM Cyclopamine for 6 hours and analyzed by qPCR. f. ChIP-qPCR analysis. In vitro differentiated cells were transduced with Wnt1-expressing retrovirus, treated with 15mM LiCl for 24 hrs and fixed for ChIP at the NP stage. ChIP fragments were immunoprecipitated with normal rabbit IgG or anti-β-catenin antibody and analyzed by qPCR. The average of three independent ChIP analyses (n=3, p<0.05) are presented. g. E11.5 VMs were dissected as illustrated without LiCl treatment and used for ChIP using β-catenin antibody (n=3, p<0.05).
It was previously shown that SHH treatment could ventralize chick intermediate midbrain explants, accompanied by induction of Lmx1a and other ventral midbrain phenotype (Andersson et al., 2006b). Thus, we tested whether Wnt1 can still induce Lmx1a in the presence of the SHH signaling inhibitor cyclopamine (Kittappa et al., 2007) and found that Wnt1 induced Lmx1a independent of SHH signaling (Suppl. Fig. 3a). We next tested whether acute treatment with SHH or cyclopamine had an immediate effect on Lmx1a expression. ES-derived NP cells were treated with 500ng/ml SHH or 1μM cyclopamine for 6 hours and analyzed by qPCR. While these treatments led to corresponding changes in Gli1 mRNA levels, there was no significant changes in Lmx1a mRNA levels (Fig. 1e) as well as TH or Nurr1 mRNA levels (Suppl. Fig. 3b), suggesting that these genes are not direct targets of the SHH signaling. However, it is still possible that SHH can indirectly regulate these genes through further downstream targets.
To address whether Wnt1 directly regulates any of these potential targets via the canonical Wnt signaling pathway, we performed chromatin immunoprecipitation (ChIP)-qPCR analysis. At day 1 of the NP stage, in vitro differentiated ES cells were transduced with retroviral Wnt1, treated with 15mM LiCl at day 3 to stabilize β-catenin, and then fixed for ChIP at day 4. ChIP was performed either with a control antibody or with anti-β-catenin antibody. qPCR analysis showed significant binding of β-catenin complex to the well conserved TCF/LEF binding site in the Lmx1a promoter, but not to another potential TCF/LEF site in the third intron of Lmx1a, showing the specificity of β-catenin binding in our assay system (Fig. 1f). For the Otx2 promoter, ChIP-qPCR analysis showed that there is direct association of the β-catenin complex to its well conserved TCF/LEF sites during mDA differentiation (Fig. 1f). We used the c-myc promoter’s TCF/LEF binding sites as positive control (Yochum et al., 2007), and observed comparable binding with the Lmx1a promoter and the Otx2 promoter (Fig. 1f). In the absence of LiCl treatment, the ChIP experiment yielded comparable results (Suppl. Fig. 4), suggesting that Wnt1 expression alone is sufficient for stabilizing the β-catenin complex in this system. Furthermore, this binding of β-catenin complex was Wnt1-dependent (Suppl. Fig. 4). For the Pitx3 promoter, the regulation by Wnt1 appears to be indirect, since we could not find well-conserved TCF/LEF binding sites on the Pitx3 promoter, even though there is a possibility of regulation by a long-range enhancer.
To further test whether these direct downstream targets are bound by the β-catenin complex in vivo during embryonic development, we performed the ChIP analysis using dissected VM of E11.5 embryo. This analysis confirmed that the Lmx1a and Otx2 promoters are physically associated with the β-catenin complex (Fig. 1g), supporting our in vitro data that Lmx1a and Otx2 are direct targets of the Wnt1 signaling pathway during mDA development.
Lmx1a directly regulates Wnt1 expression during mDA differentiation
Since Lmx1a showed the most robust effect by Wnt1 overexpression, we attempted to identify Lmx1a’s downstream targets. J1 ES cells were differentiated in vitro, infected with empty or Lmx1a-expressing retrovirus at the NP stage, and analyzed at ND3 after further differentiation. Interestingly, qPCR analysis showed that Lmx1a dramatically increased expression of Wnt1, but not that of SHH or Wnt5a (Fig. 2a). We also overexpressed Lmx1a using episomal vector and observed similar results (data not shown). In addition, immunocytochemical analysis showed that exogenous Lmx1a expression robustly increased the numbers of Wnt1+ cells (Fig. 2b–c).
Fig. 2.
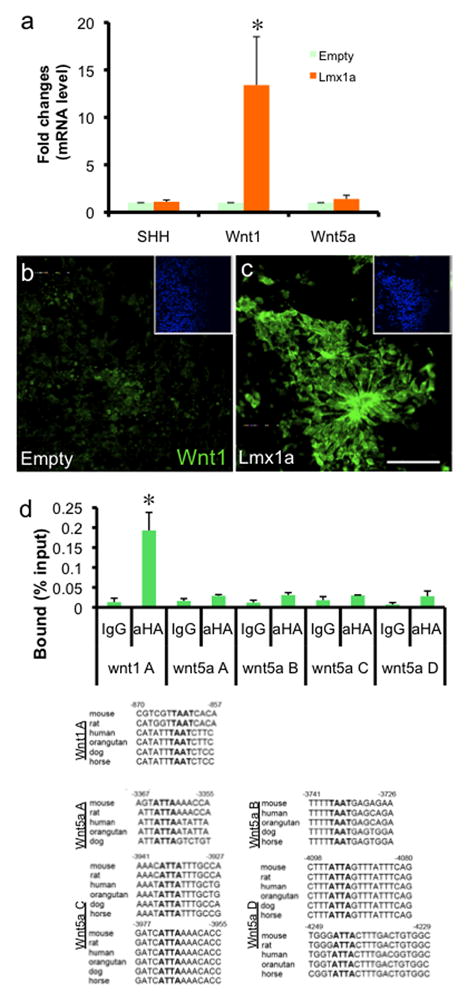
Lmx1a directly regulates Wnt1 expression. a. qPCR analysis on in vitro differentiated ES cells with empty or Lmx1a-expressing retrovirus (ND3; n=4, p<0.05). b–c. Immunocytochemistry on the same cells. Scale bar represents 50μm. Inset shows Hoechst staining. d. ChIP-qPCR analysis on Wnt1 and Wnt5a promoter region (n=3, p<0.05). In vitro differentiated ES cells transduced with retrovirus expressing HA-tagged Lmx1a was fixed for ChIP at ND3. ChIP fragments were immunoprecipitated either with normal rabbit IgG or anti-HA antibody and analyzed by qPCR. Results represent the average of three independent ChIP experiments.
We next tested the possibility that Lmx1a directly regulates the expression of Wnt1 by ChIP-qPCR analysis. In vitro differentiated J1 cells at the NP stage were transduced with retrovirus expressing HA-tagged Lmx1a, and harvested for ChIP at ND3. Crosslinked chromatin complex was immunoprecipitated using anti-HA antibody or control IgG, and analyzed by qPCR. There was significant Lmx1a binding to the well-conserved homeodomain binding site in the Wnt1 promoter, but not to 6 well-conserved sites contained in 4 PCR fragments on the Wnt5a promoter, demonstrating the specificity of in vivo Lmx1a binding (Fig. 2d). Importantly, this ChIP data is consistent with the overexpression data that Lmx1a regulates Wnt1, but not Wnt5a (Fig. 2a–c), further supporting the validity of our ChIP analysis. We confirmed the binding of Lmx1a to the Wnt1 promoter by an independent method (electrophoretic mobility shift assay (EMSA)), and observed specific DNA-protein complex formation which was supershifted by anti-Lmx1a antibody (Suppl. Fig. 5a). Taken together, our results reveal the presence of a tight autoregulatory loop between Wnt1 and Lmx1a during mDA differentiation of ES cells.
Next, we tested whether Lmx1a regulates the expression of Wnt1 during mouse embryonic midbrain development in vivo, using the wildtype (wt) and dreher (dr/dr) mice (Millonig et al., 2000). In situ hybridization analysis of littermate wt vs. dr/dr embryos showed that Wnt1 expression is compromised by Lmx1a mutation in developing midbrain (Fig. 3a–d). At E11.5, this defect was more evident, although Wnt1 expression was partially spared in the ventral most part (Fig. 3c–d). One possible explanation of this residual Wnt1 expression is the functional compensation by Lmx1b, which is expressed in the entire mDA domain at E10.5 (Fig. 3e and g) and in the ventral most part at E11.5 (Fig. 3f and h), which will be further discussed later. The specificity of the antibodies against Lmx1a and Lmx1b is shown in Suppl. Fig. 6. To further test whether there is a direct interaction between Lmx1a and the Wnt1 promoter during embryonic development, we performed ChIP analysis using dissected VM of E11.5 embryo and found that the Wnt1 promoter is physically associated with Lmx1a in developing VM (Fig. 3i), confirming the presence of the Wnt1-Lmx1a autoregulatory loop in the embryo as well as during ES cell differentiation.
Fig. 3.
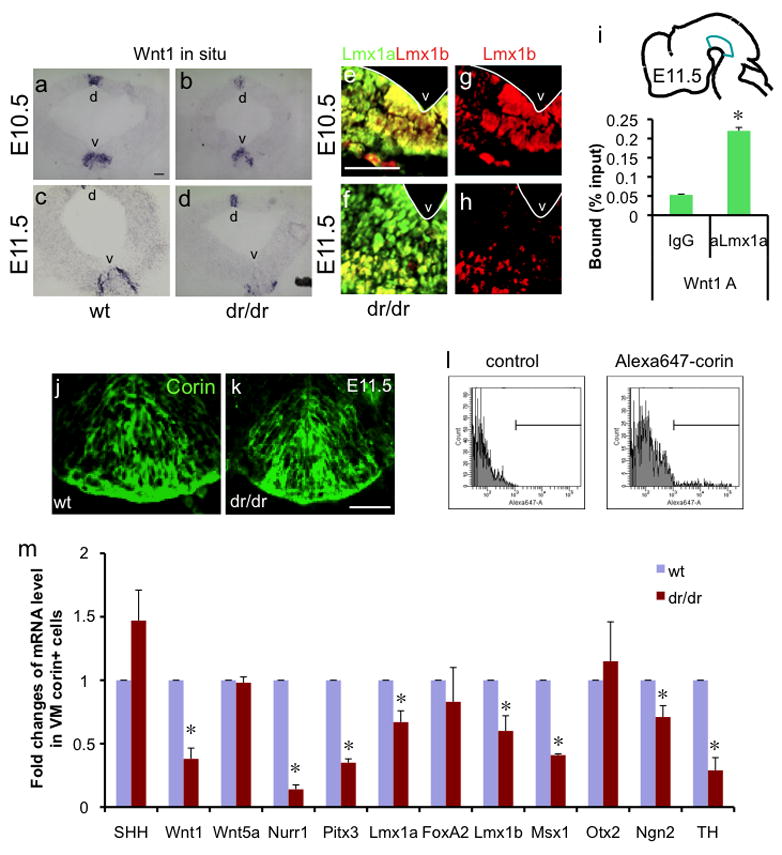
Lmx1a regulates Wnt1 expression during embryonic midbrain development. a–d. In situ hybridization analysis of Wnt1 expression. Coronal mesencephalic section of E10.5 (a, b) and E11.5 (c, d) littermate wt or dr/dr embryos. d marks dorsal mesencephalon and v marks VM. e–h. Lmx1b is expressed in the entire ventral midbrain of E10.5 embryos but is restricted to the ventral most part in E11.5 embryos. Coronal midbrain sections were stained using Lmx1b or Lmx1a antibody. The white line marks ventricle. Scale bar represents 50μm. i. E11.5 VMs were dissected as illustrated and used for ChIP using Lmx1a antibody. Binding of Lmx1a to the Wnt1 promoter was assayed by qPCR (n=3, p<0.05). j–k. Anti-corin antibody used for FACS purification marks the mDA domain, as shown in E11.5 VM of littermate wt or dr/dr embryo. Scale bar represents 50μm. l. FACS purification of mDA domain cells of littermate wt and dr/dr after staining with anti-corin antibody and Alexa-647-conjugated secondary antibody. The corin+ population is marked. m. qPCR analysis of purified mDA domain cells on the expression of regulators of mDA neuronal development. The result is the average from three independent FACS purifications (n=3, p<0.05).
To quantitatively analyze the effect of Lmx1a mutation on gene expression in vivo, we purified E11.5 mesencephalic floor plate (mFP) cells, which generate mDA neurons (Kittappa et al., 2007; Ono et al., 2007), from littermates wt and dr/dr embryos. To purify mFP cells, we did fluorescent activated cell sorting (FACS; Fig. 3l) using antibody against corin, a cell surface marker specifically expressed in developing FP cells (Fig. 3j–k) (Ono et al., 2007). mRNA analysis showed that Lmx1a mutation caused a significant decrease (approximately 60%) in expression of Wnt1, but not that of Wnt5a (Fig. 3m), consistent with the result from ES cell differentiation (Fig. 2a). We observed mild decrease in Lmx1a, Lmx1b and Ngn2 mRNA levels in the dr/dr embryos, consistent with the previous study (Ono et al., 2007).
Lmx1a directly binds the promoter element(s) and regulates expression of Nurr1 and Pitx3
In addition to Wnt1 gene regulation by Lmx1a mutation, there was significant reduction in the expression of Nurr1 and Pitx3 (Fig. 3m). In the dr/dr embryo, this downregulation of Nurr1 and Pitx3 could be an indirect effect of defective DA neuron differentiation. Alternatively, it may be caused by direct regulation of Lmx1a. To address these possibilities, we tested if Lmx1a directly regulates the expression of Pitx3 and Nurr1 during ES cell in vitro differentiation. Retroviral Lmx1a expression increased the expression of Pitx3, Nurr1 and Lmx1b, whereas it failed to significantly affect the expression of Otx2, FoxA2 or En1 at ND3 (Fig. 4a). We also observed significant increase in Msx1 and Ngn2 mRNA levels, consistent with a previous study (Andersson et al., 2006b). We also repeated this experiment using an episomal Lmx1a expression system and obtained similar results (data not shown). Immunocytochemical analysis showed that exogenous Lmx1a expression increased the number of Nurr1+ (Fig. 4b–c; from 2.07+0.38 to 3.88±0.46 %Nurr1+ cells/Hoechst+ cells) and Pitx3+ cells (Fig. 4e–f; from 0.58±0.40 to 3.50±0.33 %Pitx3+ cells/Hoechst+ cells) as well as Lmx1b+ cells (Suppl. Fig. 7a–b). Interestingly, many Pitx3+ cells and Nurr1+ cells were not yet positive for TH at ND3 (Fig. 4c and f), suggesting that the increase in Pitx3 and Nurr1 gene expression is a direct effect, but not the byproduct of increased mDA neurons. At a later time point (ND7), the majority of Pitx3+ cells became TH+, suggesting that Lmx1a precociously induced Pitx3 expression in immature DA neurons (Suppl. Fig. 7c–f). In addition, Lmx1a expression significantly increased the %TH+ cells/β-tubulin+ cells from 0.87±0.21 to 2.98±0.84 without supplementing the culture with SHH, unlike the previous report where the effect of Lmx1a on DA induction was strictly dependent upon addition of SHH to the culture (Andersson et al., 2006b). Endogenous SHH expression at the NP stage may explain such difference.
Fig. 4.
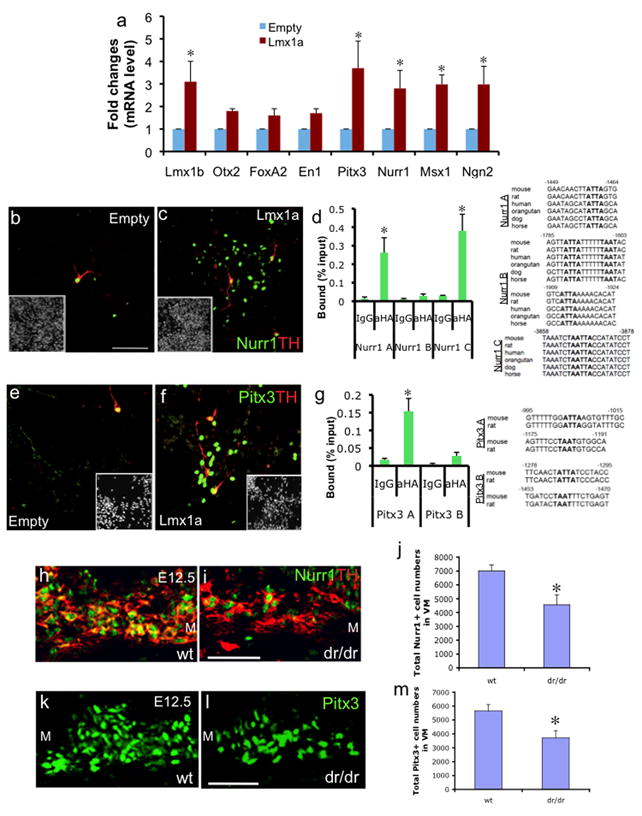
Lmx1a directly regulates Nurr1 and Pitx3. a. qPCR analysis on in vitro differentiated ES cells with empty or Lmx1a-expressing retrovirus (ND3; n=4, p<0.05). b–c. Immunocytochemistry on the same cells. Scale bar represents 50μm. d. ChIP-qPCR analysis on Nurr1 promoter region (n=3, p<0.05), performed as described above. e–f. Immunocytochemistry on the same cells. g. ChIP-qPCR analysis on Pitx3 promoter region (n=3, p<0.05), performed as described above. h–i. Immunohistochemistry analysis of VM in E12.5 littermates’ wt and dr/dr embryos using anti-Nurr1 and anti-TH antibody. M denotes medial VM. Scale bar represents 50μm. j. Cell counting analysis of Nurr1+ cells in ventral midbrain of E12.5 littermates’ wt and dr/dr embryos (n=4, p<0.05). Cell numbers were counted from every 6th sections using the StereoInvestigator image capture equipment and software. The estimated total cell numbers based on counting every 6th section are shown. k–l. Immunohistochemistry analysis of ventral midbrain in E12.5 littermates wt and dr/dr embryos using anti-Pitx3 antibody. m. Cell counting analysis of Pitx3+ cell numbers as described above (n=4, p<0.05).
To further address whether Lmx1a directly regulates gene expression of the mDA regulators, Nurr1 and Pitx3, we performed ChIP analysis. We found Lmx1a significantly bound to Nurr1A and Nurr1C PCR fragments, but not the Nurr1B fragment (Fig. 4d), and confirmed the specific binding of Lmx1a by supershift EMSA (Suppl. Fig. 5b). For the Pitx3 promoter, we observed significant Lmx1a binding to Pitx3A, but not Pitx3B PCR fragment (Fig. 4g), and also confirmed it by supershift EMSA (Suppl. Fig. 5c). We also performed ChIP to test whether Ngn2 is directly regulated by Lmx1a, but observed no significant binding (Suppl. Fig. 8b).
To further confirm the regulation of Nurr1 and Pitx3 by Lmx1a during embryonic midbrain development, we performed immunohistochemistry and stereological analysis on littermate wt and dr/dr embryos. We counted the number of Nurr1+ and Pitx3+ cells in the entire mDA domain in every 6th coronal VM section, using the StreoInvestigator image capture equipment and software. We found significant decreases in Nurr1+ and Pitx3+ cell numbers in dr/dr embryos compared to littermate wt embryos (Fig. 4h–m), whereas there was no significant difference in the FoxA2+ or Otx2+ cell numbers between wt and dr/dr embryos (Suppl. Fig. 9a–b). Taken together, our results strongly suggest that Lmx1a directly regulates Nurr1 and Pitx3, but not FoxA2 or Otx2 both in mDA differentiation of ES cells and in embryonic midbrain development.
Lmx1a and Lmx1b have overlapping functions in regulating mDA regulators
Compared to the robust induction of mDA differentiation in ES cells by Lmx1a, dreher mice displayed only mild dysregulation of mDA development. This could be explained either by lack of functional significance of Lmx1a during embryonic mDA development or by the presence of another gene with redundant function. For the latter possibility, Lmx1b is one such candidate, because (1) it is expressed in the same domain as Lmx1a during mDA development and (2) it is highly related to Lmx1a with 61% overall amino acid identity (Hobert and Westphal, 2000). Thus, to explore whether Lmx1b and Lmx1a share some redundant functions in mDA differentiation, we compared the effect of Lmx1a and Lmx1b’s overexpression during in vitro differentiation of ES cells. J1 ES cells were differentiated in vitro, infected with Lmx1a- or Lmx1b-expressing retrovirus at the NP stage, and analyzed at ND3. In line with Wnt1’s residual expression pattern in dr/dr embryos (Fig. 3a–h), both Lmx1a and Lmx1b upregulated Wnt1 expression (Fig. 5a). SHH expression was unaffected by either gene, while both Pitx3 and Nurr1 expression were upregulated by Lmx1a or Lmx1b (Fig. 5a), showing the redundant function of Lmx1a and Lmx1b in target gene regulation. Interestingly, Lmx1b expression mildly but significantly upregulated Lmx1a expression (Fig. 5a). ChIP analysis showed that Lmx1b binds to the Lmx1a promoter and also Lmx1a binds to the Lmx1b promoter, indicating cross-regulation between these two genes (Suppl. Fig. 8b). In addition, we also examined whether there is any self-regulation of Lmx1a or Lmx1b. qPCR analysis using endogenous message-specific primers revealed that Lmx1a regulates itself, but Lmx1b does not (Suppl. Fig. 8a). Consistent with this, we observed that Lmx1a but not Lmx1b specifically binds to the well conserved binding site within its own promoter (Suppl. Fig. 8b).
Fig. 5.
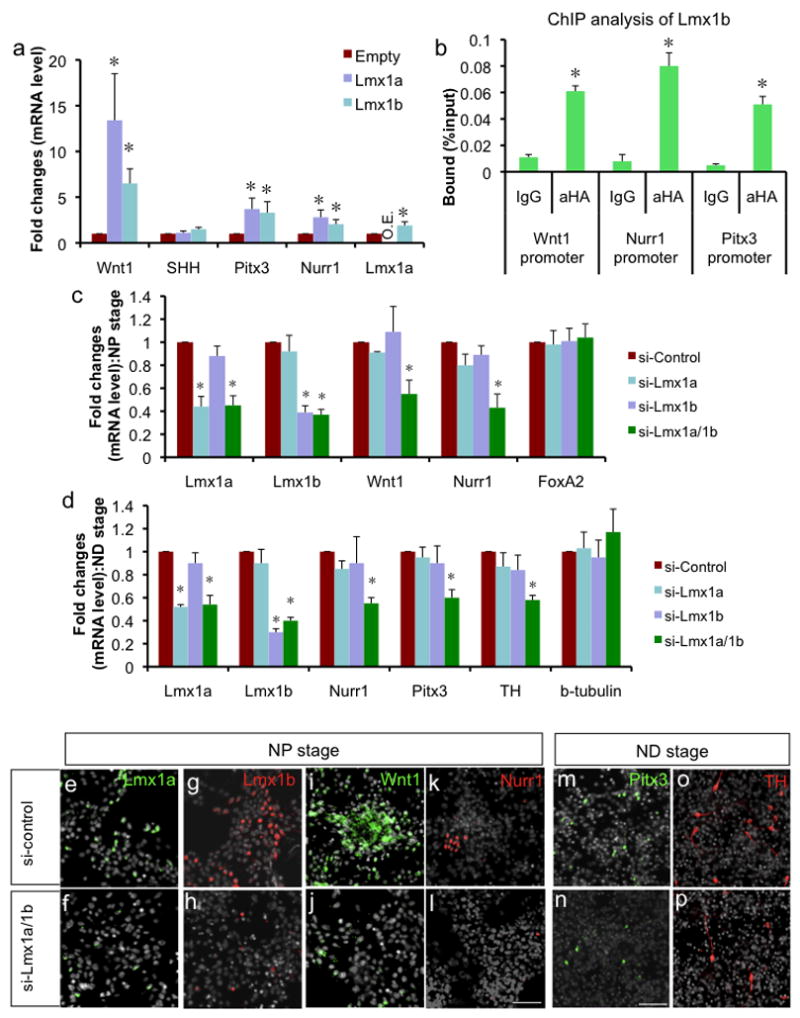
a. qPCR analysis on in vitro differentiated cells transduced with empty, Lmx1a- or Lmx1b-expressing retrovirus at ND3 (n=4, p<0.05). O.E. denotes overexpression of Lmx1a (20.6±8.2). b. ChIP-qPCR analysis of Lmx1b (n=3, p<0.05). In vitro differentiated ES cells transduced with retrovirus expressing HA-tagged Lmx1b were fixed for ChIP at ND3. ChIP fragments were immunoprecipitated either with normal rabbit IgG or anti-HA antibody and analyzed by qPCR. Binding of Lmx1b to the Lmx1a target sites in the Wnt1, Nurr1 or Pitx3 promoters were tested using the same primer sets. c. qPCR analysis of siRNA-treated NP cells. ES cell-derived NP cells were treated with SHH and FGF8 for 4 days for induction/proliferation of mDA NPs and then transfected with control siRNA, Lmx1a siRNA, Lmx1b siRNA or Lmx1a/1b siRNAs, and analyzed 30 hours after transfection (n=4, p<0.05). d. qPCR analysis of siRNA-treated ND cells. ES cell-derived NP cells were treated with SHH and FGF8 for 4 days for induction/proliferation of mDA NPs, further differentiated until day 2 of ND stage, transfected with control siRNA, Lmx1a siRNA, Lmx1b siRNA or Lmx1a/1b siRNAs, and analyzed 30 hours after transfection (n=4, p<0.05). e–l. Immunocytochemistry on NP cells treated with control siRNA or Lmx1a/1b siRNAs one day after transfection. Scale bar represents 50μm. m–p. Immunocytochemistry on ND cells treated with control siRNA or Lmx1a/1b siRNAs one day after transfection. Scale bar represents 50μm.
Observed cross-regulation between Lmx1a and Lmx1b raised the possibility that Lmx1b regulates target genes indirectly through Lmx1a. Thus, to test whether Lmx1b can directly regulate target genes, we did ChIP-qPCR analysis following transduction with retrovirus expressing HA-tagged Lmx1b. We found that Lmx1b significantly bound to the promoters of Wnt1, Nurr1 and Pitx3 (Fig. 5b), though milder than Lmx1a. We also tested the binding of Lmx1a or Lmx1b to the Msx1 promoter, and found that they both bind to the well conserved homeodomain binding sites residing at −3.5kb upstream of the Msx1 gene (Suppl. Fig. 10).
To further study the redundant function between Lmx1a and Lmx1b, we attempted to knock down these genes using gene-specific siRNA approach. ES cell-derived NP cells were treated with SHH and FGF8 for 4 days to induce/proliferate mDA NPs, and then transfected with control siRNA, Lmx1a siRNA, Lmx1b siRNA or both Lmx1a/1b siRNAs using Nucleofector (Amaxa) and analyzed after 30 hours. Transfection of each siRNA treatment significantly reduced the mRNA level of Lmx1a or Lmx1b (Fig. 5c and e–h). Transfection of single siRNA did not have significant effect on Wnt1 or Nurr1 gene expression. This insignificant effect is in contrast with the robust induction effect observed in overexpression experiment (Fig. 5a). This can be explained by incomplete knockdown by siRNA and/or nonphysiological overexpression effect caused by retroviral transduction. However, when both genes were knocked down, there was significant decrease in the target gene expression (Fig. 5c and i–l, Suppl. Fig. 11), suggesting that Lmx1a and Lmx1b compensate each other’s function in regulating mDA regulator genes. Since Pitx3+ cells were not yet detectable at this NP stage, we repeated the gene knockdown experiment at ND stage cells. ES cell-derived cells were treated with SHH and FGF8 for 4 days, differentiated in N3 media for 2 days, transfected with siRNA and analyzed by qPCR analysis 30 hrs after transfection. siRNA treatment to each genes significantly reduced the mRNA level of Lmx1a or Lmx1b (Fig. 5d). Only when both Lmx1a and Lmx1b genes were knocked down, there was significant decrease in Nurr1 and Pitx3 gene expression (Fig. 5d and m–n). Furthermore, knock down of both genes also downregulated TH mRNA level and TH+ cell numbers (Fig. 5d and o–p).
Wnt1-Lmx1 autoregulatory loop induces mDA differentiation synergistically with the SHH signaling pathway
The most salient finding of this study is the tight autoregulatory loop between Wnt1 and Lmx1a during mDA differentiation of ES cells as well as during embryonic midbrain development. This autoregulatory loop, in turn, directly regulates Otx2 expression, through the canonical Wnt signaling pathway, and Nurr1 and Pitx3 expression, through Lmx1a. This finding about the Wnt1 pathway together with prior knowledge about the SHH-FoxA2 pathway in mDA differentiation, prompted us to hypothesize that activation of both signaling pathways by exogenous expression of direct downstream targets of these pathways (i.e., Otx2, Lmx1a and FoxA2) may synergistically induce mDA differentiation. To test such a hypothesis, we transduced ES-derived NPs with FoxA2-, Lmx1a- or Otx2-expressing retroviruses, either alone or together. Indeed, when all three key mediators (Lmx1a, Otx2 and FoxA2) were overexpressed, we observed a robust synergistic induction of the mDA marker genes, TH, Pitx3 and Nurr1 (Fig. 6a), as examined by qPCR analysis. Immunocytochemical analysis also showed significant increase in mDA neurons as shown by increase in the number of cells expressing both TH and Pitx3 (Fig. 6b–e). However, there was no significant change in β-tubulin+ neuronal cell numbers or GFAP+ astrocyte cell numbers (Fig. 6f–g). Further analysis showed that TH+ neurons generated by activation of both signaling pathways represent mature DA neuronal phenotype assessed by coexpression of DAT and DDC, but not by empty vector-transduction (Fig. 6h–o). In the three factor-transduced cells, the majority of TH+ neurons also coexpressed Lmx1b and Nurr1, confirming their mDA phenotype, but not in the empty-vector-transduced cells (Fig. 6p–w). Three factor-transduced cells contained both A9-like (Aldh1a1+) and A10-like (Calbindin+) mDA neurons (Suppl. Fig. 12a–b). In addition, other non-DA neurons such as serotonergic (5HT+), cholinergic (ChAT+) or GABAergic (GABA+) neurons were similarly observed after in vitro differentiation of both empty vector-transduced and three factor-transduced cells (Suppl. Fig. 12c–e; data not shown). Cell counting analysis showed that there was a significant increase in %TH+ cells/β-tubulin+ cells by three-factor transduction from 4.85±024 to 26.30±0.49 (from 2.35±0.11 to 13.22±0.57 %TH+/Hoechst+ cells).
Fig. 6.
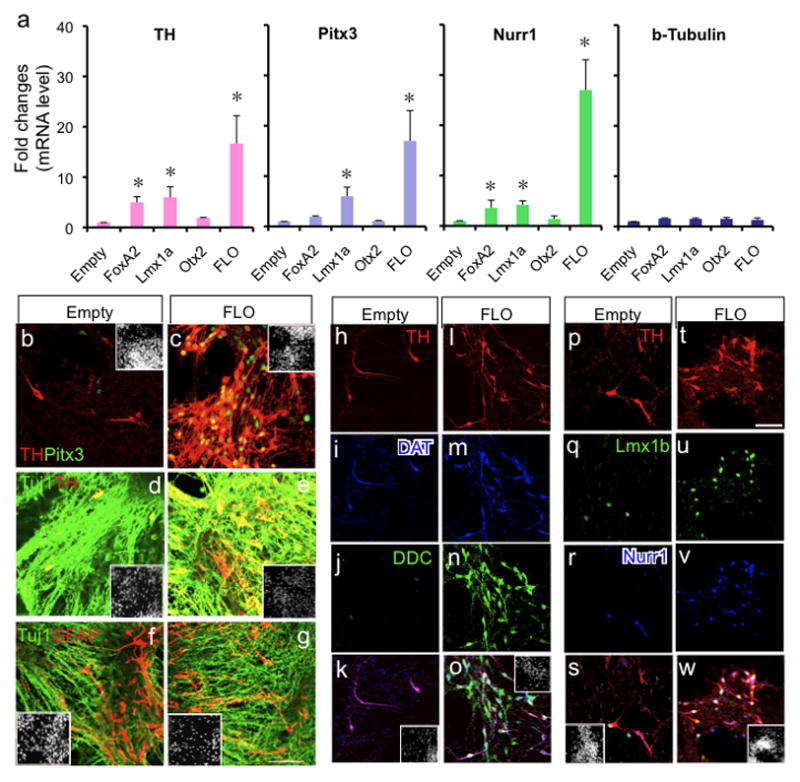
The Wnt1 signaling pathway induces mDA differentiation of ES cells synergistically with the SHH pathway. a. qPCR analysis on in vitro differentiated cells transduced with empty, FoxA2-, Lmx1a-, or Otx2-expressing retrovirus at ND6 (n=4, p<0.05). FLO designates cells transduced with all three viruses that express FoxA2, Lmx1a or Otx2. b–c. Co-transduction of three factors (FLO) leads to a significant increase in Pitx3+ TH+ mDA neurons compared to empty virus-transduced cells. d–g. Cell transduction with three factors does not significantly alter the proportion of neurons (Tuj1+) or astrocytes (GFAP+). h–o. Three factor transduction increases the cells with mature DA phenotype, shown by coexpression of DAT and DDC with TH. p–w. Three factor transduction increases cells with mDA phenotype, shown by coexpression of Lmx1b and Nurr1 with TH. Immunocytochemistry on in vitro differentiated ES cells at stage ND6. Inset shows Hoechst staining. Scale bar represents 50mm.
Discussion
Midbrain dopaminergic (mDA) neurons critically control voluntary movement, emotion, and reward through specific neuronal circuits (Bjorklund and Lindvall, 1984), and their selective degeneration and/or dysregulation is associated with major neurological and psychiatric disorders. Especially, selective loss of mDA neurons in the substantia nigra is associated with PD (Lang and Lozano, 1998). Successful cell replacement therapy for PD requires generation of optimal cell sources. There has been extensive effort to generate mDA neurons from stem cells (Chung et al., 2002; Kawasaki et al., 2000; Kim et al., 2002), but optimal differentiation of stem cells to authentic mDA neurons requires further understanding of the molecular mechanisms underlying mDA neuronal development. Toward this goal, numerous laboratories investigated mDA neuron development, resulting in identification of important signaling molecules (e.g., SHH and Wnt1) and key transcription factors (e.g., FoxA2, Lmx1a, Lmx1b, Msx1, Ngn2, Nurr1, and Pitx3) (reviewed in (Ang, 2006; Smidt and Burbach, 2007)). However, molecular interactions/networks between these extrinsic factors and intrinsic transcription factors are not well understood.
In this study, by analyzing molecular networks involving Wnt1 during mDA differentiation of ES cells, we showed that Wnt1 directly regulates Lmx1a, a key intrinsic factor for mDA differentiation, eliciting functional cascades that lead to mDA differentiation. The early function of Wnt1 based on its expression in the isthmus (<E9.5) is well established (McMahon et al., 1992), but how Wnt1 functions in mDA differentiation through its ventral midbrain expression (E9.5–E12.5) has not been fully understood. The Wnt1-regulated molecular network revealed in this study explains the functional role of Wnt1 in mDA phenotype specification (Castelo-Branco et al., 2004; Prakash et al., 2006) apart from its well-established role in NP proliferation (Megason and McMahon, 2002). Furthermore, we identified the extrinsic signaling molecule Wnt1 as a major target of Lmx1a during mDA differentiation, forming an autoregulatory loop between Wnt1 and Lmx1a. Our study further demonstrates that Lmx1a directly regulates two critical regulators of mDA neuron differentiation, the Nurr1 and Pitx3 genes as well as Wnt1 and that Wnt1 directly regulates Otx2 as well as Lmx1a through the canonical Wnt signaling pathway during mDA differentiation (Fig. 7). This Otx2 regulation by Wnt1 is consistent with previous observations shown by ectopic expression of Wnt1 (Prakash et al., 2006) and conditional knock out (cKO) of β-catenin (Joksimovic et al., 2009) during mouse embryonic development. It is also worthwhile to note that cKO of Otx2 resulted in reduction of Wnt1 expression in midbrain neurons (Prakash et al., 2006), but this mechanism may be indirect, considering the role of Otx2 as a transcriptional repressor . For Lmx1a regulation by the β-catenin complex, two recent studies showed conflicting results; one reported that SHH-cre;β-catenin cKO shows significant decrease in Lmx1a expression (Joksimovic et al., 2009), whereas the other reported that SHH-cre;β-catenin cKO shows only mild changes (Tang et al., 2009). Such a controversy raised the need for more direct in vitro analysis of Wnt1-Lmx1a regulation. Our in vivo and in vitro analyses described in this study show that the β-catenin complex indeed directly associates with the Lmx1a promoter and regulates its expression.
Fig. 7.
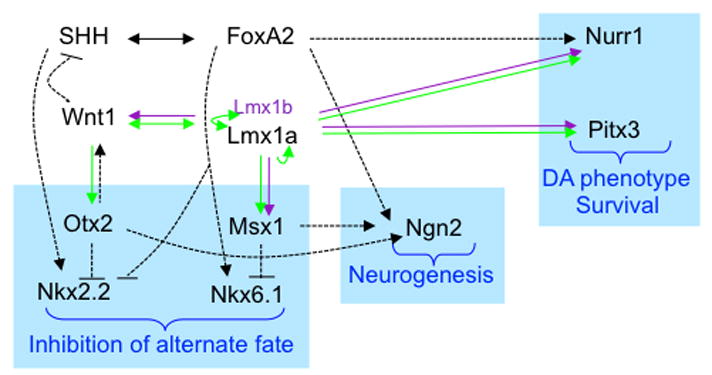
The emerging genetic network of the Wnt1 signaling pathway reveals the interaction between the Wnt1 and SHH pathways at three major steps of mDA development; i) ventralization and inhibition of alternate fates, ii) promotion of neurogenesis, and iii) DA phenotype specification and survival. Arrow indicates positive regulation and -| indicates negative regulation. Black arrows indicate the regulation previously shown. Green and purple arrows indicate the regulations observed in this study. Overlapping function of Lmx1b is indicated by purple arrows. Dotted lines represent regulations that are not shown to be direct yet. Solid lines represent regulation that has been shown to be direct. Please refer to the Discussion section for details and related references.
Our new finding that Pitx3 and Nurr1 are the direct downstream targets of the Wnt1-Lmx1a autoregulatory loop links a key signaling pathway of mDA differentiation to the major molecular regulators of terminal differentiation/survival of mDA neurons. A recent study using FoxA2 KO mice revealed that FoxA2 regulates Nurr1 expression in developing midbrain (Ferri et al., 2007), and in the present study we observed that Lmx1a directly binds to the Nurr1 promoter in vivo and activates Nurr1 expression. Thus, it seems that regulation of Nurr1 is one of the converging points of the SHH-FoxA2 pathway and the Wnt1-Lmx1a pathway. However, Lmx1a did not affect SHH or FoxA2 expression, showing the independent nature of these two pathways. In addition, our data show that Lmx1a is a link between a major signaling molecule Wnt1 and an important mDA-specific transcription factor, Pitx3. Interestingly, Prakash et al. observed that in Wnt1 KO mice, the few surviving TH+ neurons never express Pitx3 (Prakash et al., 2006). Furthermore, Smidt et al. observed that in Lmx1b KO mice the few observed TH+ neurons also failed to express Pitx3 (Smidt et al., 2000). Taken together, these previous studies corroborate our results and suggest the existence of a cascade linking Wnt1, Lmx1a/1b and Pitx3 for mDA differentiation.
Lmx1a was first identified for its roof plate phenotype (Millonig et al., 2000), but until recently its role in ventral midbrain development remained unknown. This is likely due to its mild ventral phenotype caused by compensation by Lmx1b, as shown here. This is different from the chick system where Lmx1b cannot compensate for the loss of Lmx1a in midbrain DA development and where disruption of Lmx1a leads to complete absence of midbrain DA neurons even in the presence of Lmx1b (Andersson et al., 2006b). In contrast, our data strongly suggest that, in the mouse system, Lmx1a and Lmx1b co-operatively regulate mDA neuron development by sharing redundant functions. First, our gene expression analyses of mDA domains and developing corin+ mFP cells showed that mDA phenotype is only mildly affected in Lmx1a mutant dr/dr embryo. Notably, the defect of target gene (Wnt1) expression was modest in the ventral most part where Lmx1b is still expressed, suggesting its compensating function. Second, our siRNA-based single and double knock down experiments of Lmx1a and Lmx1b in in vitro-differentiated ES cells showed that knocking down a single gene has no or marginal effect on target gene expression, whereas knocking down both genes significantly affected target gene expression. Third, our extensive ChIP analyses indicate that both Lmx1a and Lmx1b directly bind to the promoters of target genes during mDA differentiation of ES cells, again supporting their redundant functions during mDA differentiation. In line with our results, Dr. Siew-Lan Ang’s laboratory observed that whereas Lmx1adreher/dreher or ShhCre/+, Lmx1bflox/flox embryos show partial or no reduction in the number of mDA neurons, respectively, there was a much more severe loss of mDA neurons in Lmx1adreher/dreher;ShhCre/+, Lmx1bflox/flox double mutant embryos at E12.5 (personal communication). The severe mDA phenotype of Lmx1b KO mice (Smidt et al., 2000) compared to the no phenotype in ShhCre/+, Lmx1bflox/flox mice could be caused by its early role in isthmus formation (Guo et al., 2007; Matsunaga et al., 2002) and/or by differences in genetic backgrounds. Despite the observed overlapping function of Lmx1a and Lmx1b in regulating downstream targets, Lmx1b is not regulated by Wnt1 (Matsunaga et al., 2002). In line with this, we could not find well conserved TCF/LEF binding sites in the 50kb Lmx1b promoter.
In summary, as opposed to the previously known SHH-FoxA2 pathway, this study identified an important complementary pathway for mDA development, the Wnt1-Lmx1a autoregulatory loop, during mDA differentiation of ES cells as well as during mouse embryonic midbrain development. Notably, this Wnt1-Lmx1a pathway appears to be formed independent of the SHH-FoxA2 pathway, although they functionally interact with each other during mDA development. In support of this notion, our data show that overexpression of SHH or its blocking by cyclopamine did not affect Lmx1a expression. In addition, induction of Lmx1a gene expression by Wnt1 during in vitro differentiation of mES cells was not affected by cyclopamine. Based on our work along with previous studies, we propose that these two major signaling pathways, once formed, functionally interact with each other at three major steps of mDA development (Fig. 7). First, the SHH-FoxA2 pathway induces the ventral phenotype, but also induces the alternate ventral phenotype characterized by the markers Nkx2.2 and Nkx6.1 (Ferri et al., 2007; Pabst et al., 2000; Vokes et al., 2007), which is inhibited by the Lmx1a-Wnt1 pathway through Otx2 (Prakash et al., 2006) and Msx1 (Andersson et al., 2006b). FoxA2 also inhibits the Nkx2.2 phenotype, which is induced by SHH (Ferri et al., 2007). Furthermore, Joksimovic et al. recently demonstrated the importance of more direct inhibition of SHH expression by Wnt1 at early time point (<E10.5) of mDA differentiation (Joksimovic et al., 2009), adding another layer of regulation to the functional crosstalk between SHH and Wnt1 pathway for mDA differentiation. Second, another major step of mDA development is the conversion of non-neuronal FP cells to NPs, in which proneural genes such as Ngn2 could play an important role (Andersson et al., 2006a; Kele et al., 2006). Two signaling pathways collaborate on this step, as FoxA2, Otx2 and Msx1 activate Ngn2 expression (Andersson et al., 2006b; Ferri et al., 2007; Vernay et al., 2005), thus promoting neurogenesis. Thirdly, these two pathways also collaborate on the expression of functional DA genes such as TH and DAT. The SHH-FoxA2 pathway, directly or through Nurr1, plays an important role in expression of these genes, and the Wnt1-Lmx1a pathway, through Pitx3 and Nurr1, regulates these genes’ expression (Ang, 2006; Smidt and Burbach, 2007) (Fig. 7). These intimate functional interactions between these two pathways predict that activation of key mediators of both signaling pathways may facilitate ES cell differentiation to mDA neurons by efficiently providing the proper cellular environment for each other. Indeed, activation of both pathways by exogenous expression of three key mediators resulted in synergistic induction of mDA differentiation, compared to the induction of a single pathway. These studies demonstrate the usefulness of ES cell differentiation to investigate the molecular network of mDA differentiation and also in turn, show that knowledge gained from such mechanistic studies can facilitate the generation of cell sources for cell replacement therapy for PD.
Experimental Procedures
ES Cell culture and in vitro differentiation
ES cells were maintained and differentiated as described previously (Chung et al., 2002), for more details, see Suppl. Information.
For transgenic expression studies, we intentionally used suboptimal conditions to clearly see the effect of transgene expression without masking its effect by stimulating its upstream events by culture conditions. So we did not add any signaling molecules such as SHH, FGF8, Ascorbic Acids nor any growth factors such GDNF nor BDNF, nor any feeders such as MS5 nor PA6 (Andersson et al., 2006b; Kawasaki et al., 2000; Kim et al., 2002).
For siRNA transfection, NP stage cells were treated with 50ng/ml FGF8 and 100ng/ml SHH for 4 days to induce/proliferate mDA NPs, followed by transfection with siRNA using the Nucleofector (Amaxa, Walkersville, MD) mouse stem cell kit with the program A-033 according to the manufacturer’s instruction. Per transfection, 5 × 106 NP cells were treated with 480 pmol of siRNA, diluted in 10ml of N3bFGF media (or N3 media for ND stage transfection) and plated in PLO/FN-coated multiwell plate, resulting in a final siRNA concentration of 48nM. For ND stage transfection, cells were further differentiated in N3 media for 2 days before transfection. Multiple Stealth siRNAs were purchased from Invitrogen (Carlsbad, CA), screened for gene silencing efficiency by cotransfection with Lmx1a or Lmx1b-expressing plasmids and only siRNAs showing efficient gene silencing (>95%) was used for the experiments. The sequences of the siRNAs are as follows: the Lmx1a sense strand GAGGAGAGCAUUCAAGGCCUCGUUU; the Lmx1a antisense strand AAACGAGGCCUUGAAUGCUCUCCUC; the Lmx1b sense strand GGAACGACUCCAUCUUCCACGAUAU; the Lmx1b antisense strand AUAUCGUGGAAGAUGGAGUCGUUCC. Thirty hours after transfection, cells were fixed for immunocytochemistry or harvested for RNA preparation.
Cell counting and statistical analysis
Cells were counted from blind-coded samples using an integrated Axioskop 2 microscope (Carl Zeiss, Thornwood, NY) and the StereoInvestigator image capture equipment and software (Microbright Field, Williston, VT). For statistical analysis, we used the Statview software and performed analysis of variance (ANOVA) with an alpha level of 0.05 to determine possible statistical differences between group means. When significant differences were found, post hoc analysis was performed using Fisher’s PLSD (alpha=0.05).
ChIP-qPCR analysis
ChIP-qPCR analysis was done as described previously (Yochum et al., 2007), and for details see Suppl. Information.
Mouse
Heterozygous dreher mice were purchased from the Jackson Laboratory (Bar Harbor, ME). B6C3Fe a/a-Lmx1adr-J/J mouse harbors a point mutation, which makes Lmx1a protein nonfunctional (Millonig et al., 2000). For details on embryo analysis, see Suppl. Information.
FACS purification
FACS purification was done as described previously (Pruszak et al., 2007), and for details, see Suppl. Information.
Supplementary Material
Acknowledgments
This work was supported by NIH grants P50 NS39793 and MH48866, and international grants from the Korean Brain Research Center and Dongyang Corporation Co. in Korea. The authors also thank Dr. Siew-Lan Ang at MRC, UK, for sharing her unpublished work as a personal communication. We appreciate Dr. German at UCSF, Dr. Birchmeier at Max Delbrück Center for Molecular Medicine, Dr. Morgan at MGH and Dr. Duester at Burnham Institute for kindly providing us with valuable antibodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson E, Jensen JB, Parmar M, Guillemot F, Bjorklund A. Development of the mesencephalic dopaminergic neuron system is compromised in the absence of neurogenin 2. Development. 2006a;133:507–516. doi: 10.1242/dev.02224. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006b;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Arenas E. Foxa2: the rise and fall of dopamine neurons. Cell Stem Cell. 2008;2:110–112. doi: 10.1016/j.stem.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Bjorklund A, Lindvall O. Handbook of Chemical Neuroanatomy. Amsterdam: Elsevier; 1984. Dopamine-containing systems in the CNS. [Google Scholar]
- Castelo-Branco G, Rawal N, Arenas E. GSK-3beta inhibition/beta-catenin stabilization in ventral midbrain precursors increases differentiation into dopamine neurons. J Cell Sci. 2004;117:5731–5737. doi: 10.1242/jcs.01505. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Chung S, Sonntag KC, Andersson T, Bjorklund LM, Park JJ, Kim DW, Kang UJ, Isacson O, Kim KS. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Guo C, Qiu HY, Huang Y, Chen H, Yang RQ, Chen SD, Johnson RL, Chen ZF, Ding YQ. Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development. 2007;134:317–325. doi: 10.1242/dev.02745. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Epstein DJ. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development. 2003;130:3891–3902. doi: 10.1242/dev.00590. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Yun BA, Kittappa R, Anderegg AM, Chang WW, Taketo MM, McKay RD, Awatramani RB. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Kim JH, Auerbach JM, Rodriguez-Gomez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sanchez-Pernaute R, Bankiewicz K, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- Lee SM, Danielian PS, Fritzsch B, McMahon AP. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development. 1997;124:959–969. doi: 10.1242/dev.124.5.959. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL. EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development. 2001;128:181–191. doi: 10.1242/dev.128.2.181. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Katahira T, Nakamura H. Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development. 2002;129:5269–5277. doi: 10.1242/dev.129.22.5269. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Millonig JH, Millen KJ, Hatten ME. The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Pabst O, Herbrand H, Takuma N, Arnold HH. NKX2 gene expression in neuroectoderm but not in mesendodermally derived structures depends on sonic hedgehog in mouse embryos. Development Genes and Evolution. 2000;210:47–50. doi: 10.1007/pl00008188. [DOI] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, et al. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Pruszak J, Sonntag KC, Aung MH, Sanchez-Pernaute R, Isacson O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimaki-Vire J, Peltopuro P, Lahti L, Naserke T, Blak AA, Vogt Weisenhorn DM, Yu K, Ornitz DM, Wurst W, Partanen J. Fibroblast growth factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci. 2007;27:8581–8592. doi: 10.1523/JNEUROSCI.0192-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Tang M, Miyamoto Y, Huang EJ. Multiple roles of beta-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development. 2009;136:2027–2038. doi: 10.1242/dev.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL. Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci. 2005;25:4856–4867. doi: 10.1523/JNEUROSCI.5158-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Yochum GS, McWeeney S, Rajaraman V, Cleland R, Peters S, Goodman RH. Serial analysis of chromatin occupancy identifies beta-catenin target genes in colorectal carcinoma cells. Proc Natl Acad Sci U S A. 2007;104:3324–3329. doi: 10.1073/pnas.0611576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


